A virus-encoded protein suppresses methylation of the viral genome through its interaction with AGO4 in the Cajal body
Figures
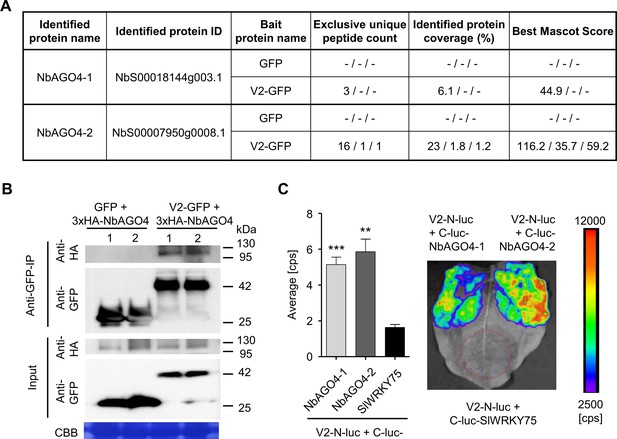
V2 interacts with AGO4 from N. benthamiana.
(A) Unique peptide count, protein coverage, and best Mascot Score of NbAGO4-1 and NbAGO4-2 co-immunoprecipitated with V2-GFP, as identified by affinity purification followed by mass spectrometry (AP-MS). Results from three independent biological repeats are shown. "-” indicates no peptide was detected. (B) 3xHA-NbAGO4-1 and 3xHA-NbAGO4-2 specifically interact with V2-GFP in co-immunoprecipitation (co-IP) assays upon transient expression in N. benthamiana. Free GFP was used as negative control. CBB, Coomassie brilliant blue staining. Three independent biological replicates were performed with similar results. (C) NbAGO4-1 and NbAGO4-2 interact with V2 in split-luciferase assays. V2-N-luc and C-luc-NbAGO4-1/2 were transiently co-expressed in N. benthamiana; C-luc-SlWRKY75 is used as negative control. The luciferase bioluminescence from at least three independent leaves per experiment was imaged 2 days after infiltration. The average bioluminescence, measured in counts per second (cps), as well as an image of a representative leaf are shown. Values represent the mean of three independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference (according to Student’s t-test, **: p<0.01, ***: p<0.001) compared to the negative control.
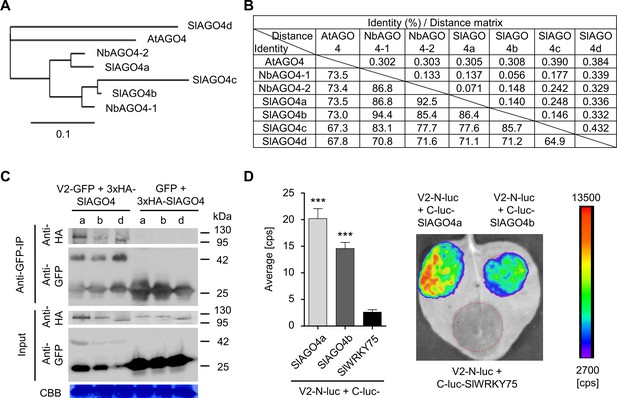
V2 interacts with AGO4 from tomato.
(A) Phylogenetic tree of AtAGO4, NbAGO4, and SlAGO4 proteins. The phylogenetic analysis was performed with phylogeny.fr (Dereeper et al., 2010; Dereeper et al., 2008). (B) Pairwise identity and genetic distance matrix among AtAGO4, NbAGO4 and SlAGO4 proteins. The analysis was performed by Geneious (https://www.geneious.com). (C) 3xHA-SlAGO4a, 3xHA-SlAGO4b, and 3xHA-SlAGO4d specifically interact with V2-GFP in co-immunoprecipitation (co-IP) assays upon transient expression in N. benthamiana. Free GFP was used as negative control. CBB, Coomassie brilliant blue staining. Three independent biological replicates were performed with similar results. (D) SlAGO4a and SlAGO4b interact with V2 in split-luciferase assays. V2-N-luc and C-luc-SlAGO4a/b were transiently co-expressed in N. benthamiana; C-luc-SlWRKY75 is used as negative control. The luciferase bioluminescence from at least three independent leaves per experiment was imaged two days after infiltration. The average bioluminescence, measured in counts per second (cps), as well as an image of a representative leaf are shown. Values represent the mean of three independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference (according to Student’s t-test, ***: p<0.001) compared to the negative control.
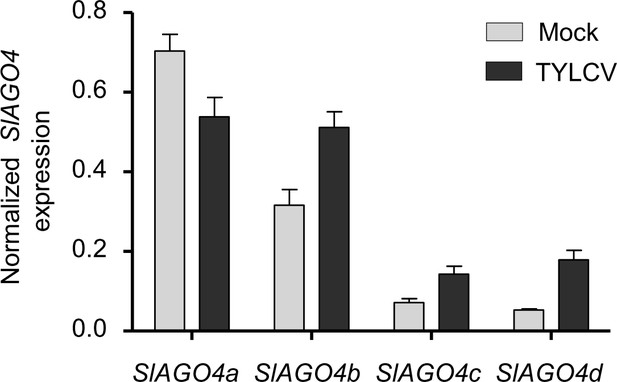
SlAGO4 expression in TYLCV-infected and control tomato plants.
SlAGO4a/b/c/d expression in TYLCV-infected or control (mock-inoculated) tomato plants at 3 weeks post-inoculation (wpi), as measured by qRT-PCR. Gene expression was normalized to SlActin. Values are the mean of three independent biological replicates; error bars indicate SEM.
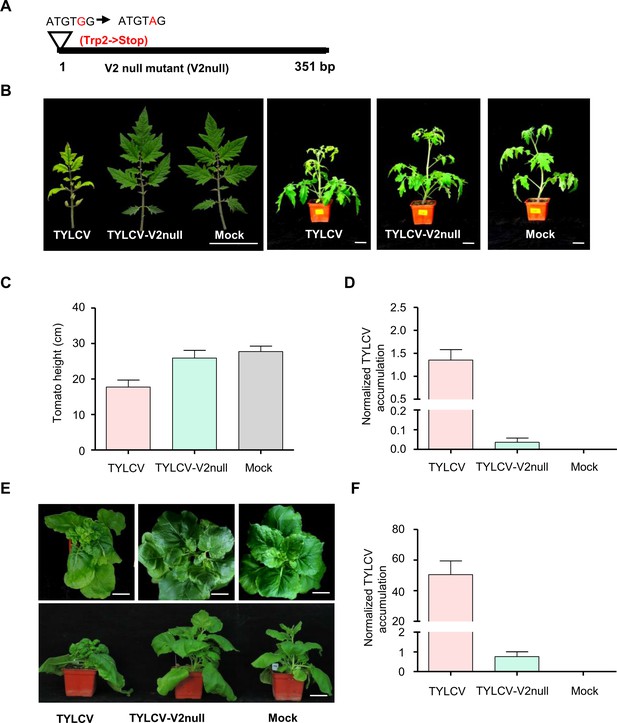
V2 is essential for systemic TYLCV infection in tomato and N. benthamiana.
(A) Design of the V2 null TYLCV mutant used in this work. The second codon of the V2 ORF, originally encoding a Trp (TGG), is mutated to a STOP codon (TAG). (B) Representative pictures of tomato plants infected with TYLCV wild-type or V2 null mutant (TYLCV-V2null) or mock-inoculated. Photographs were taken at 3 weeks post-inoculation (wpi). Bar, 5 cm. (C) Height of tomato plants infected with TYLCV wild-type or V2 null mutant (TYLCV-V2null) or mock-inoculated at 3 wpi. Values are the mean of five independent biological replicates; error bars indicate SEM. (D) Viral (TYLCV) accumulation in tomato plants infected with TYLCV wild-type or V2 null mutant (TYLCV-V2null), or mock-inoculated at 3 wpi, measured by qPCR. Each sample corresponds to the apical leaves from six plants. The accumulation of viral DNA is normalized to the 25S ribosomal RNA interspacer (ITS). Values are the mean of six independent biological replicates; error bars indicate SEM. (E) Representative pictures of N. benthamiana plants infected with TYLCV wild-type or V2 null mutant (TYLCV-V2null) or mock-inoculated. Photographs were taken at 3 weeks post-inoculation (wpi). Bar, 5 cm. (F) Viral (TYLCV) accumulation in N. benthamiana plants infected with TYLCV wild-type or V2 null mutant (TYLCV-V2null) or mock-inoculated at 3 wpi, measured by qPCR. Each sample corresponds to the apical leaves from six plants. The accumulation of viral DNA is normalized to the 25S ribosomal RNA interspacer (ITS). Values are the mean of six independent biological replicates; error bars indicate SEM.
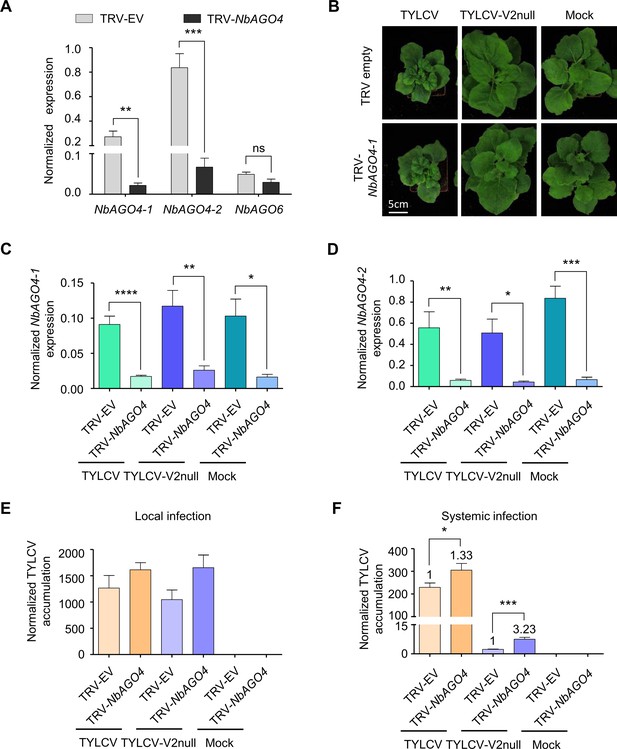
V2 counters the AGO4-dependent antiviral defence to promote virulence.
(A) Expression of NbAGO4-1, NbAGO4-2, and NbAGO6 in N. benthamiana plants infected with TRV-EV (empty vector) or TRV-NbAGO4, measured by reverse transcription quantitative real-time PCR (RT-qPCR). Gene expression was normalized to NbTubulin. Values are the mean of four independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. **: p<0.01, ***: p<0.001, ns: not significant. (B) Representative pictures of N. benthamiana plants infected with the indicated combinations of viruses. Photographs were taken at 3 weeks post-inoculation (wpi). (C) NbAGO4-1 expression in NbAGO4-silenced plants and control plants infected with TYLCV, TYLCV-V2null, or mock-inoculated at 3 wpi measured by RT-qPCR. Gene expression was normalized to NbTubulin. Values are the mean of six independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. *: p<0.05, **: p<0.01, ****: p<0.0001. (D) NbAGO4-2 expression in NbAGO4-silenced plants and control plants infected by TYLCV, TYLCV-V2null, or mock-inoculated at 3 wpi measured by RT-qPCR. Gene expression was normalized to NbTubulin. Values are the mean of six independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. *: p<0.05, **: p<0.01, ***: p<0.001. (E) Viral (TYLCV) accumulation in local infections in NbAGO4-silenced or control plants, measured by qPCR. Infiltrated leaf patches from different plants were collected at 4 dpi. The experimental design is shown in Figure 3—figure supplement 1A. The accumulation of viral DNA is normalized to the 25S ribosomal RNA interspacer (ITS). Values are the mean of eight independent biological replicates; error bars indicate SEM. (F) Viral (TYLCV) accumulation in systemic infections in NbAGO4-silenced or control plants, measured by qPCR. Apical leaves from six plants were collected at 3 wpi. The experimental design is shown in Figure 3—figure supplement 1B. The accumulation of viral DNA is normalized to the 25S ribosomal RNA interspacer (ITS). Four independent biological replicates were performed with similar results; one representative result is shown. Values are the mean of six independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. *: p<0.05, ***: p<0.001. The relative fold change of viral accumulation between NbAGO4-silenced plants and control plants is shown above each column.
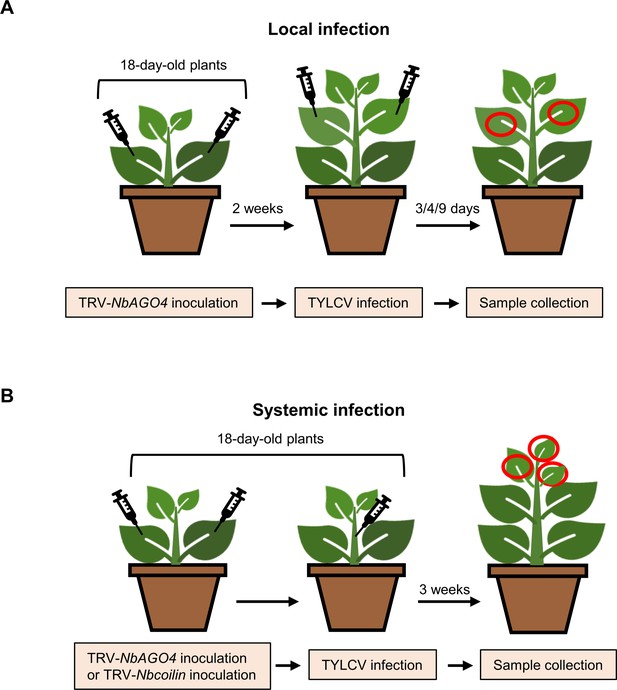
Experimental design for local and systemic TYLCV infection assays in NbAGO4- or Nbcoilin-silenced N. benthamiana plants.
(A) Experimental design for local TYLCV infection assays. A. tumefaciens carrying the TRV-EV or TRV-NbAGO4 infectious clones were inoculated on 18-day-old N. benthamiana cotyledons. Two weeks later, young leaves were infiltrated with A. tumefaciens carrying the TYLCV infectious clone (wild-type or V2null) and leaf patches were collected at 3, 4, or 9 days post-inoculation (dpi). (B) Experimental design for systemic TYLCV infection assays. A. tumefaciens carrying the TRV-EV, TRV-NbAGO4, or TRV-Nbcoilin infectious clones were inoculated on 18-day-old N. benthamiana cotyledons. At the same time, A. tumefaciens carrying the TYLCV infectious clone (wild-type or V2null) were injected into plant stems. The top three leaves were collected at 3 weeks post-inoculation (wpi).
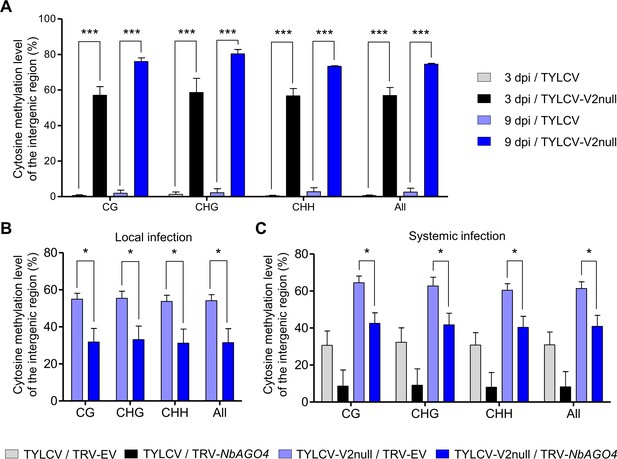
V2 suppresses the AGO4-dependent methylation of viral DNA.
(A) Percentage of methylated cytosines in the intergenic region (IR) of TYLCV in local infection assays with TYLCV wild-type or V2 null mutant (TYLCV-V2null) in N. benthamiana at 3 or 9 days post-inoculation (dpi), as detected by bisulfite sequencing. The original single-base resolution bisulfite sequencing data are shown in Figure 4—figure supplement 1A. Values are the mean of three independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. ***: p<0.001. The values of cytosine methylation in each biological replicate are shown in Supplementary file 1. (B) Percentage of methylated cytosines in the intergenic region (IR) of TYLCV in local infection assays with the V2 null mutant TYLCV (TYLCV-V2null) in AGO4-silenced (TRV-NbAGO4) or control (TRV-EV) N. benthamiana plants at 4 dpi, as detected by bisulfite sequencing. Samples come from the same plants used in Figure 3E. The original single-base resolution bisulfite sequencing data are shown in Figure 4—figure supplement 1B. Values are the mean of four independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. *: p<0.05. The values of cytosine methylation in each biological replicate are shown in Supplementary file 1. (C) Percentage of methylated cytosines in the intergenic region (IR) of TYLCV in systemic infection assays with TYLCV wild-type or V2 null mutant (TYLCV-V2null) in AGO4-silenced (TRV-NbAGO4) or control (TRV-EV) N. benthamiana plants at 3 weeks post-inoculation (wpi), as detected by bisulfite sequencing. Samples come from the same plants used in Figure 3F. The original single-base resolution bisulfite sequencing data are shown in Figure 4—figure supplement 2. Values are the mean of four independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. *: p<0.05. The values of cytosine methylation in each biological replicate are shown in Supplementary file 1.
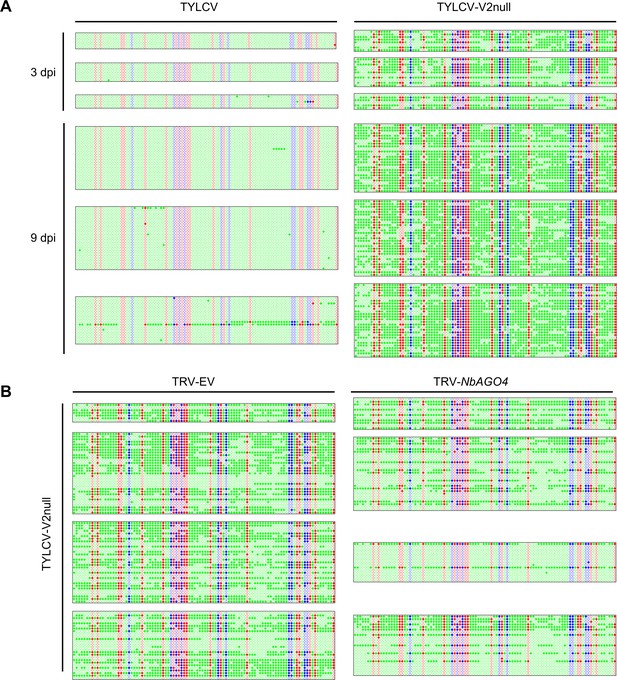
Original single-base resolution bisulfite sequencing data of the intergenic region (IR) of TYLCV (wild-type and V2 null mutant) in local infection assays, related to Figure 4A,B.
(A) Original single-base resolution bisulfite sequencing data for Figure 4A. At least five individual clones were sequenced per replicate and sample at 3 dpi, and >18 individual were sequenced per replicate and sample at 9 dpi. Each single circle, corresponding to a cytosine, is colored in blue, red, or green, representing the CHG, CG, or CHH contexts, respectively. Methylated cytosines are represented by filled circles, while unmethylated cytosines are represented by empty circles. Values of IR methylation in independent biological replicates is shown in Supplementary file 1. (B) Original single-base resolution bisulfite sequencing data for Figure 4B. At least seven individual clones were sequenced per sample at 4 dpi in the first replicate, and >15 individual clones were sequenced per sample at 4 dpi in replicates second to fourth. Each single circle, corresponding to a cytosine, is colored in blue, red, or green, representing the CHG, CG, or CHH contexts, respectively. Methylated cytosines are represented by filled circles, while unmethylated cytosines are represented by empty circles. Values of IR methylation in independent biological replicates are shown in Supplementary file 1.

Original single-base resolution bisulfite sequencing data of the intergenic region (IR) of TYLCV and TYLCV-V2null in systemic infection assays, related to Figure 4C.
Original single-base resolution bisulfite sequencing data for Figure 4C. >14 individual clones were sequenced per sample and replicate. Each single circle, corresponding to a cytosine, is colored in blue, red, or green, representing the CHG, CG, or CHH contexts, respectively. Methylated cytosines are represented by filled circles, while unmethylated cytosines are represented by empty circles. Values of IR methylation in independent biological replicates are shown in Supplementary file 1.
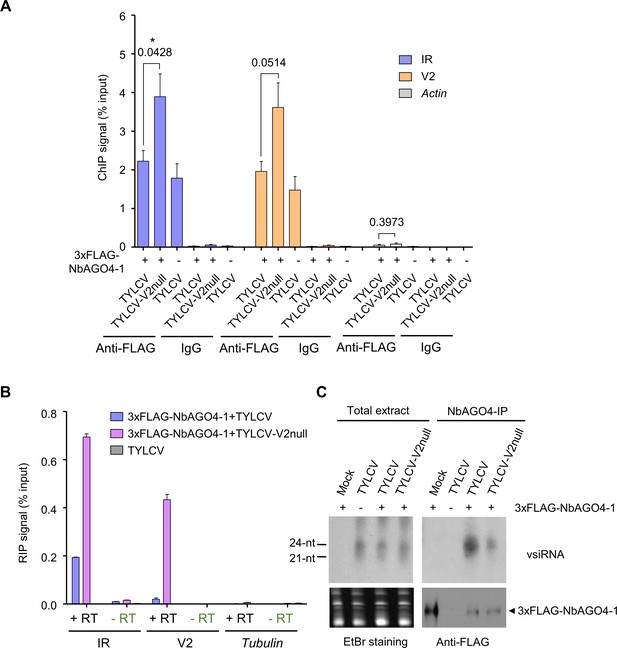
V2 interferes with AGO4 binding to the viral genome through hindering its association to the viral RNA but does not hamper production or loading of vsiRNA.
(A) 3xFLAG-NbAGO4-1 binds the viral (TYLCV) genome. Binding was detected by chromatin immunoprecipitation (ChIP) upon transient expression in N. benthamiana followed by qPCR. Two regions of the viral genome, the IR and the V2 ORF, were analyzed; Actin was used as negative control. Values represent the mean of four independent biological replicates; error bars represent SEM. Asterisks indicate a significant difference according to Student’s t-test; the P-value for the different comparisons is shown. *: p<0.05. (B) RNA immunoprecipitation (RIP)-based detection of viral RNA bound by NbAGO4-1 upon transient expression in N. benthamiana leaves infected by TYLCV or TYLCV-V2 null. Tubulin serves as an unbound loading control. Samples without reverse transcriptase (-RT) are used as control for DNA contamination. Values represent average qRT-PCR signal normalized to inputs of three technical replicates; error bars represent SEM. This experiment was repeated three times with similar results; additional independent biological replicates can be found in Figure 5—figure supplement 1. (C) NbAGO4-1 binds viral small interfering RNA (vsiRNA) independently of V2. Northern blot of vsiRNA in total extracts or 3xFLAG-NbAGO4-1 immunoprecipitates (NbAGO4-IP) of N. benthamiana leaf patches infiltrated with TYLCV wild-type or V2 null mutant infectious clones (TYLCV, TYLCV-V2null) at 2 days after infiltration. Detection was performed with a 32P-labeled DNA probe for the intergenic region (IR).
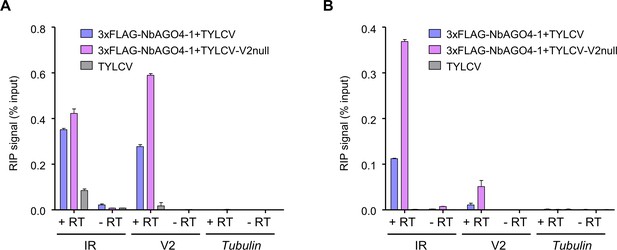
V2 interferes with the NbAGO4-1 binding to the viral RNA.
(A) and (B) show independent additional replicates of the RIP experiments shown in Figure 5B.
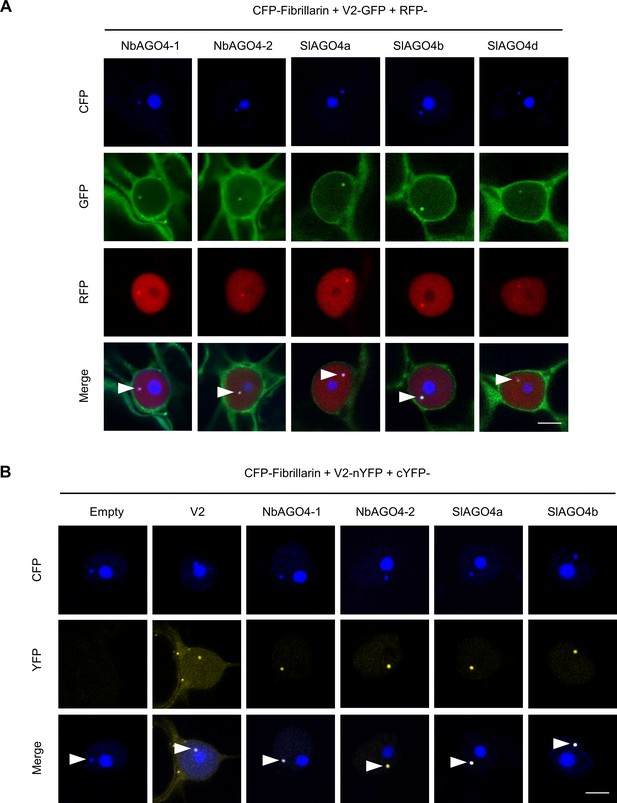
V2 interacts with AGO4 in the Cajal body.
(A) V2-GFP and RFP-AGO4 co-localize in the Cajal body. CFP-Fibrillarin, V2-GFP and RFP-NbAGO4-1/2 or RFP-SlAGO4a/b/d were transiently co-expressed in N. benthamiana epidermal cells. CFP-Fibrillarin is used as nucleolus and Cajal body marker. Confocal images were taken at two days after infiltration. Arrowheads indicate the position of the Cajal body. Bar, 5 μm. This experiment was repeated more than three times with similar results. (B) V2 interacts with AGO4 in the Cajal body. The N-terminal half of the YFP fused to V2 (V2-nYFP) was transiently co-expressed with the C-terminal half of the YFP alone (cYFP, as negative control), or cYFP-NbAGO4, cYFP-SlAGO4, or cYFP-V2 (as positive control) in N. benthamiana leaves. CFP-Fibrillarin was used as nucleolus and Cajal body marker. Confocal images were taken at two days after infiltration. Yellow fluorescence indicates a positive interaction. Arrowheads indicate the position of the Cajal body. Bar, 5 μm. This experiment was repeated more than three times with similar results.
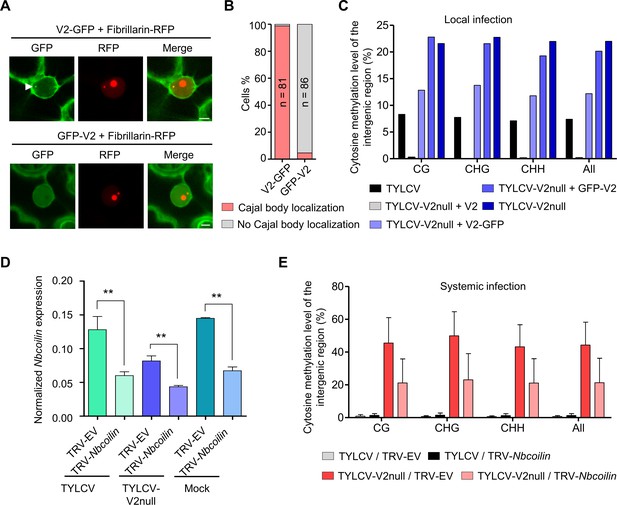
Methylation of the viral DNA and its suppression by V2 occur in a Cajal body-dependent manner.
(A) V2-GFP co-localizes with Fibrillarin-RFP (nucleolus and Cajal body marker) in the Cajal body, while GFP-V2 does not. RFP-Fibrillarin and V2-GFP or GFP-V2 were transiently co-expressed in N. benthamiana epidermal cells. Confocal images were taken at two days after infiltration. Arrowheads indicate the position of the Cajal body. Bar, 5 μm. This experiment was repeated three times with similar results. (B) Quantification of Cajal body localization of V2-GFP or GFP-V2. (C) V2-GFP, but not GFP-V2, can restore the suppression of viral DNA methylation of a V2 null TYLCV mutant in local infection assays. Cytosine methylation in the intergenic region (IR) of the V2 null mutant genome in locally infected leaf patches of N. benthamiana expressing V2, V2-GFP, or GFP-V2 was detected by bisulfite sequencing at 3 days post-inoculation (dpi). TYLCV or TYLCV-V2null alone were used as controls. The original single-base resolution bisulfite sequencing data are shown in Figure 7—figure supplement 1B. This experiment was repeated twice with similar results. (D) Nbcoilin expression in Nbcoilin-silenced (TRV-Nbcoilin) and control plants (TRV-EV) infected with TYLCV, TYLCV-V2null, or mock-inoculated at 3 wpi measured by RT-qPCR. Gene expression was normalized to NbTubulin. Values are the mean of six independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. **: p<0.01. (E) Percentage of methylated cytosines in the intergenic region (IR) of TYLCV in systemic infection assays with TYLCV wild-type or V2 null mutant (TYLCV-V2null) in Nbcoilin-silenced (TRV-Nbcoilin) or control (TRV-EV) N. benthamiana plants at 3 weeks post-inoculation (wpi), as detected by bisulfite sequencing. Values are the mean of three independent biological replicates; error bars indicate SEM. The original single-base resolution bisulfite sequencing data are shown in Figure 7—figure supplement 2D.
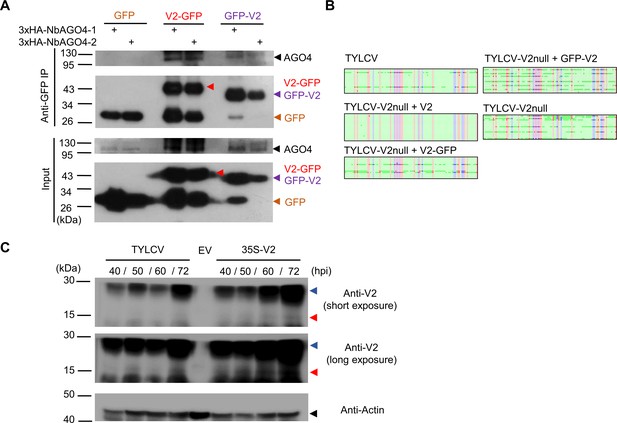
Relevance of the Cajal body localization of the V2-AGO4 interaction.
(A) 3xHA-NbAGO4-1 and 3xHA-NbAGO4-2 interact with V2-GFP and GFP-V2 in co-immunoprecipitation (co-IP) assays upon transient expression in N. benthamiana. Free GFP was used as negative control. Three independent biological replicates were performed with similar results. (B) Original single-base resolution bisulfite sequencing data for Figure 7C. >16 individual clones were sequenced per sample and replicate. Each single circle, corresponding to a cytosine, is colored in blue, red, or green, representing the CHG, CG or CHH contexts, respectively. Methylated cytosines are represented by filled circles, while unmethylated cytosines are represented by empty circles. (C) V2 protein levels in N. benthamiana leaves locally infected by TYLCV or transiently expressing V2 under the 35S promoter. Empty vector (EV) was used as negative control. Three independent biological replicates were performed with similar results. Red arrowheads indicate the predicted size of the V2 protein; blue arrowheads indicate an additional specific band of higher size. hpi, hours post-inoculation.
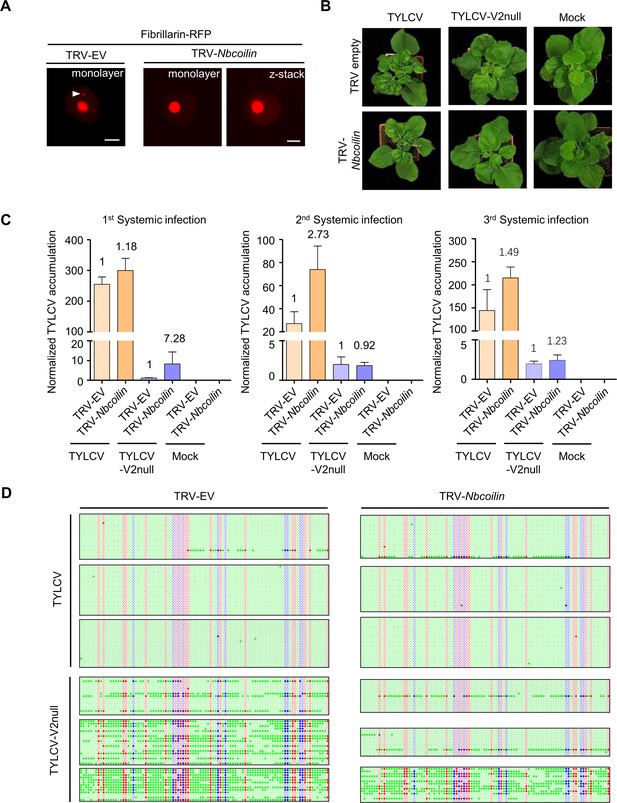
TYLCV infection in Nbcoilin-silenced plants.
(A) No Cajal body was observed in the nuclei of Nbcoilin-silenced plants, whereas in control plants at least one Cajal body was normally present in each nucleus; these results are in agreement with Shaw et al., 2014. Fibrillarin-RFP, used as a nucleolus and Cajal body marker, was transiently expressed in N. benthamiana epidermal cells of Nbcoilin-silenced (TRV-Nbcoilin) and control plants (TRV-EV). Confocal images were taken at 2 days after infiltration. Arrowheads indicate the position of the Cajal body. Bar, 5 μm. This experiment was repeated three times with similar results. (B) Representative pictures of N. benthamiana plants infected with the indicated combinations of viruses. Photographs were taken at 3 weeks post-inoculation (wpi). (C) Viral (TYLCV) accumulation in systemic infections in Nbcoilin-silenced or control plants, measured by qPCR. Apical leaves from six plants were collected at 3 wpi. The experimental design is shown in Figure 3—figure supplement 1B. The accumulation of viral DNA is normalized to the 25S ribosomal RNA interspacer (ITS). Results from three independent experiments are shown. Values are the mean of six independent biological replicates; error bars indicate SEM. The relative fold change of viral accumulation between Nbcoilin-silenced plants and control plants is shown above each column. (D) Original single-base resolution bisulfite sequencing data for Figure 7E. >11 individual clones were sequenced per sample and replicate. Each single circle, corresponding to a cytosine, is colored in blue, red, or green, representing the CHG, CG, or CHH contexts, respectively. Methylated cytosines are represented by filled circles, while unmethylated cytosines are represented by empty circles. Values of IR methylation in independent biological replicates are shown in Supplementary file 1.
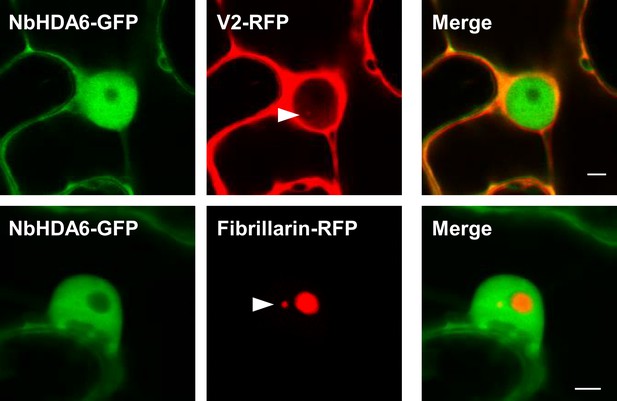
NbHDA6-GFP does not localize to the Cajal body.
NbHDA6-GFP and V2-RFP or Fibrillarin-RFP were transiently co-expressed in N. benthamiana epidermal cells. Fibrillarin-RFP is used as nucleolus and Cajal body marker. Confocal images were taken at 2 days after infiltration. Arrowheads indicate the position of the Cajal body. Bar, 5 μm. This experiment was repeated three times with similar results.
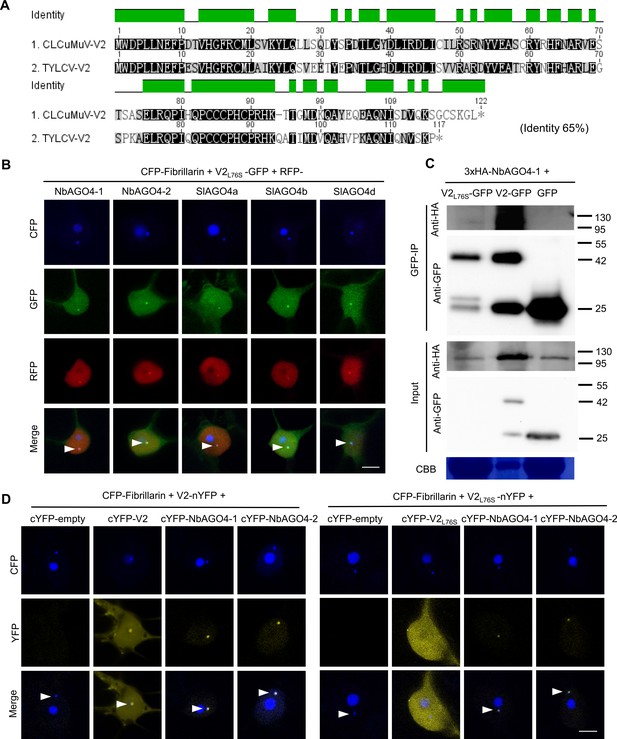
TYLCV V2L76S interacts with AGO4 in the Cajal body.
(A) Alignment of the amino acid sequences of V2 from Cotton Leaf Curl Multan virus (CLCuMuV) and V2 from TYLCV. The alignment was performed by Geneious (https://www.geneious.com). Black background indicates conservation. The identity of these two proteins is 65%. (B) V2L76S-GFP and RFP-AGO4 co-localize in the Cajal body. CFP-Fibrillarin, V2L76S-GFP, and RFP-NbAGO4-1/2 or RFP-SlAGO4a/b/d were transiently co-expressed in N. benthamiana epidermal cells. CFP-Fibrillarin is used as anucleolus and Cajal body marker. Confocal images were taken at two days after infiltration. Arrowheads indicate the position of the Cajal body. Bar, 5 μm. This experiment was repeated three times with similar results. (C) 3xHA-NbAGO4-1 interacts with V2-GFP and V2L76S-GFP in co-immunoprecipitation (co-IP) assays upon transient expression in N. benthamiana. Free GFP was used as negative control. CBB, Coomassie brilliant blue staining. The V2-GFP sample was diluted 1/20 for western blot to reach a protein amount comparable to that of V2L76S-GFP. Three independent biological replicates were performed with similar results. (D) V2L76S interacts with AGO4 in the Cajal body. The N-terminal half of the YFP fused to V2L76S (V2L76S-nYFP) was transiently co-expressed with the C-terminal half of the YFP alone (cYFP, as negative control), or cYFP-NbAGO4, cYFP-SlAGO4, or cYFP- V2L76S in N. benthamiana leaves. CFP-Fibrillarin was used as a nucleolus and Cajal body marker. V2 was used as control. Yellow fluorescence indicates a positive interaction. Arrowheads indicate the position of the Cajal body. Bar, 5 μm. This experiment was repeated three times with similar results.
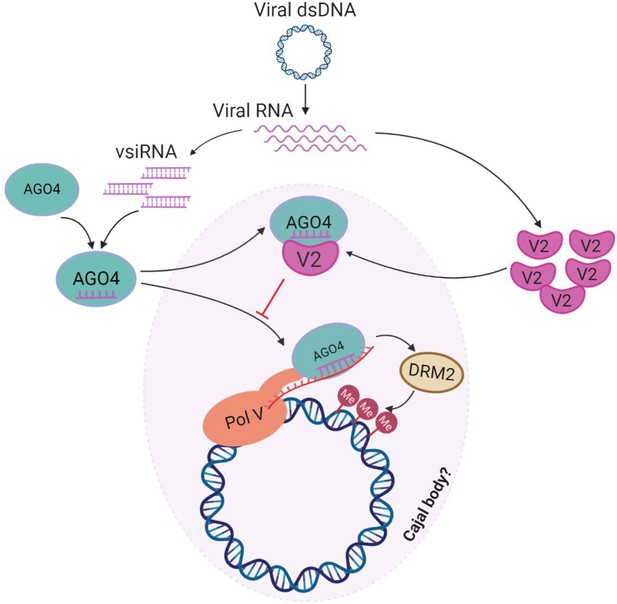
Model for the V2-mediated inhibition of the AGO4-dependent methylation of the viral DNA.
During the viral infection, the ssDNA TYLCV genome forms dsDNA replicative intermediates, which could be targeted by the host AGO4-dependent RNA-directed DNA methylation (RdDM) pathway in a Cajal body-dependent manner as an antiviral defence mechanism. Viral small interfering RNA (vsiRNA) are generated and loaded into AGO4. In the absence of the virus-encoded V2 protein, the AGO4-vsiRNA complex could be effectively guided toward the viral genome by complementary base pairing to the scaffold RNA and association with Pol V, and recruit the methyltransferase DRM2 to catalyze methylation of the viral genome. When V2 is present, however, V2 interacts with AGO4 and interferes with the binding of this protein to the viral RNA and the viral DNA, enabling viral evasion from the AGO4-dependent DNA methylation.
Z-stack video showing the co-localization of V2-GFP and Fibrillarin-RFP in the Cajal body.
Z-stack video showing the localization of GFP-V2 and Fibrillarin-RFP.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Nicotiana benthamiana) | NbAGO4-1 | GenBank | DQ321490 | |
| Gene (Nicotiana benthamiana) | NbAGO4-2 | GenBank | DQ321491 | |
| Gene (Solanum lycopersicum) | SlAGO4a | Sol Genomics | SOLYC01G008960 | |
| Gene (Solanum lycopersicum) | SlAGO4b | Sol Genomics | SOLYC06G073540 | |
| Gene (Solanum lycopersicum) | SlAGO4d | Sol Genomics | SOLYC01G096750 | |
| Antibody | anti-V2 (Rabbit polyclonal) | This work | N/A | WB (1:2000) |
| Antibody | anti-Actin (Rabbit polyclonal) | Agrisera | Cat# AS13 2640 | WB (1:5000) |
| Antibody | anti-FLAG (Mouse monoclonal) | Sigma | Cat# F3165 | IP: (5 µg per gram of tissue) WB (1:5000) |
| Antibody | anti-HA (12CA5) (Mouse monoclonal) | Roche | Cat# 11583816001 | WB (1:5000) |
| Antibody | anti-GFP (Mouse monoclonal) | Abiocode | Cat# M0802-3a | WB (1:5000) |
| Commercial assay or kit | ClonExpress MultiS One Step Cloning Kit | Vazyme | Cat# C113-01 | |
| Commercial assay or kit | pENTR/D-TOPO Cloning Kit | Invitrogen | Cat# K240020SP | |
| Commercial assay or kit | Gateway LR Clonase II Enzyme Mix | Invitrogen | Cat# 11791100 | |
| Commercial assay or kit | QuickChange Lightning Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 210518 | |
| Commercial assay or kit | Plant RNA kit | OMEGA Bio-tek | Cat# R6827 | |
| Commercial assay or kit | iScriptTM cDNA Synthesis Kit | Bio-Rad | Cat# 1708890 | |
| Commercial assay or kit | DNeasy Plant Mini Kit | QIAGEN | Cat# 69104 | |
| Commercial assay or kit | EpiTect Plus DNA Bisulfite Kit | QIAGEN | Cat# 59124 | |
| Commercial assay or kit | QIAquick PCR Purification Kit | QIAGEN | Cat# 28106 | |
| Commercial assay or kit | Superscript III first-strand synthesis system | Thermo Fisher Scientific | Cat# 18080051 | |
| Software, algorithm | Cytosine methylation analysis Kismeth | Gruntman et al., 2008 | PMID:18786255 | http://katahdin.mssm.edu/kismeth/revpage.pl |
Additional files
-
Supplementary file 1
Values of IR methylation in independent experiments and biological replicates.
- https://cdn.elifesciences.org/articles/55542/elife-55542-supp1-v1.docx
-
Supplementary file 2
Exclusive unique peptide count of NRPE1 co-immunoprecipitated with NbAGO4-1 in the presence or absence of V2 as identified by AP-MS.
- https://cdn.elifesciences.org/articles/55542/elife-55542-supp2-v1.docx
-
Supplementary file 3
List of primers used for cloning in this study.
- https://cdn.elifesciences.org/articles/55542/elife-55542-supp3-v1.docx
-
Supplementary file 4
List of plasmids used in this study.
- https://cdn.elifesciences.org/articles/55542/elife-55542-supp4-v1.docx
-
Supplementary file 5
List of primers used for qPCR and qRT-PCR in this study.
- https://cdn.elifesciences.org/articles/55542/elife-55542-supp5-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55542/elife-55542-transrepform-v1.docx





