Start codon disruption with CRISPR/Cas9 prevents murine Fuchs’ endothelial corneal dystrophy
Figures
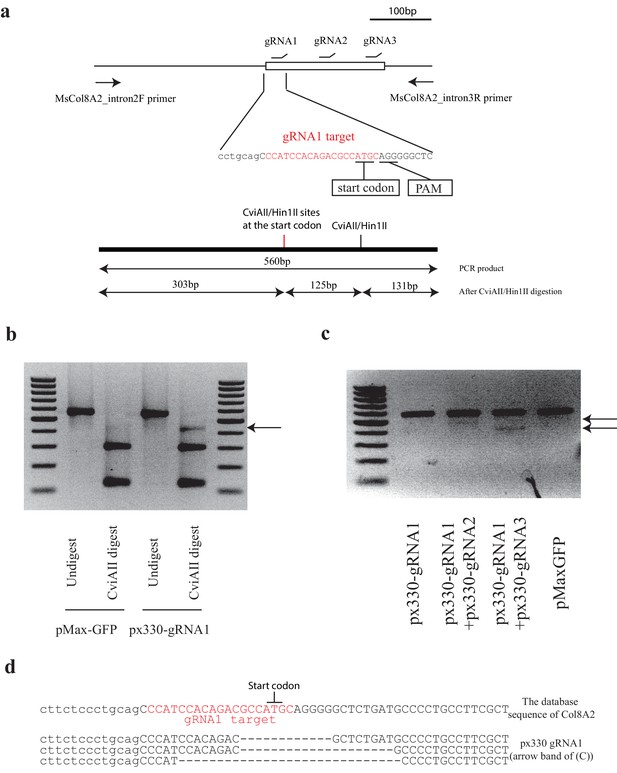
Design of Col8a2 guide RNA and indel confirmation in vitro.
(a) Design of guide RNAs (gRNAs) for mouse Col8a2 gene and the schematic diagram of indel detection by restriction enzyme digestion of the PCR product. gRNA1, which is used for Ad-Cas9-Col8a2gRNA, was designed to disrupt the Col8a2 start codon. PCR primers were designed to flank the start codon and gRNA-targeting sites. PCR product from the intact DNA sequence was of 560 bp, which was digested to 303 bp, 131 bp, and 126 bp by CviAII/Hin1II restriction enzymes. (b) In px330-gRNA1-transfected NIH3T3 cells, the PCR product showed an extra band (~430 bp, arrow) after CviAII digestion. pMax-GFP was used as a control. (c) A combination of two plasmids (px330-gRNA1 + px330-gRNA2 and px330-gRNA1 + px330-gRNA2) yields lower bands (arrow), reflecting the deletion between the targeted sites. (d) Deletion of the start codon by px330-gRNA1 was confirmed by Sanger sequencing after cloning.
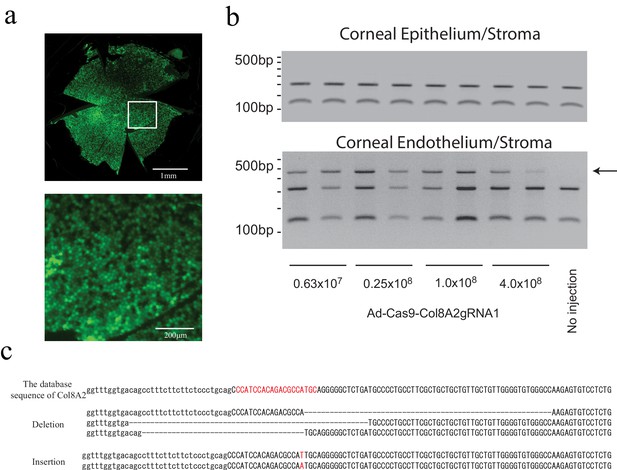
Intracameral injection of Ad-Cas9-Col8a2gRNA1 induces indel at the Col8a2 start codon in corneal endothelium.
(a) Adenovirus infection to corneal endothelium via intracameral injection was confirmed by adenovirus GFP. Top: whole mouse cornea flatmount. Bottom: the magnified section of the image. (b) Ad-Cas9-Col8a2gRNA1 induced an insertion/deletion (indel) at the Col8a2 start codon in the corneal endothelium but not in the corneal epithelium/stroma. Genomic DNA of corneal endothelium/stroma and corneal epithelium/stroma was PCR amplified with primers flanking the Col8a2 start site and digested with CviAII, which recognizes the intact Col8a2 start codon (5’-CATG-3’). The CviAII undigested band (arrow) demonstrates the indel at the Col8a2 start codon. (c) Sanger sequencing of the cloned PCR product from genomic DNA purified from corneal endothelium/stroma confirming indels at the Col8a2 start codon.
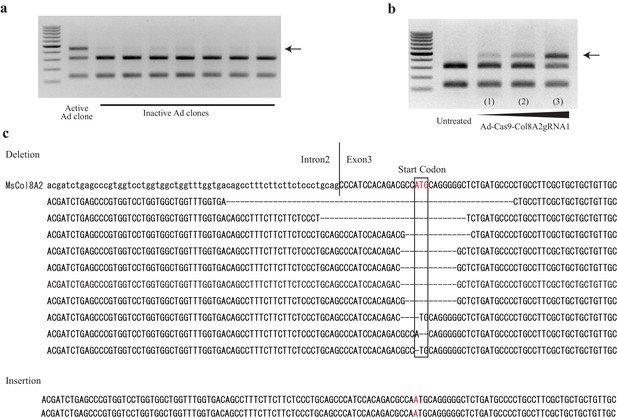
Ad-Cas9-Col8a2gRNA cloning and its indel activity in vitro.
(a) Cloning of Ad-Cas9-Col8a2gRNA by its indel activity in AD293 cells. The method was the same as described for Figure 1. The extra band (arrow) demonstrates the indel at the start codon. We examined 30 different clones and found only one clone with indel activity. (b) As Ad-Cas9-Col8a2gRNA titer increased, indel activity also increased. (c) Results from Sanger sequencing of cloned PCR products.
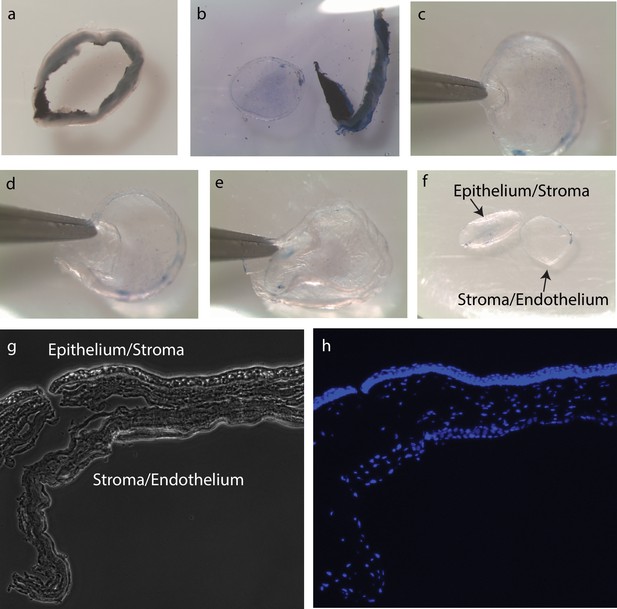
Procedure of peeling off mouse corneal endothelium.
Mouse corneal endothelium was peeled off mechanically. (a) Mouse cornea after excision from the rest of the eye. (b) Mouse cornea was stained with 0.4% trypan blue for visualization, and the limbus/sclera was removed. (c–e) Mechanical peeling of corneal endothelium. (f) Epithelium/stroma and stroma/endothelium after complete separation. (g, h) Cryosection image of the cornea with endothelium peeled. 4′,6-diamidino-2-phenylindole (DAPI) staining showed incomplete separation of corneal endothelium and stroma.
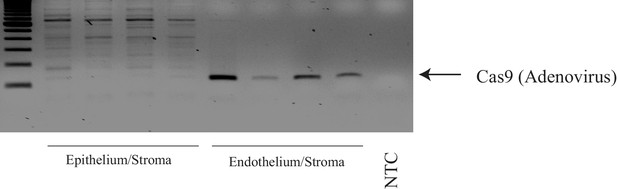
Adenovirus genome was detected in corneal endothelium but not epithelium.
1 week following adenovirus injection, we purified DNA from corneal endothelium and epithelium/stroma, respectively. Then, PCR was conducted using primers to detect Cas9.
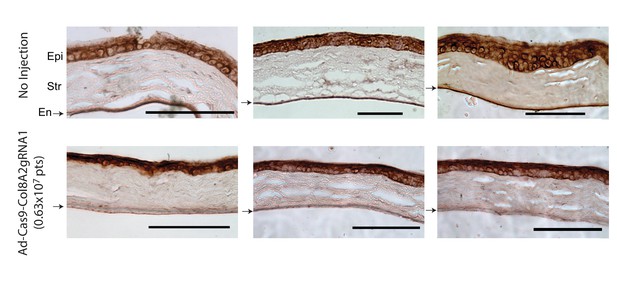
Ad-Cas9-Col8a2gRNA reduces COL8A2 expression in mouse corneal endothelium but not epithelium.
COL8A2 protein immunostaining from the cornea 2 months after injection with Dulbecco’s phosphate-buffered saline (DPBS) (4 µl, upper figures) or Ad-Cas9-Col8a2gRNA (0.63 × 107 vg in 4 µl, lower figures). In Ad-Cas9-Col8a2gRNA-injected corneas, lower COL8A2 protein expression was seen in corneal endothelium, but not in epithelium. Epi: epithelium, Str: stroma, En (arrow): endothelium. Scale bar = 100 µm.
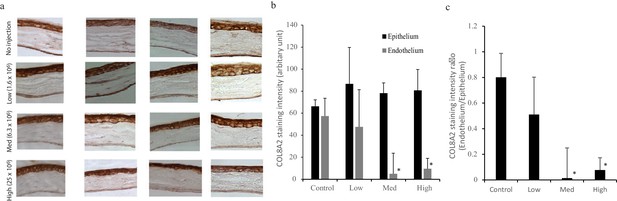
COL8A2 reduction in corneal endothelium is correlated with the amount of Ad-Cas9-Col8a2gRNA.
(a) COL8A2 immunostaining in each amount of Ad-Cas9-Col8a2gRNA injection. (b) COL8A2 expression in corneal endothelium and corneal epithelium was quantified from the images. The staining intensity in isotype control was subtracted as a background. (c) The ratio of COL8A2 staining between corneal endothelium and epithelium. *p<0.01 by Student’s t-test. The source data is Figure3-figure supplement1_source data.xlsx.
-
Figure 3—figure supplement 1—source data 1
COL8A2 staining intensity.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig3-figsupp1-data1-v2.xlsx
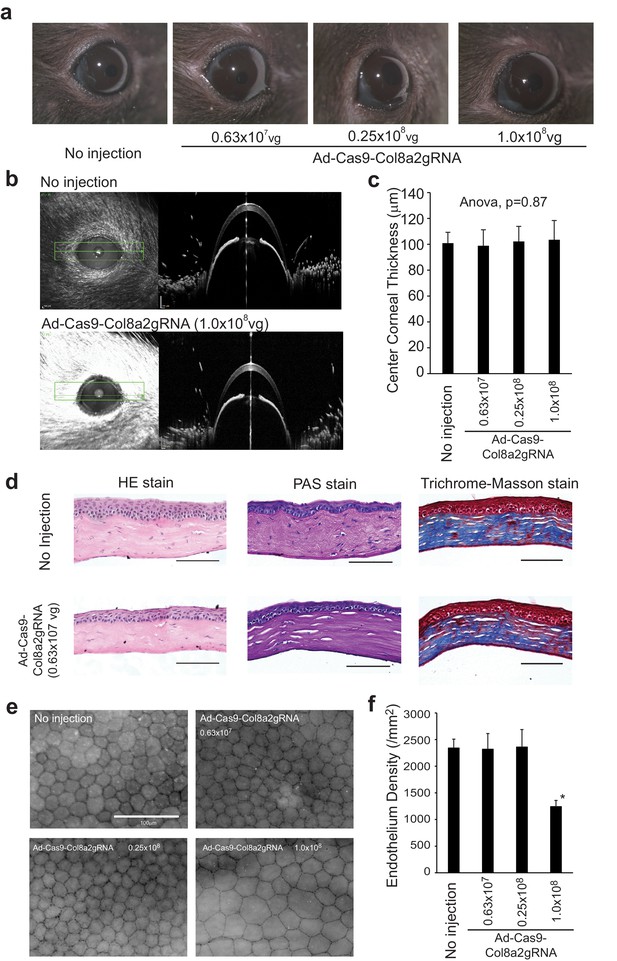
Low doses of Ad-Cas9-Col8a2gRNA did not show toxicity.
(a) Injection of Ad-Cas9-Col8a2gRNA at 0.63 × 107, 0.25 × 108, and 1.0 × 108 vg did not result in corneal edema or opacity. (b) Representative corneal optical coherence tomography (OCT) images captured by Heidelberg Spectralis microscope with/without Ad-Cas9-Col8a2gRNA injection. (c) The average of central corneal thickness in each condition. Significant differences among groups were not observed (analysis of variance (ANOVA), p = 0.78). n = 8–12. Error bars show standard deviation. (d) Hematoxylin-eosin (HE), Periodic Acid-Schiff (PAS), and trichrome Masson staining showed no apparent phenotypes in Ad-Cas9-Col8A2gRNA-injected corneas compared to non-injected corneas. Scale bar = 50 µm. (e) Representative images of corneal flat mounts immunolabeled with ZO-1 antibody for each condition. Scale bar = 100 µm. (f) Average corneal endothelium densities. 1.0 × 108 vg Ad-Cas9-Col8A2gRNA reduced corneal endothelium density significantly; n = 6–9. *p<0.001 by Student’s t-test. Error bars show standard deviation. The source data is (c) Figure4-source data 1.xlsx and (f) Figure4-source data 2.xlsx.
-
Figure 4—source data 1
Central corneal thickness.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Corneal endothelial cell density.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig4-data2-v2.xlsx
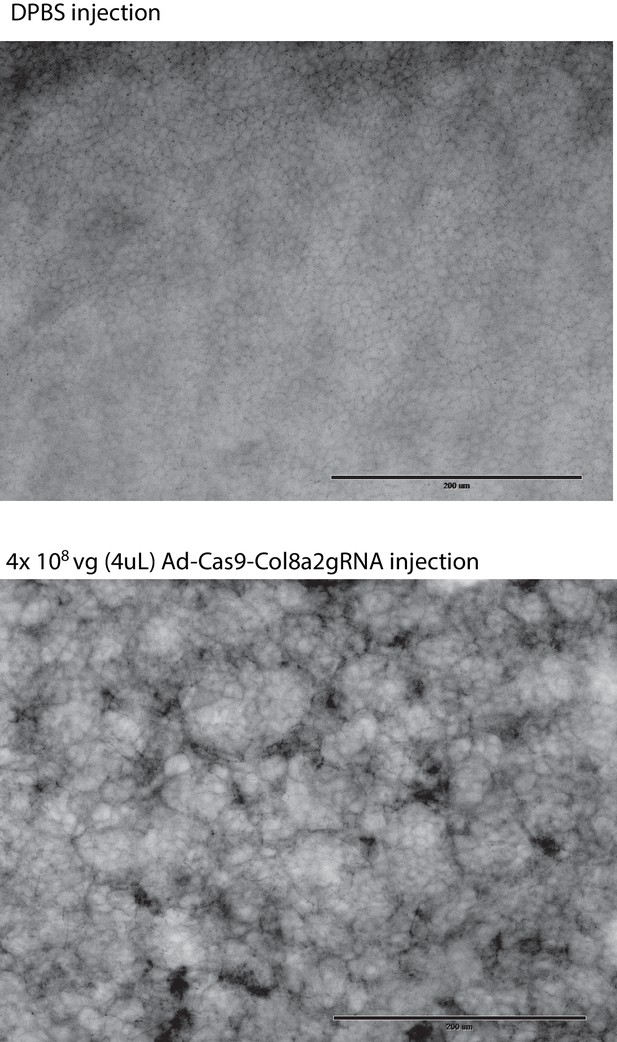
High levels of Ad-Cas9-Col8a2gRNA (4 × 108) are toxic to corneal endothelium in C57BL/6J mice.
2 weeks following intracameral injection of Dulbecco’s phosphate-buffered saline (DPBS) or Ad-Cas9-Col8a2gRNA (4 × 108 vg), corneas were harvested to examine endothelial integrity with anti-ZO-1 antibody. This high titer of Ad-Cas9-Col8a2gRNA led to widespread devastation of the corneal endothelium. Scale bar = 200 µm.
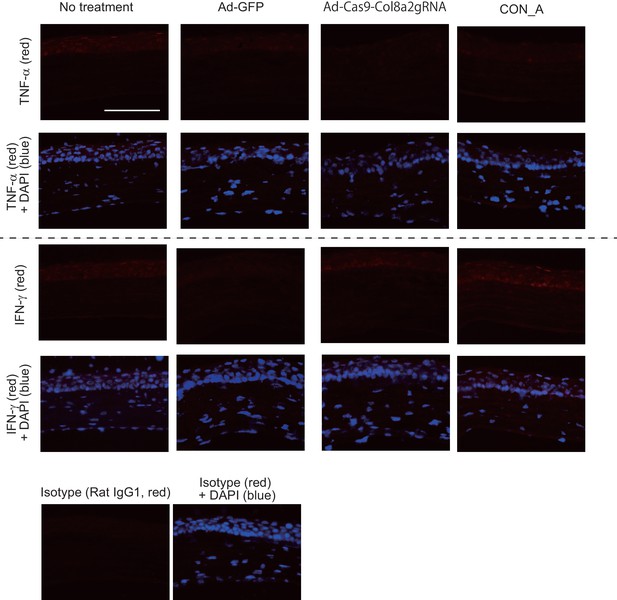
Intracameral injection of 0.25 × 108 Ad-Cas9-Col8a2gRNA does not show significant induction of inflammation markers.
Tumor necrosis factor alpha (TNFα) and interferon gamma (IFNγ) were stained 4 weeks post Ad-GFP, Ad-Cas9-Col8a2gRNA, or concanavalin A (1 μg) intravitreal injection. Scale bar is 100 μm.
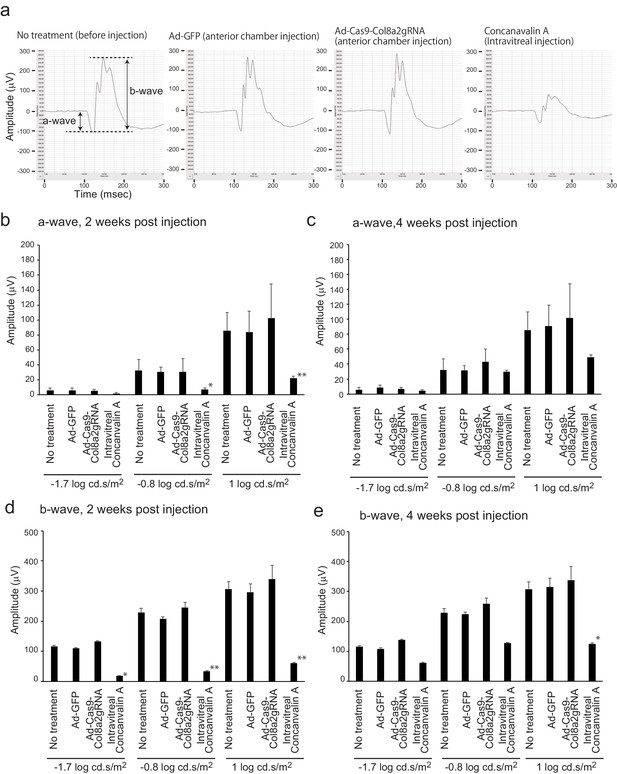
Ad-Cas9-Col8a2gRNA does not induce retinal disfunction.
Dark-adapted electroretinography (ERG) was used for evaluation of retinal function. (a) Representative ERG of no treatment (prior to injection), Ad-GFP (anterior chamber injection), Ad-Cas9-Col8a2gRNA (anterior chamber injection), and concanavalin A (intravitreal injection). Intravitreal injection of concanavalin A was used as a positive control by inducing retinal inflammation. (b, c) a-wave of no treatment and each treatment 2 and 4 weeks post injection. We used three different stimulus light intensities (−1.7,–0.8, and 1 log cd.s/m2). (d, e) b-wave of no treatment and each treatment 2 and 4 weeks post injection. n = 14 (no treatment), 6 (Ad-GFP), 6 (Ad-Cas9-Col8a2gRNA), and 2 (concanavalin A, 1 μg). *p<0.05 and **p<0.01 by Student’s t-test compared to no-treatment control. The source data is Figure4-figure supplement3_source data.xlsx.
-
Figure 4—figure supplement 3—source data 1
ERG.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig4-figsupp3-data1-v2.xlsx
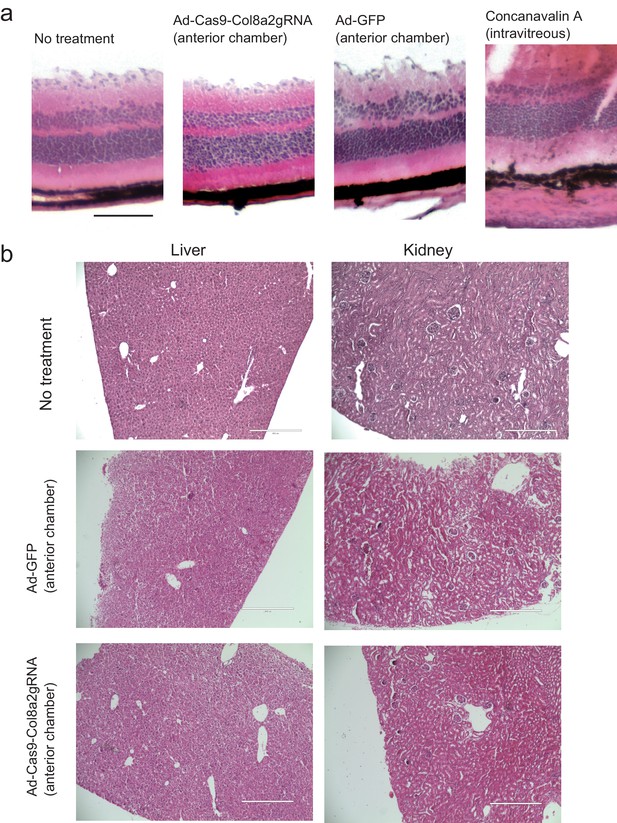
Anterior chamber injection of 0.25 × 108 Ad-Cas9-Col8a2gRNA does not show retina, liver, and kidney toxicity.
4 weeks post injection, we observed each tissue by hematoxylin-eosin (HE) staining. (a) Retina. Scale bar is 100 μm. (b) Liver and kidney. Scale bar is 400 μm.
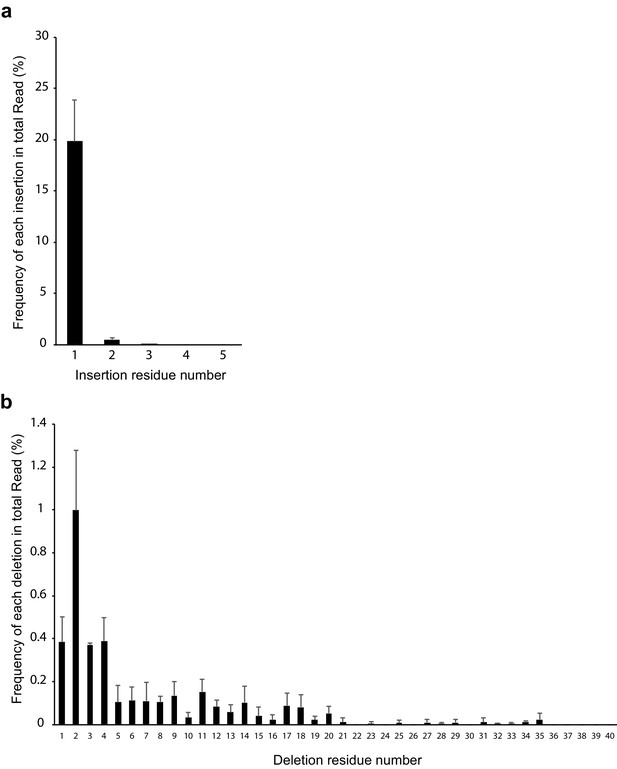
Distribution of inserted and deleted residue number.
(a) Frequency of insertion. 1 bp insertion was most frequent. (b) Frequency of deletion. 2 bp deletion was most frequent. n = 4. Error bar represents standard deviation. The source data is Figure5-source data.xlsx.
-
Figure 5—source data 1
Counts of insertion/deletion.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig5-data1-v2.xlsx
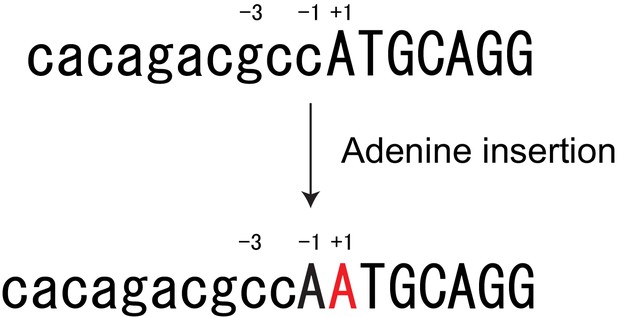
Single adenine insertion at the mouse Col8a2 start codon.
An adenine insertion produced a cryptic ATG codon that resulted in disruption of Kozak sequence (G to C at −3 position).
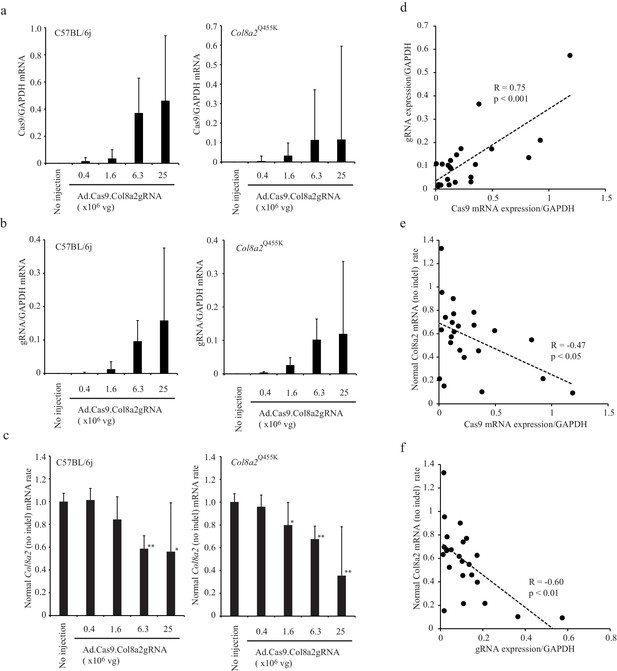
The indel rate was correlated to Cas9 and gRNA expression.
(a and b) Cas9 mRNA and gRNA expression in corneal endothelium 1 week following anterior chamber injection of Ad-Cas9-Col8a2gRNA. (c) The expression ratio of mouse Col8a2 mRNA without indels and total Col8a2 mRNA with/without indels were determined by real-time reverse transcription-PCR (RT-PCR). *p<0.05 and **p<0.01 by Student’s t-test. (d) gRNA and Cas9 mRNA expression are positively correlated. (e) Normal Col8a2 mRNA (no indel) rate and Cas9 mRNA are negatively correlated. (f) Normal Col8a2 mRNA (no indel) rate and gRNA expression are negatively correlated. The source data is Figure6_source data.xlsx.
-
Figure 6—source data 1
Quantification by real-time PCR.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig6-data1-v2.xlsx
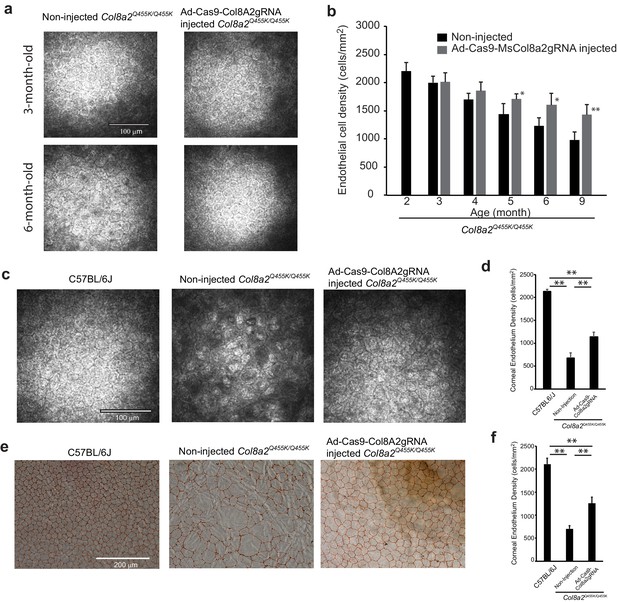
Ad-Cas9-Col8a2gRNA intracameral injection rescues corneal endothelium loss in the early-onset Fuchs’ dystrophy mice (Col8a2Q455K/Q455K) model.
(a) Representative in vivo corneal endothelium images using the Heidelberg Rostock microscope at 3 and 6 months post injection. Ad-Cas9-Col8a2gRNA was injected intracamerally into Col8a2Q455K/Q455K mice at 2 months of age. Scale bar = 100 μm. (b) Time course change in corneal endothelial cell density of Col8a2Q455K/Q455K mice, n = 5. Ad-Cas9-Col8a2gRNA slows loss of corneal endothelial cells compared to no-injection group. (c) Representative in vivo corneal endothelium image at 12 months of age. Age-matched C57BL/6J and non-injected Col8a2Q455K/Q455K mice were used for comparison. Ad-Cas9-Col8a2gRNA qualitatively improved endothelial cell density. Scale bar = 100 μm. (d) Average corneal endothelium densities: C57BL/6J: 2134 ± 45 cells/mm2, non-injected Col8a2Q455K/Q455K: 677 ± 110 cells/mm2, and Ad-Cas9-Col8a2gRNA-injected Col8a2Q455K/Q455K: 1141 ± 102 cells/mm2, n = 4. Error bars show standard deviation. (e) Representative corneal endothelium from each group stained with Alizarin red. Scale bar = 200 μm. (f) Average corneal endothelium densities calculated from Alizarin red-stained corneas: C57BL/6J: 2108 ± 134 cells/mm2, non-injected Col8a2Q455K/Q455K: 696 ± 70 cells/mm2, and Ad-Cas9-Col8a2gRNA-injected Col8a2Q455K/Q455K: 1256 ± 135 cells/mm2, n = 4. Error bars show standard deviation. The source data is Figure7_source data.xlsx.
-
Figure 7—source data 1
Corneal endothelial cell density in vivo and ex vivo.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig7-data1-v2.xlsx
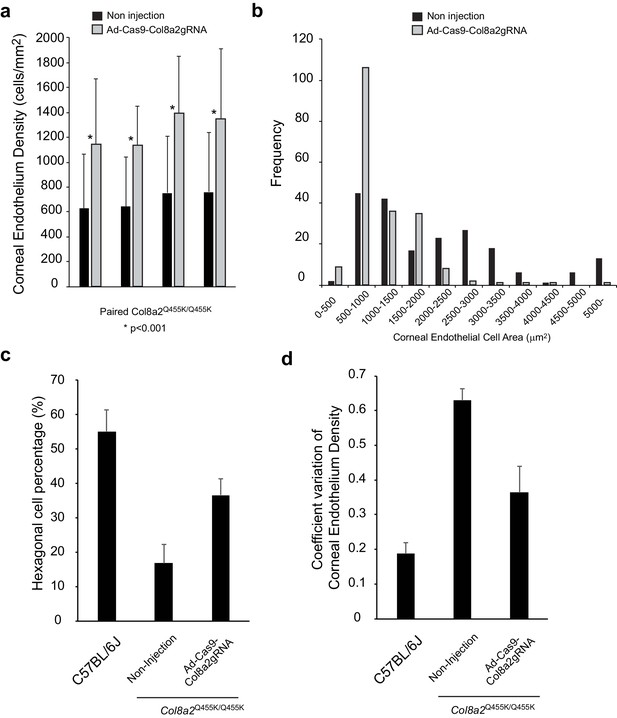
Ad-Cas9-Col8A2gRNA improves various characteristics of corneal endothelium in Col8a2Q455K/Q455K mice.
(a) Corneal endothelium density in each cornea was calculated using Alizarin red staining. A total of 50 different cell areas were measured in each cornea. Injected (Ad-Cas9-Col8a2gRNA) and non-injected corneas in the same mouse were compared by Student’s paired t-test. (b) Histogram of corneal endothelial cell area in Ad-Cas9-Col8A2gRNA-injected cornea and non-injected cornea quantitatively demonstrates left-shifting in cell size, that is, enhanced density, in the former. N = 200 in each group from four different corneas. (c) Hexagonality and (d) coefficient of variation (COV) of corneal endothelium were significantly improved by Ad-Cas9-Col8A2gRNA intracameral injection in Col8a2Q455K/Q455K mice. N = 200 from four different corneas in each group. The source data is Figure8_source data.xlsx.
-
Figure 8—source data 1
The area, hexagonality, and COV of corneal endothelial cells.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig8-data1-v2.xlsx
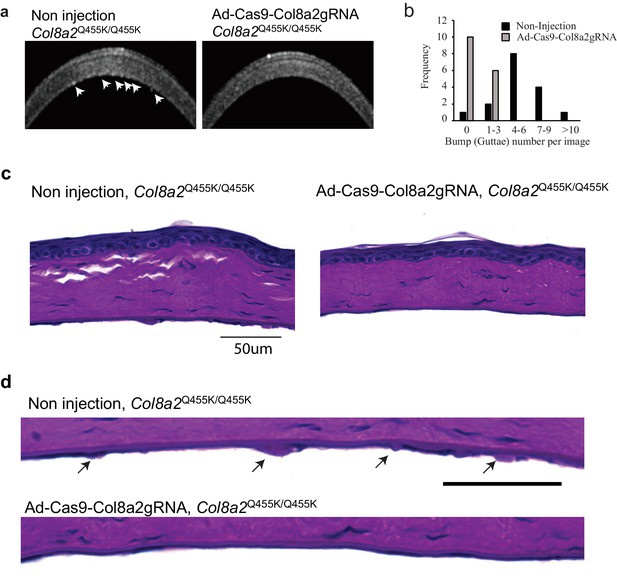
Ad-Cas9-Col8A2gRNA reduced guttae-like structures on the corneal endothelium in Col8a2Q455K/Q455K mice.
(a) Corneal optical coherence tomography (OCT) revealed numerous guttae-like excrescences (arrows) in 1-year-old Col8a2Q455K/Q455K mice, but far fewer in Ad-Cas9-Col8a2gRNA-injected Col8a2Q455K/Q455K mice. (b) Histogram showing the number of guttae-like structures in each group. Non-injected Col8a2Q455K/Q455K: 5.2 ± 3.4 excrescences/image and Ad-Cas9-Col8a2gRNA-injected Col8a2Q455K/Q455K: 0.5 ± 0.73 excrescences/image. n = 16. P-value by Mann-Whitney U-test is <0.001. (c, d) Periodic Acid-Schiff (PAS)-stained corneas from non-injected and Ad-Cas9-Col8a2gRNA-injected Col8A2Q455K/Q455K mice. The arrows indicate guttae-like structures (excrescences). The source data is Figure9_source data.xlsx.
-
Figure 9—source data 1
Number of guttae-like structures.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig9-data1-v2.xlsx
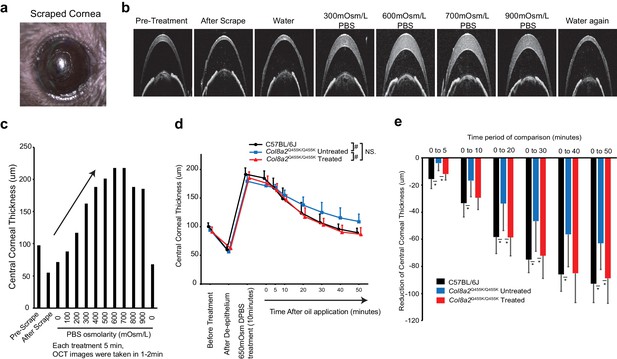
Ad-Cas9-Col8a2gRNA rescued corneal endothelium pumping function in Col8a2Q455K/Q455K mouse.
(a) Stereomicroscopic images of scraped mouse cornea. (b) Corneal optical coherence tomography (OCT) images of pre-treatment, after scrape, and after treatment with 0 mOsm/l (water), 300, 600, 700, and 900 mOsm/l Dulbecco’s phosphate-buffered saline (DPBS) application followed by water again. (c) Changes in corneal thickness in response to variance in DPBS osmolality demonstrate that maximal swelling occurred at 600–700 mOsm/l DPBS. (d) Repeated measurements of central corneal thickness were taken using corneal OCT after application of 650 mOsm/l PBS. To prevent evaporation, 4 µl of silicone oil was applied at t = 0 (n = 6). #p<0.001 by regression analysis. NS: not significant. (e) De-swelling of central corneal thickness was measured from 0 min to 5, 10, 20, 30, 40, and 50 min. Non-injected Col8a2Q455K/Q455K corneas showed significantly delayed de-swelling compared to C57BL/6J corneas. In contrast, Ad-Cas9-Col8a2gRNA injection significantly improved corneal de-swelling rate similar to that of C57BL/6J controls (n = 6). *p<0.05 by Student’s t-test. The source data is Figure10_source data.xlsx.
-
Figure 10—source data 1
Time course change of corneal thickness.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig10-data1-v2.xlsx
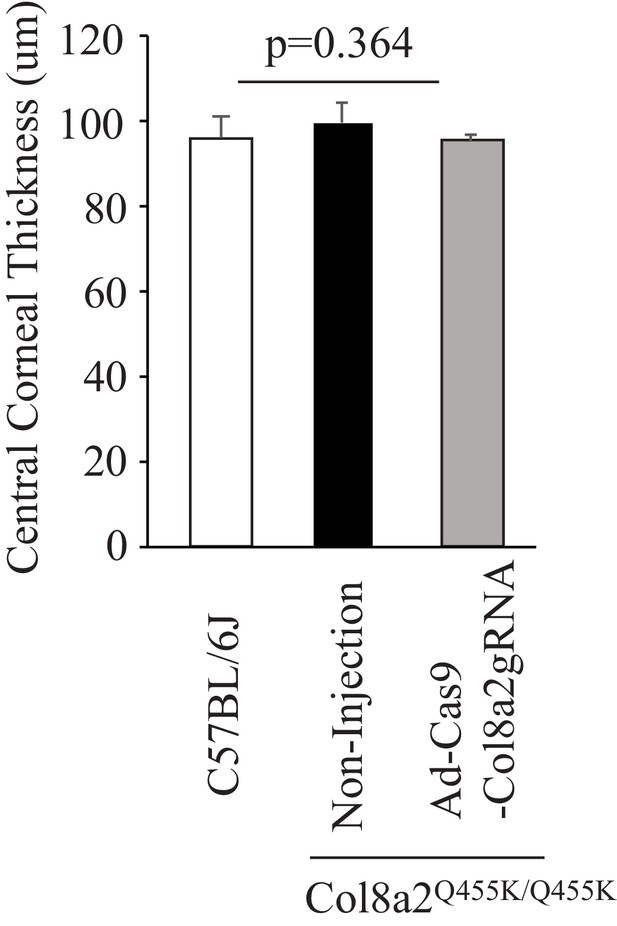
Col8a2Q455K/Q455K mice (12 months old) did not show a significant difference in central corneal thickness.
Central corneal thickness was measured by corneal optical coherence tomography (OCT); p-value was calculated by analysis of variance (ANOVA). The source data is Figure10-figure supplement1_source data.xlsx.
-
Figure 10—figure supplement 1—source data 1
Central corneal thickness.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig10-figsupp1-data1-v2.xlsx
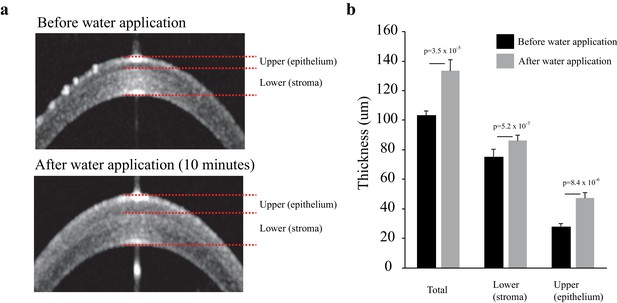
Water applied to the corneal surface expands the thickness of corneal epithelium rather than the stroma.
(a) Representative corneal optical coherence tomography (OCT) images before and after water was applied for 10 min. (b) The average thickness of total, upper (epithelium), and lower (stroma) corneal surface before and after water application for 10 min. n = 5. Error bars show standard deviation; p-value was calculated by Student’s t-test. The source data is Figure10-figure supplement2_source data.xlsx.
-
Figure 10—figure supplement 2—source data 1
Thickness of epithelium and stroma.
- https://cdn.elifesciences.org/articles/55637/elife-55637-fig10-figsupp2-data1-v2.xlsx
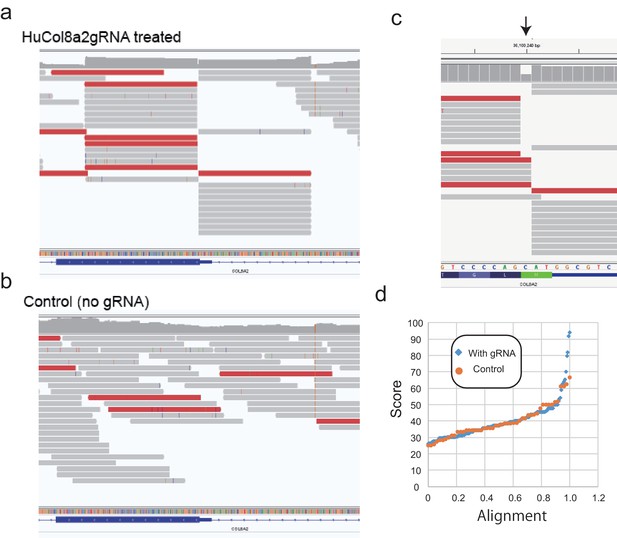
Modified digenome analysis for potential off-targets.
(a, b) Mapping of reads to human COL8A2 target site from HuCol8a2gRNA-treated genomic DNA (gDNA) and control gDNA. (c) A gap was observed in in vitro digestion of genomic DNA. (d) Modified digenome score alignment (0–1.0) of control gDNA (no gRNA) and HuGol8a2gRNA-treated gDNA.
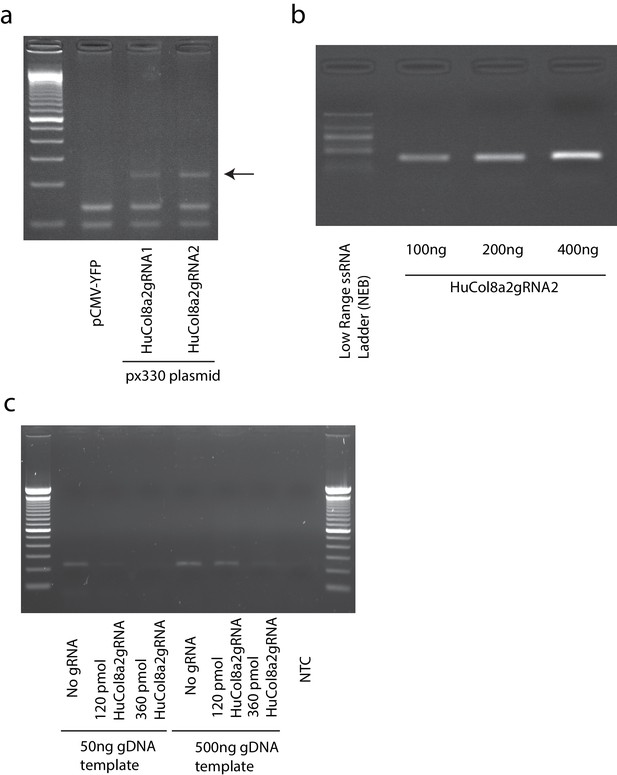
In vitro digestion by Cas9/HuCol8a2gRNA.
(a) Plasmid-based Cas9/HuCol8a2gRNA induced (insertions/deletions) indels in AD293 cells. Arrow indicates an indel band. (b) Gel electrophoresis image of in vitro transcription of HuCol8a2gRNA. (c) PCR confirmed in vitro digestion of purified AD293 genomic DNA by Cas9/HuCol8a2gRNA. PCR primers were designed to target the digestion site.
Tables
Indel rate at mouse Col8a2 target site by Ad-Cas9-Col8a2gRNA from corneal endothelium.
| Total read | No change | Insertion | Deletion | Indel | |
|---|---|---|---|---|---|
| Cornea1 | 87,554 | 68,228 | 16,378 | 2948 | 19,326 |
| (77.9%) | (18.7%) | (3.4%) | (22.1%) | ||
| Cornea2 | 97,749 | 69,455 | 24,202 | 4092 | 28,294 |
| (77.1%) | (24.8%) | (4.2%) | (28.9%) | ||
| Cornea3 | 87,908 | 71,664 | 13,508 | 2736 | 16,244 |
| (81.5%) | (24.8%) | (3.1%) | (18.5%) | ||
| Cornea4 | 93,234 | 69,747 | 19,831 | 3656 | 23,487 |
| (74.8%) | (21.3%) | (3.9%) | (25.2%) | ||
| Average of ratio | 76.3 ± 4.5% | 20.0 ± 4.0% | 3.6 ± 0.5% | 23.7 ± 4.5% | |
-
The source data is Table1-source data.xlsx.
-
Table 1—source data 1
Indel rate in Col8a2 gene.
- https://cdn.elifesciences.org/articles/55637/elife-55637-table1-data1-v2.xlsx
Ratio of A:T:G:C in 1 bp insertions.
| Total read number of single insertions in the start codon (between A and T) | A | T | G | C | |
|---|---|---|---|---|---|
| Cornea1 | 15,655 | 7925 (50.6%) | 6703 (42.8%) | 230 (1.5%) | 797 (5.1%) |
| Cornea2 | 23,315 | 10,877 (46.7%) | 10,890 (46.7%) | 294 (1.3%) | 1254 (5.4%) |
| Cornea3 | 13,083 | 6035 (46.1%) | 6013 (46.0%) | 320 (2.4%) | 715 (5.5%) |
| Cornea4 | 18,829 | 9706 (51.5%) | 8088 (43.0%) | 356 (1.9%) | 679 (3.6%) |
| Average of ratio | 48.7 ± 2.7% | 44.6 ± 2.0% | 1.8 ± 0.5% | 4.9 ± 0.9% | |
-
The source data is Table2-source data.xlsx.
-
Table 2—source data 1
Number of inserted DNA residues.
- https://cdn.elifesciences.org/articles/55637/elife-55637-table2-data1-v2.xlsx
Normalized indel rate by the purified genomic DNA amount.
| Concentration (ng/ul) | gDNA amount (ng, 16 ul elution) | Cell number from gDNA amount | Intact indel rate (%) | Normalized indel rate (%) | |
|---|---|---|---|---|---|
| Cornea1 | 14.5 | 232 | 38,744 | 22.1 | 118.6 |
| Cornea2 | 10.5 | 168 | 28,056 | 28.9 | 112.3 |
| Cornea3 | 12 | 192 | 32,064 | 18.5 | 82.1 |
| Cornea4 | 10.4 | 166.4 | 27,789 | 25.2 | 97.0 |
HuCol8a2gRNA off-target sites with homology.
| Chr | Gap | Start | Gene | Plus | Depth | Perc | Minus | Depth | Perc | Total | Sequence with PAM* | Identity (% including PAM) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0 | 36100241 | COL8A2, coding (target site) | 13 | 13 | 100 | 5 | 6 | 83.33 | 94.74 | CGTCCACGGACGCCATGCTGGG | 100 |
| 1 | 1 | 36100241 | 13 | 13 | 100 | 9 | 15 | 60 | 78.57 | ||||
| 1 | 1 | 36100241 | 11 | 11 | 100 | 7 | 11 | 63.64 | 81.82 | ||||
| 2 | 1 | -1 | 143388988 | Intergenic | 21 | 30 | 70 | 26 | 29 | 89.66 | 79.66 | CGTCCATGGACCCCAAGCTAGG | 81.8 |
| 1 | 0 | 143388989 | 29 | 59 | 49.15 | 26 | 29 | 89.66 | 62.5 | ||||
| 3 | 1 | -1 | 144214582 | Intergenic | 21 | 30 | 70 | 26 | 31 | 83.87 | 77.05 | CGTCCATGGACCCCAAGCTAGG | 81.8 |
| 1 | 0 | 144214583 | 29 | 59 | 49.15 | 26 | 31 | 83.87 | 61.11 | ||||
| 4 | 1 | -1 | 144751794 | SRGAP2-AS1 | 26 | 31 | 83.87 | 20 | 29 | 68.97 | 76.67 | CGTCCATGGACCCCAAGCTAGG | 81.8 |
| 1 | 0 | 144751794 | 26 | 31 | 83.87 | 29 | 58 | 50 | 61.8 | ||||
| 5 | 2 | -1 | 89549893 | Intergenic | 21 | 30 | 70 | 17 | 20 | 85 | 76 | CGTCCATGGACCCCAAGCTAGG | 81.8 |
| 2 | 0 | 89549894 | 29 | 59 | 49.15 | 17 | 20 | 85 | 58.23 | ||||
| 6 | 2 | -1 | 91624245 | Intergenic | 21 | 30 | 70 | 26 | 29 | 89.66 | 79.66 | CGTCCATGGACCCCAAGCTAGG | 81.8 |
| 2 | 0 | 91624246 | 29 | 59 | 49.15 | 26 | 29 | 89.66 | 62.5 | ||||
| 7 | 4 | -1 | 3707175 | Intergenic | 7 | 8 | 87.5 | 10 | 10 | 100 | 94.44 | TGCCCACGGGCACCATGTTGGG | 77.3 |
| 4 | -1 | 3707175 | 7 | 8 | 87.5 | 9 | 9 | 100 | 94.12 | ||||
| 8 | 4 | -1 | 4185990 | Intergenic | 15 | 21 | 71.43 | 19 | 28 | 67.86 | 69.39 | AGTCCATGGACCACAAGCTAGG | 72.7 |
| 4 | 0 | 4185990 | 15 | 21 | 71.43 | 26 | 54 | 48.15 | 54.67 | ||||
| 9 | 5 | -1 | 76221510 | SV2C, intron | 9 | 11 | 81.82 | 10 | 17 | 58.82 | 67.86 | TGTCCAC-AACGTCATGCTTGG | 72.7 |
| 5 | -1 | 76221510 | 9 | 11 | 81.82 | 7 | 14 | 50 | 64 | ||||
| 10 | 10 | -1 | 74854844 | KAT6B, intron | 12 | 20 | 60 | 21 | 21 | 100 | 80.49 | CGTACACAGAAACCATGCTGGG | 81.8 |
| 10 | -1 | 74854844 | 12 | 20 | 60 | 19 | 19 | 100 | 79.49 | ||||
| 11 | 10 | -1 | 130741051 | Intergenic | 7 | 10 | 70 | 10 | 16 | 62.5 | 65.38 | AGTCCA-GGAGGCCATGCTTGG | 81.8 |
| 10 | -1 | 130741051 | 7 | 10 | 70 | 10 | 16 | 62.5 | 65.38 | ||||
| 12 | 13 | -1 | 75612825 | LMO7-AS1 | 8 | 14 | 57.14 | 13 | 17 | 76.47 | 67.74 | GGTCCAC-GCCGCCATGCCCGG | 77.3 |
| 13 | -1 | 75612825 | 8 | 14 | 57.14 | 13 | 16 | 81.25 | 70 | ||||
| 13 | 15 | 1 | 88847941 | ACAN, coding | 16 | 16 | 100 | 18 | 21 | 85.71 | 91.89 | AGCCCCCGGACCCCATGCGTGG | 77.3 |
| 15 | 1 | 88847941 | 16 | 16 | 100 | 17 | 20 | 85 | 91.67 |
-
*Red characters indicate mismatched DNA residues.
HuCol8a2gRNA off-target sites without homology.
| Location | Chr | Gap | Start | Plus | Depth | Perc | Minus | Depth2 | Perc2 | Total | Sequence (50 bp around the detection site) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 1 | 189206630 | 5 | 10 | 50 | 6 | 9 | 66.67 | 57.89 | gaacctcccacctcagcctaccgagtagctgagactatgggcacattccg |
| 3 | 1 | 189206630 | 5 | 9 | 55.56 | 5 | 7 | 71.43 | 62.5 | ||
| 2 | 4 | 0 | 83023938 | 5 | 7 | 71.43 | 6 | 11 | 54.55 | 61.11 | acacatggacacagggagggggacatcactgtgtgatgtggggggcaagg |
| 3 | 8 | 1 | 1351347 | 6 | 11 | 54.55 | 12 | 16 | 75 | 66.67 | ggccgtgcgggtcctgagtgtggaacggccgtgcgggtcctgactgtgtg |
| 4 | 8 | 0 | 143167239 | 15 | 26 | 57.69 | 11 | 13 | 84.62 | 66.67 | ggaagtggagaaggggaaggaaggtcgtctagggaggaagtggagagggg |
| 5 | 9 | 1 | 64082996 | 6 | 11 | 54.55 | 5 | 7 | 71.43 | 61.11 | tatatatatatatatatatatatatatatatatatatatatatatatata |
| 6 | 10 | 1 | 3085303 | 15 | 17 | 88.24 | 7 | 13 | 53.85 | 73.33 | cccccactccactctccagcacagtcccccactccactctccagcacagt |
| 7 | 16 | -1 | 19382526 | 5 | 8 | 62.5 | 5 | 8 | 62.5 | 62.5 | agttctcatctggaatttctataatagacccagagtcaacagccaggttc |
| 8 | 16 | -1 | 34625947 | 46 | 57 | 80.7 | 8 | 26 | 30.77 | 65.06 | caaagctatccaaatatccacttgtagattatattcgagtgcattcgatg |
Detected sites with digenome scores >60 in the control genomic DNA.
| Location | Chr | Gap | Start | Plus | Depth | Perc | Minus | Depth2 | Perc2 | Total | Sequence (50 bp around the detection site) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 112180048 | 5 | 10 | 50 | 5 | 6 | 83.33 | 62.5 | aaaagaaagtatcaaaggagtaaacagacaacctacagaatgggagaaaa |
| 2 | 8 | 0 | 58814608 | 6 | 9 | 66.67 | 5 | 9 | 55.56 | 61.11 | atagttttaggatttcaggatgccttctgttcagtttagtttatattgtt |
| 3 | 12 | 1 | 74918031 | 5 | 7 | 71.43 | 5 | 8 | 62.5 | 66.67 | tacctagaaagcaagcagaatactcttagccaagaaaacaatatgtactc |
| 4 | 18 | -1 | 49878347 | 5 | 10 | 50 | 6 | 8 | 75 | 61.11 | ttaaaaatacttttttttttcctgcatctgatttggctgtcagtgtgaaa |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus, C57BL/6J) | C57BL/6J | Jackson laboratories | Stock # 000664 RRID:IMSR_JAX:000664 | |
| Strain, strain background (Mus musculus, 129S6/SvEvTac and C57BL/6J) | Col8a2Q455K | Johns Hopkins Medical Institutions | PMID:22002996 RRID:MGI:5305276 | |
| Antibody | a-COL8A2, rabbit polyclonal, | Thermo Fisher Scientific | Cat# PA5-35077 RRID:AB_2552387 | (5 μg/ml) |
| Antibody | Isotype, rabbit polyclonal | Thermo Fisher Scientific | Cat# 02–6102 RRID:AB_2532938 | (5 μg/ml) |
| Antibody | a-ZO1, mouse monoclonal | Thermo Fisher Scientific | Cat# 339188 RRID:AB_2532187 | (2.5 μg/ml) |
| Antibody | a-TNFa, rat monoclonal | BioLegend | Cat# 506301, clone: MP6-XT22 RRID:AB_315422 | (5 μg/ml) |
| Antibody | a-IFNg, rat monoclonal | BioLegend | Cat# 505801, clone: XMG1.2 RRID:AB_315395 | (5 μg/ml) |
| Antibody | Isotype, rat monoclonal | BioLegend | Cat# 400401, clone RTK2071 RRID:AB_326507 | (5 μg/ml) |
| Antibody | Secondary to rat IgG, conjugated with AlexaFluor647, goat polyclonal | Thermo Fisher Scientific | Cat# A-21247 RRID:AB_141778 | (2 μg/ml) |
| Cell line (human) | AD-293 | Agilent Technologies | Cat# 240085 | |
| Cell line (mouse) | NIH3T3 | ATCC | Cat# CRL-1658 RRID:CVCL_0594 | |
| Recombinant DNA reagent | px330 | Addgene | Cat# 42230 RRID:Addgene_42230 | Plasmid |
| Recombinant DNA reagent | pShuttle | Addgene | Cat# 16402 RRID:Addgene_16402 | Plasmid |
| Strain, strain background (Escherichia coli) | BJ5183-AD-1 | Agilent Technologies | Cat# 200157 | Competent cells |
| Strain, strain background (Escherichia coli) | XL10-Gold | Agilent Technologies | Cat# 200314 | Competent cells |
| Strain, strain background (Escherichia coli) | DH5a | NEB | Cat# C2987H | Competent cells |
| Sequence-based reagent | MsCol8a2_intron2F | This paper | PCR primer | cggtggtaggtggtaattgg |
| Sequence-based reagent | MsCol8a2_intron3R | This paper | PCR primer | tgtggtctggagtgtctgga |
| Sequence-based reagent | gRNAcloneF_EcoRV | This paper | PCR primer | TAGATATCgagggcctatttcccatgattc |
| Sequence-based reagent | gRNAcloneR_XbaI | This paper | PCR primer | TATCTAGAagccatttgtctgcagaattggc |
| Sequence-based reagent | Forward PCR primer for DNAseq | This paper | PCR primer | TTCTTCTTCTCCCTGCAGCC |
| Sequence-based reagent | Reverse PCR primer for DNAseq | This paper | PCR primer | GCACATACTTTACCGGGGCA |
| Sequence-based reagent | HuCol8a2_F | This paper | PCR primer | tgatcttttggtgaccccgg |
| Sequence-based reagent | HuCol8a2_R | This paper | PCR primer | GGATGTACTTCACTGGGGCA |
| Sequence-based reagent | Forward PCR primer for gRNA template of in vitro transcription | This paper | PCR primer | TAATACGACTCACTATAGCGTCCACGGACGCCATG |
| Sequence-based reagent | Reverse PCR primer for gRNA template of in vitro transcription | This paper | PCR primer | AAAAGCACCGACTCGGTGCCA |
| Sequence-based reagent | Cas9_Forward | This paper | PCR primer | CCGAAGAGGTCGTGAAGAAG |
| Sequence-based reagent | Cas9_Reverse | This paper | PCR primer | GCCTTATCCAGTTCGCTCAG |
| Sequence-based reagent | gRNA_Forward | This paper | PCR primer | AGACGCCATGCGTTTTAGAG |
| Sequence-based reagent | gRNA_Reverse | This paper | PCR primer | CGGTGCCACTTTTTCAAGTT |
| Sequence-based reagent | Mouse GAPDH_Forward | This paper | PCR primer | AACTTTGGCATTGTGGAAGGGCTC |
| Sequence-based reagent | Mouse GAPDH_Reverse | This paper | PCR primer | ACCAGTGGATGCAGGGATGATGTT |
| Sequence-based reagent | Mouse Col8a2_Forward1 at Indel site | This paper | PCR primer | CCACCTACACGTACGACGAA |
| Sequence-based reagent | Mouse Col8a2_Reverse1 | This paper | PCR primer | ACTCGGTGGAGTAGAGACCA |
| Sequence-based reagent | Mouse Col8a2_Forward2 | This paper | PCR primer | CCATCCACAGACGCCATG |
| Sequence-based reagent | Mouse Col8a2_Reverse2 | This paper | PCR primer | GGGCTGCACATACTTTACCG |





