Convergent evolution of small molecule pheromones in Pristionchus nematodes
Figures
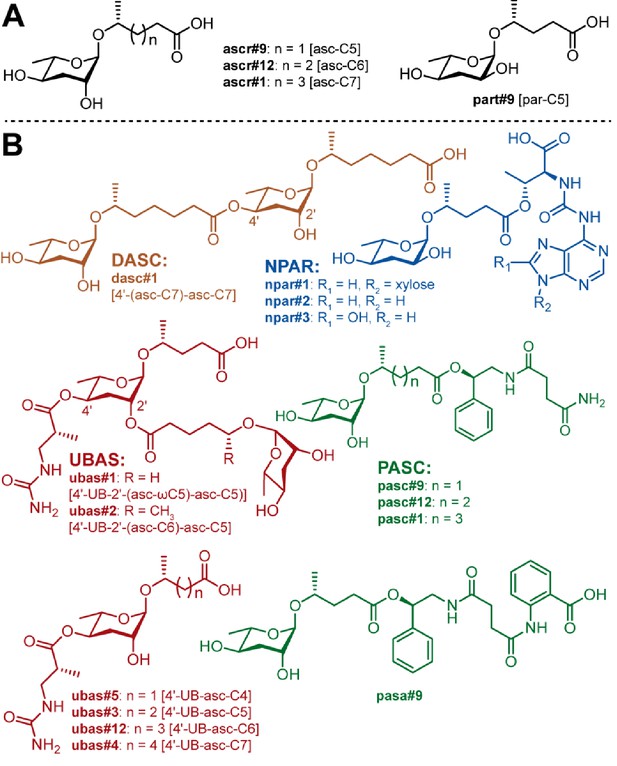
Known NDMMs from P. pacificus.
(A) Simple ascarosides, and the paratoside part#9 [par-C5]. (B) Modular NDMMs (DASC, NPAR, PASC, and UBAS chemicals) from P. pacificus.

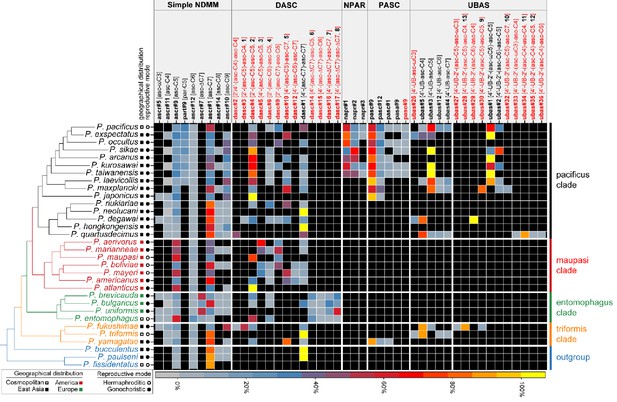
Phylogeny of the genus Pristionchus adapted from Rödelsperger et al., 2018.
Around 40 Pristionchus species have been described from world-wide samplings. Species are divided into four major clades (pacificus, maupasi, entomophagus and triformis) plus an outgroup. Nematodes from the pacificus and triformis clades are mostly found in East Asia, whereas nematodes from the maupasi and entomophagus clades are largely restricted to America and Europe, respectively. Note that the outgroup species were also isolated from East Asia. Red color highlights species that are investigated in this study. These 32 species cover all major clades. Dioecious outbreeding species usually have similar numbers of males and females in laboratory cultures. We use the term ‘gonochorists’ for these species to follow common nematode nomenclature. Androdioecious species consist of self-fertilizing hermaphrodites and males but are commonly referred to simply as hermaphroditic because males are found only rarely under standard laboratory culture conditions.
Comparative analysis of the exo-metabolomes of 32 Pristionchus nematodes revealed evolutionary diversity of NDMMs.
Three biological replicates for each species were grown in 100 ml S-medium for comparative LC-MS/MS analysis (see 'Materials and methods'). Simple NDMMs and complex NDMMs including DASC, NPAR, PASC, and UBAS chemicals (Supplementary file 3—Tables 1–3) are summarized; compounds found at trace levels (including the newly described UPAS class) are not included. Values were standardized within classes to help to visualize variation in relative production levels within rare NDMM classes, as overall production levels were dominated by simple NDMMs (see Figure 2—source data 1). NDMM abundances were standardized by dividing mean peak area for each NDMM by the total for the respective NDMM class then expressing the abundance as a percentage for each species; for clarity, monomeric and dimeric UBAS compounds were standardized separately. Black cells represent the apparent absence of the chemical. Black and red compound labels represent known and novel NDMMs, respectively.
-
Figure 2—source data 1
Quantification of NDMMs in 32 Pristionchus nematodes.
- https://cdn.elifesciences.org/articles/55687/elife-55687-fig2-data1-v2.xlsx
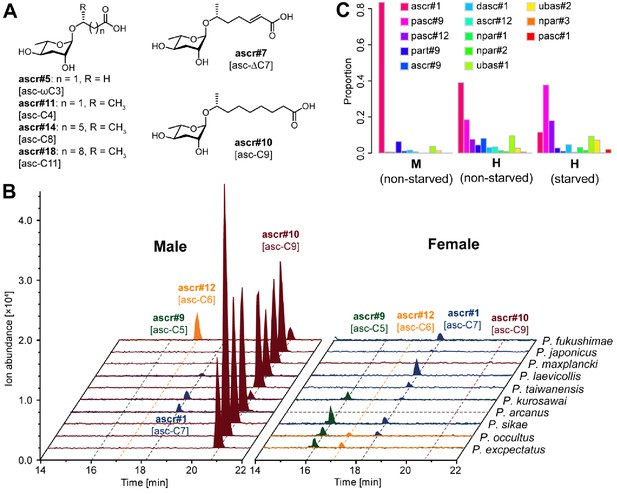
Simple ascarosides detected from Pristionchus nematodes.
(A) Chemical structures of newly detected simple ascarosides. (B) Males of 10 gonochoristic species release large amounts of ascr#10 [asc-C9] in a sex-specific manner. Dashed gray line indicates that nothing was detected in the culture, probably because of contamination of the P. arcanus females. (C) Males and hermaphrodites of P. pacificus released ascr#1 [asc-C7] and pasc#9, respectively, but no ascr#10 [asc-C9]. M, male; H, hermaphrodite.
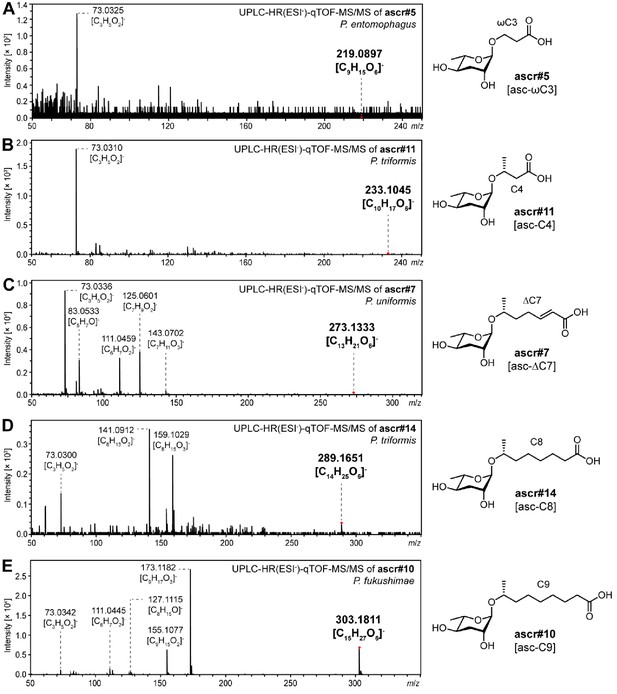
Simple ascarosides detected from various Pristionchus nematodes.
MS/MS spectral data of simple ascarosides detected from Pristionchus species, including (A) ascr#5 [asc-ωC3], (B) ascr#11 [asc-C4], (C) ascr#7 [asc-ΔC7], (D) ascr#14 [asc-C8], and (E) ascr#10 [asc-C9].
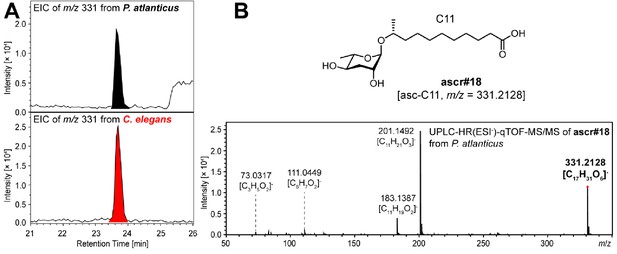
ascr#18 [asc-C11] detected from P. atlanticus.
The chemical identity of ascr#18 [asc-C11] was confirmed through a comparison of (A) the HPLC retention time and (B) the MS/MS fragmentation pattern with those of ascr#18 previously reported from C. elegans. No traces of ascr#18 were detected from other Pristionchus species.
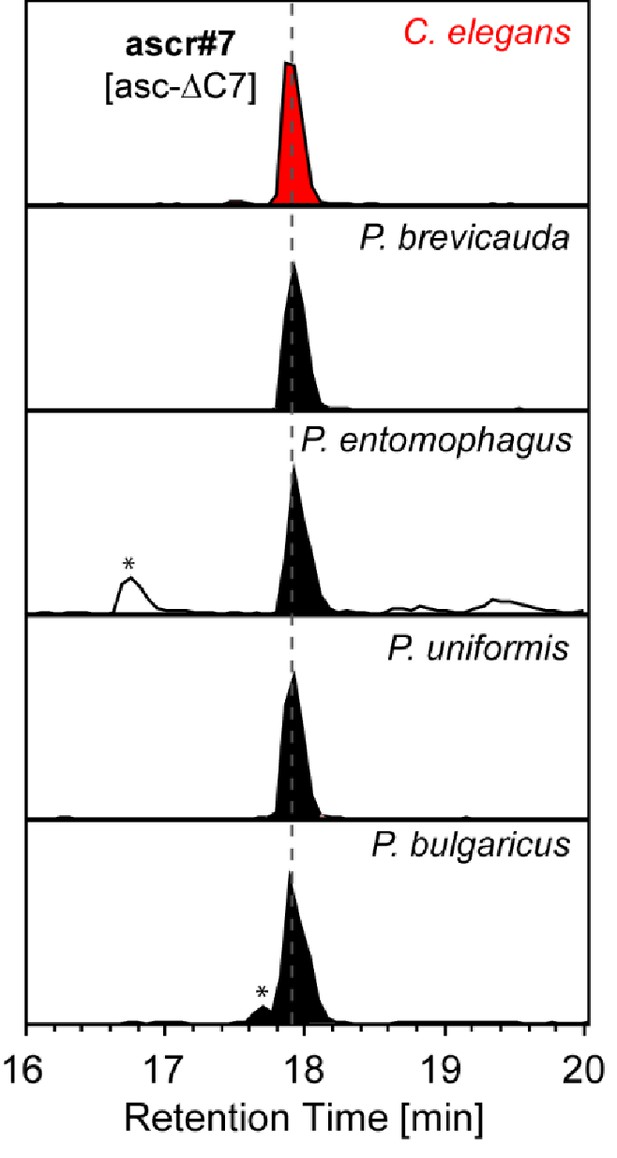
ascr#7 [asc-ΔC7] was specifically detected from the nematodes of the entomophagus clade.
Asterisks (*) denote non-ascarosides.
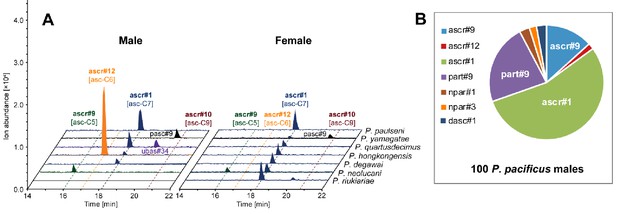
Chemical composition of different sex-derived exo-metabolomes.
Other simple ascarosides, but no ascr#10 [asc-C9], were detected from (A) males and (B) females of more distantly related gonochoristic species. (C) Hermaphroditic P. pacificus males produced ascr#1 [asc-C7] as the most abundant compound. 100 males and females were selected and cultured in 100 µl M9 buffer for LC-MS analysis (see 'Materials and methods').
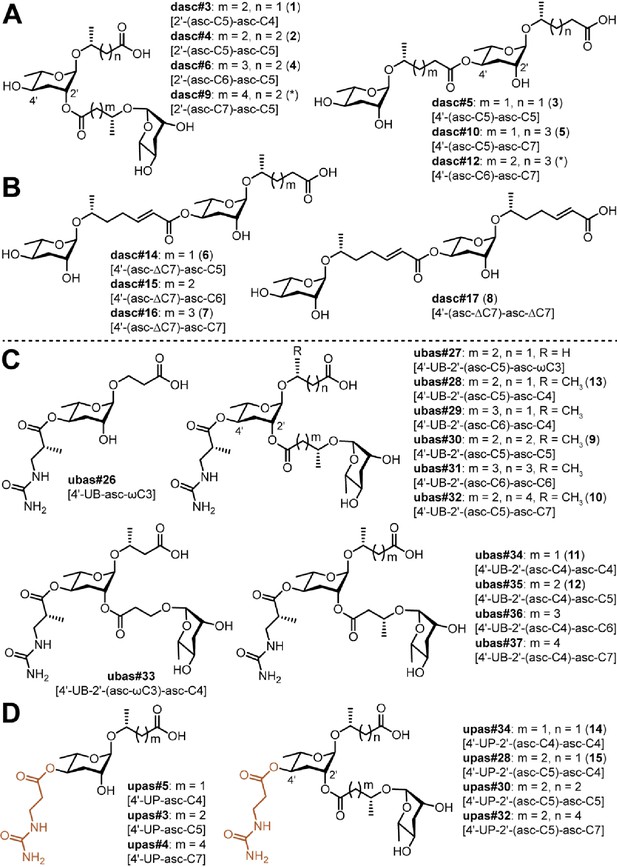
Novel DASC, UBAS, and UPAS chemicals detected from Pristionchus species.
(A) Structures of new DASC chemicals identified from Pristionchus nematodes with linkage at the 2'- or 4'-position. Note that the asterisk-marked dasc#9 [2'-(asc-C7)-asc-C5] and dasc#12 [4'-(asc-C6)-asc-C7] were tentatively assigned as being linked at the 2'- or 4'-position, respectively. (B) Nematodes from the entomophagus clade specifically produced four novel DASC chemicals harboring one or two units of asc#7 [asc-ΔC7]. (C) Structures of novel UBAS chemicals identified from Pristionchus nematodes. (D) Newly identified UPAS chemicals carrying a ureidopropionic acid group at the 4'-position of ascarylose. For further details on NDMM nomenclature, see 'Materials and methods'.
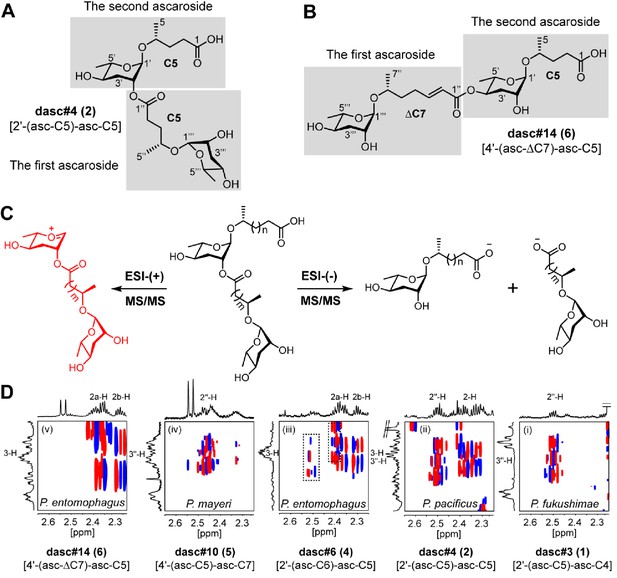
Structure elucidation of DASC chemicals.
(A, B) To describe the LC-MS/MS fragmentation of DASC chemicals, the lower and upper simple ascaroside units were defined as the first and second ascarosides, respectively. For example, two ascr#9 [asc-C5] units were defined as the first and the second ascarosides in dasc#4 [2'-(asc-C5)-asc-C5, 2], respectively (A), whereas the ascr#7 [asc-ΔC7] and the ascr#9 [asc-C5] units were defined as the first and the second ascarosides in dasc#14 [4'-(asc-ΔC7)-asc-C5], respectively (B). (C) MS/MS fragmentation pattern of DASC chemicals using negative or positive ion modes. Fragment ion signals derived from two simple ascaroside units were produced when the negative ion mode was applied. However, through cleavage of the terminal fatty acid side chain, LC-ESI-(+)-MS/MS fragmentation of DASC chemicals generated a fragment ion signal consisting of two ascarylose moieties connected by their aglycone linker (red color fragment), which was used for the identification of the carbon length of the first fatty acid side chain. Thus, both simple ascaroside units in DASC chemicals were assigned on the basis of HRMS and MS/MS analyses. Further clarification of the hydroxylation style (ω) or (ω−1) of the fatty acid side chains and the linkage at the 2'- or 4'-position required NMR data (dqf-COSY) analysis. (D) Panels (i–v) show the dqf-COSY spectra of major DASC chemicals harboring (ω−1)-oxygenated C5 fatty acid side chains, highlighting the correlations of H-2/H-3 and H-2ʹʹ/H-3ʹʹ. The dashed box in panel (iii) signals impurities.
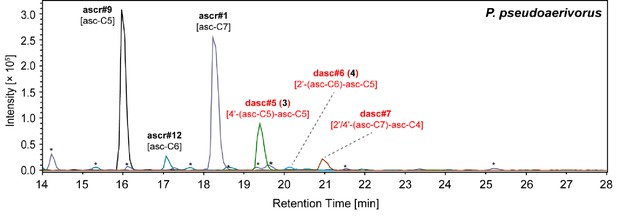
NDMMs detected from the P. pseudoaerivorus exo-metabolome.
P. pseudoaerivorus produced three simple ascarosides and several other DASC chemicals, including the maupasi-clade-specific dasc#5 [4'-asc-C5-asc-C5, 3]. Red color highlights newly identified compounds. Asterisks (*) denote non-ascarosides.

DASC chemicals detected from nematodes in the maupasi clade.
dasc#5 [4'-(asc-C5)-asc-C5, 3] harbors two identical ascr#9 [asc-C5] units with a linkage at the 4'-position. This chemical is specific to species in the maupasi clade, including P. aerivorus, P. marianneae, P. boliviae, and P. mayeri. Asterisks (*) denote non-ascarosides.
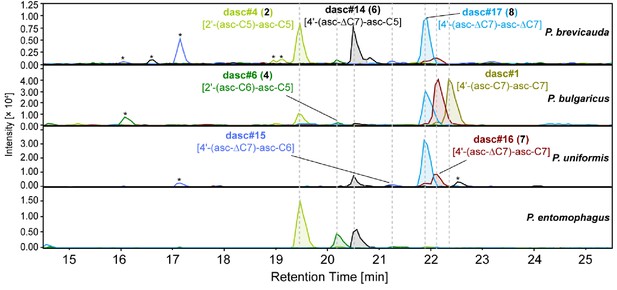
DASC chemicals detected from nematodes in the entomophagus clade.
Four novel DASC chemicals, including dasc#14 [4'-(asc-ΔC7)-asc-C5, 6], dasc#15 [4'-(asc-ΔC7)-asc-C6], dasc#16 [4'-(asc-ΔC7)-asc-C7, 7], and dasc#17 [4'-(asc-ΔC7)-asc-ΔC7, 8] carry one or two ascr#7 [asc-ΔC7] units, and are specific to species of the entomophagus clade. Asterisks (*) denote non-ascarosides.
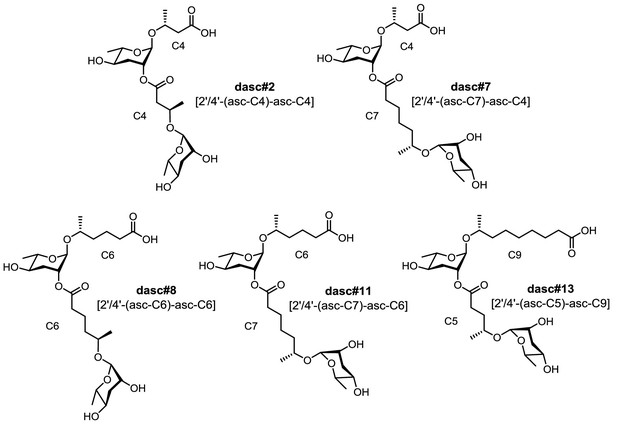
Structures of five unquantifiable DASC chemicals detected from Pristionchus species.
MS/MS fragmentation of these DASC chemicals (see Supplementary file 2a—Figures 12–16) confirmed the carbon length of the two simple ascaroside units. Note that the fatty-acid-derived side chains are assumed to be (ω−1)-hydroxylated, given the fact that all the identified ascarosides in this study were found to share (ω−1) hydroxylation with the exception of ascr#5 [asc-ωC3] (and the ascr#5-containing UBAS derivatives) and ubas#1 [4'-UB-2'-(asc-ωC5)-asc-C5]. Note that these DASC chemicals might harbor a linkage at the 2'- or 4'-position; chemical synthesis is required to characterize their exact structures.
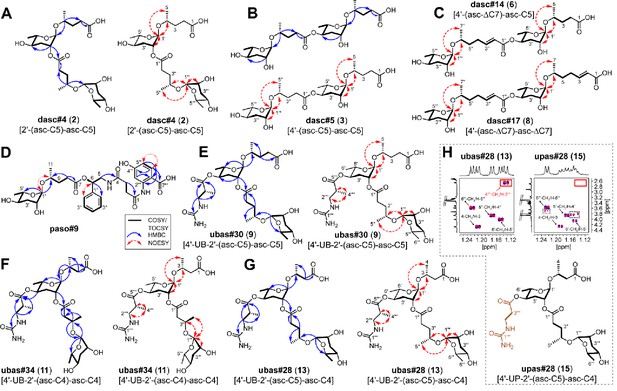
Elucidation of the structures of novel NDMMs.
(A) dasc#4 [2'-(asc-C5)-asc-C5, 2], (B) dasc#5 [4'-(asc-C5)-asc-C5, 3], (C) dasc#14 [4'-(asc-ΔC7)-asc-C5, 6], dasc#17 [4'-(asc-ΔC7)-asc- ΔC7, 8], (D) paso#9, (E) ubas#30 [4'-UB-2'-(asc-C5)-asc-C5, 9], (F) ubas#34 [4'-UB-2'-(asc-C4)-asc-C4, 11], (G) ubas#28 [4'-UB-2'-(asc-C5)-asc-C4, 13], and (H) upas#28 [4'-UP-2'-(asc-C5)-asc-C4, 15]. Key COSY/TOCSY, NOESY, and/or HMBC correlations of each compound are summarized. Comparison of selected dqf-COSY spectra between ubas#28 (13) and upas#28 (15), highlighting correlations with methyl groups, indicates that upas#28 (15) contains a ureidopropionic acid moiety (H). The dashed boxes in panel (H) signal impurities, and the red-lined box highlights the absence of a methyl group in upas#28 (15). For details, see Supplementary file 1d—Figure 38 and Supplementary file 1e—Figure 2.
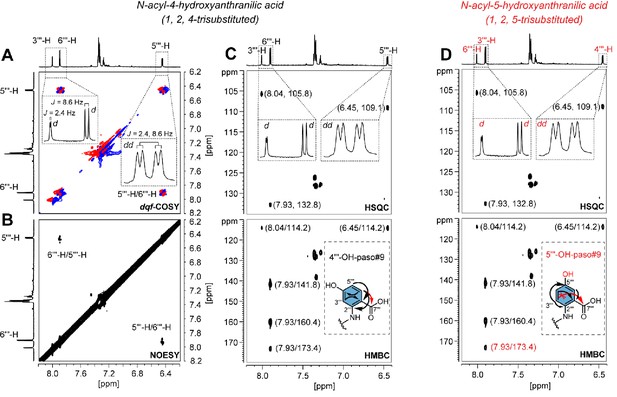
Structural characterization of paso#9.
paso#9 (Artyukhin et al., 2018) was previously proposed as a hydroxylated pasa#9 derivative (Artyukhin et al., 2018; also see Supplementary file 2b—Figure 1), but the exact position of the hydroxyl group at the anthranilic acid was unknown. The analysis of NMR data for purified paso#9 demonstrated that H-5ʹʹʹ (J = 2.4, 8.6 Hz) at 6.45 ppm showed strong (A) 1H, 1H-COSY correlation (J5ʹʹʹ, 6ʹʹʹ=8.6 Hz) and (B) NOE correlation with H-6ʹʹʹ at 7.93 ppm, which further showed (C) HMBC correlation with C-7ʹʹʹ at 173.4 ppm (Supplementary file 1c—Figures 5–13 and Supplementary file 3—Table 14). These findings indicate that C-5ʹʹʹ was free of any substituent. Further observation of a weak 1H, 1H-COSY correlation between H-5ʹʹʹ and H-3ʹʹʹ at 8.04 ppm (J3ʹʹʹ, 5ʹʹʹ=2.4 Hz) suggests a 4-hydroxyanthranilic acid moiety in paso#9 (Miyagawa et al., 1996; Ishihara et al., 1998). The possibility that paso#9 contains a 5-hydroxyanthranilic acid moiety can be excluded because of the difficulty in observing a (long-range J4) HMBC correlation from H-3ʹʹʹ to C-7ʹʹʹ (D).
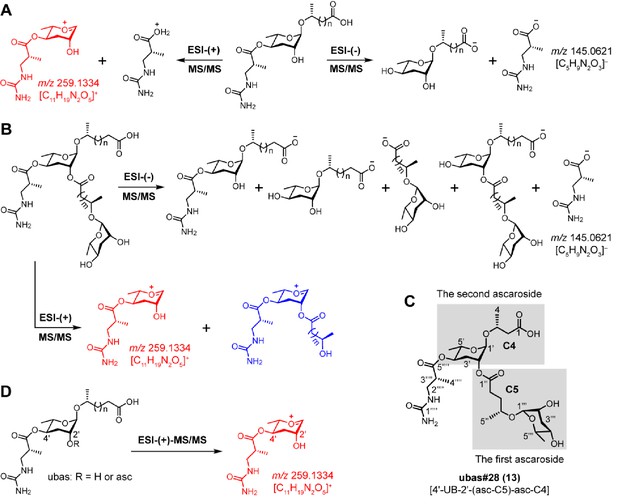
MS/MS fragmentation pattern of UBAS chemicals.
(A) ESI-(-)-MS/MS fragmentation of simple UBAS chemicals yielded a fragment ion signal of C5H9N2O3–(m/z 145.0621 [M – H]-) that was derived from the ureidoisobutyric acid moiety. ESI-(+)-MS/MS fragmentation generated a characteristic fragment ion signal of C11H19N2O5+ (m/z 259.1334 [M + H]+) (red color fragment) (Supplementary file 2c—Figures 1–4). (B) ESI-(-/+)-MS/MS fragmentation of complex UBAS chemicals with dimeric ascaroside moieties also yielded the characteristic fragment ion signals of C5H9N2O3– and C11H19N2O5+ (red color fragment) (Supplementary file 2c—Figures 5–6). Another fragment ion signal (blue color fragment) containing C11H19N2O5+ plus the first side chain enabled the clarification of the carbon length of the first fatty acid side chain, which ultimately helped to determine the identities of two ascaroside units in the complex UBAS chemicals. (C) To describe the LC-MS/MS fragmentation patterns of the complex UBAS chemicals, the lower and upper simple ascaroside units were defined as the first and second ascarosides, respectively. (D) Targeted MS/MS method for screening UBAS chemicals by profiling the characteristic ion fragment of m/z 259.1334.

NDMMs detected from the P. hoplostomus exo-metabolome.
Some major UBAS chemicals (e.g. ubas#5 [4'-UB-asc-C4]) could be detected from the P. hoplostomus exo-metabolome, although this species grew very slowly in liquid culture. Red color highlights a newly identified compound. Asterisks (*) denote non-ascarosides.
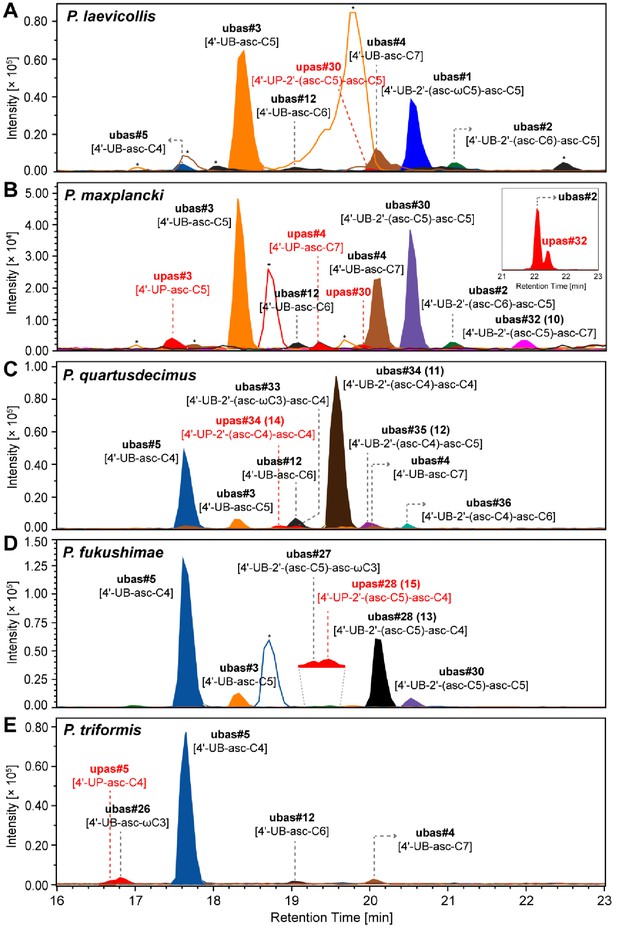
UPAS chemicals detected from five Pristionchus species.
(A–E) Trace amounts of UPAS chemicals (together with major UBAS) were detected from 100 ml of liquid culture media of five Pristionchus species: P. laevicollis (A), P. maxplancki (B), P. quartusdecimus (C), P. fukushimae (D), and P. triformis (E). Inserted EIC traces of upas#32 [4'-UP-2'-(asc-C5)-asc-C7] and ubas#2 were detected from an enriched SPE fraction of a 2 l liquid culture of P. maxplancki. MS/MS fragmentation pattern and structural assignments of UPAS chemicals are described in Supplementary file 2d—Figures 1–7. Traces of UPAS chemicals are labeled in red. Asterisks (*) denote non-ascarosides.

MS/MS fragmentation pattern of UPAS chemicals.
(A) ESI-(-)-MS/MS fragmentation of simple UPAS chemicals yielded a fragment ion signal of C4H7N2O3–(m/z 131.0468 [M – H]–) that was derived from the ureidopropionic acid moiety. ESI-(+)-MS/MS fragmentation generated a characteristic fragment ion signal of C10H17N2O5+ (m/z 245.1136 [M + H]+) (red color fragment) (Supplementary file 2d—Figures 1–3). (B) ESI-(-/+)-MS/MS fragmentation of complex UPAS chemicals with dimeric ascaroside moieties also yielded characteristic fragment ion signals of C4H7N2O3– and C10H17N2O5+ (red color fragment) (Supplementary file 2d—Figures 4–7). Another ion signal (blue color fragment) containing C10H17N2O5+ plus the first side chain enabled the identification of the carbon length of the first side chain, which ultimately helped to determine the identities of two ascaroside units in the complex UPAS chemicals. (C) To describe the MS/MS fragmentation patterns of complex UPAS chemicals, the lower and upper simple ascaroside units were defined as first and second ascarosides, respectively. (D) Targeted MS/MS method for screening UPAS chemicals by profiling the characteristic ion fragment of m/z 245.1136.
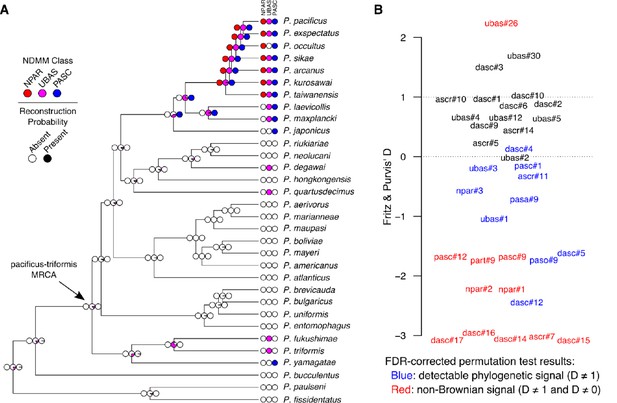
Comparative analysis of NDMMs in Pristionchus.
(A) Convergent evolution of PASC and UBAS-type NDMMs in the pacificus and triformis clades. The evolution of complex NDMMs was reconstructed via Bayesian stochastic character mapping. Colored circles at the tips of the phylogeny denote the presence (dark color) or absence (white) of NPAR (red), UBAS (magenta), and PASC (blue) NDMMs; those at the internal nodes denote the probability of presence versus absence. Results indicate that NPAR compounds evolved only once (within the pacificus clade), whereas both PASC and UBAS compounds evolved convergently within the pacificus and triformis clades. The estimated probability of being present at the node representing the most recent common ancestor (MRCA) of the pacificus and triformis clades is less than 0.01 for both NPAR and PASC, and 0.12 for UBAS. Ancestral character mapping results for individual NDMMs are provided in Table 1 and Figure 6—figure supplement 1. (B) Variation in phylogenetic signal strength among Pristionchus NDMMs. Binary-trait phylogenetic signal (Fritz and Purvis' D) was estimated for all non-constant, non-singleton NDMMs. D values of 0 and 1 correspond to expectations under Brownian evolution and non-phylogenetic randomness, respectively. Red labels denote NDMMs with D significantly different from both 0 and 1, whereas blue labels indicate those with D significantly different from 1 but not 0, on the basis of the results of permutation test-derived FDR corrected (10% threshold) P values. The remainder (black) represent cases in which we were unable to reject either baseline hypothesis. D-value estimates and permutation P values are provided in Table 1.
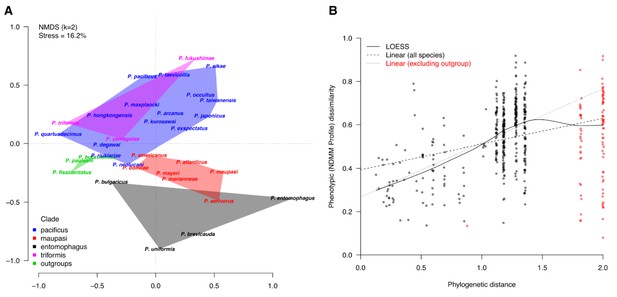
Comparative analysis of Pristionchus NDMM profiles.
(A) Ordination of Pristionchus NDMM profiles. Non-metric multidimensional scaling of NDMM profiles revealed clustering patterns that are indicative of phylogenetic signal (the minimal overlap seen for the entomophagus clade, the maupasi clade, and the outgroup set) and of convergence (the considerable overlap between the distantly related pacificus and triformis clades). (B) Evolutionary divergence of NDMM profiles. More distantly related species tended to have more dissimilar NDMM profiles (Bray-Curtis dissimilarity measures), although there was clear evidence of a plateau at extreme evolutionary distances (smoothed LOESS Curve; solid line). Linear fits to the data, both with and without the outgroup species (dashed and dotted lines, respectively), confirmed a positive relationship between phenotypic and evolutionary divergence for Pristionchus NDMM profiles. In both cases, the intercept term was significantly positive, implying rapid divergence of NDMM profiles during or shortly after speciation (i.e., when phylogenetic distance is at or near zero), after which phenotypic changes continued to accumulate at increasingly slower rates.
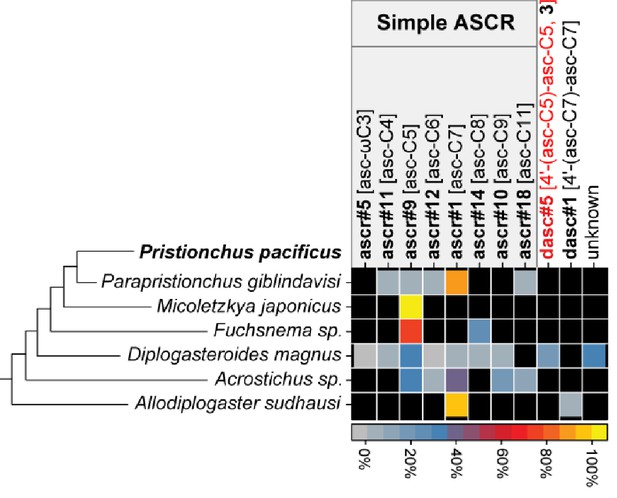
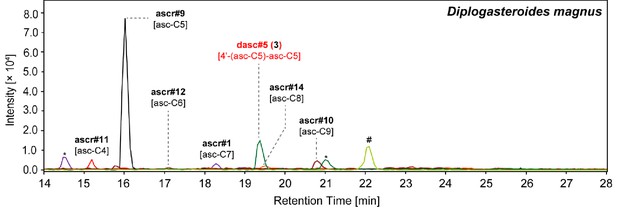
Summary of NDMMs in six (non-Pristionchus) Diplogastridae species that represent distant relatives of P. pacificus.
Three biological replicates for each species were grown in 100 ml S-medium for LC/MS analysis (see 'Materials and methods'). Mean peak area for each compound from each species was divided by total peak area, thereby representing the abundance of each compound as a percentage (Figure 7—source data 1). Black color represents the apparent absence of the chemical. Black and red compound labels are known and novel NDMMs, respectively. 'Unknown' represents uncharacterized ascaroside.
-
Figure 7—source data 1
Quantification of NDMMs in six (non-Pristionchus) Diplogastridae species.
- https://cdn.elifesciences.org/articles/55687/elife-55687-fig7-data1-v2.xlsx
Tables
Mode of evolution of major Pristionchus NDMMs.
| NDMM | Class | Conservation Pattern | fitMk model | SIMMAP changes | D | P (Ho: D = 0) | P (Ho: D = 1) | Phylogenetic interpretation |
|---|---|---|---|---|---|---|---|---|
| ascr#1 | Simple NDMM | Constant | – | – | – | – | – | Strictly conserved |
| ascr#10 | Simple NDMM | Variable | ARD | 17.50 | 0.9645 | 0.2524 | 0.8989 | Variable |
| ascr#11 | Simple NDMM | Variable | ER | 15.71 | −0.3263 | 0.6697 | 0.0051 | Somewhat conserved (Brownian?) |
| ascr#12 | Simple NDMM | Constant | – | – | – | – | – | Strictly conserved |
| ascr#14 | Simple NDMM | Variable | ER | 26.25 | 0.4424 | 0.6116 | 0.2027 | Variable |
| ascr#5 | Simple NDMM | Variable | ER | 8.54 | 0.2276 | 0.8670 | 0.1933 | Variable |
| ascr#7 | Simple NDMM | Variable | ER | 1.10 | −3.0185 | 0.0854 | 0.0000 | Highly conserved |
| ascr#9 | Simple NDMM | Constant | – | – | – | – | – | Strictly conserved |
| part#9 | Simple NDMM | Variable | ER | 2.72 | −1.7518 | 0.0854 | 0.0000 | Highly conserved |
| dasc#1 | DASC | Variable | ER | 15.77 | 0.9714 | 0.2487 | 0.8989 | Variable |
| dasc#10 | DASC | Variable | ARD | 24.71 | 1.0261 | 0.1928 | 0.9866 | Variable |
| dasc#12 | DASC | Variable | ER | 1.64 | −2.4439 | 0.2335 | 0.0035 | Somewhat conserved (Brownian?) |
| dasc#14 | DASC | Variable | ER | 1.17 | −3.0560 | 0.0854 | 0.0000 | Highly conserved |
| dasc#15 | DASC | Variable | ER | 1.16 | −3.0805 | 0.0854 | 0.0000 | Highly conserved |
| dasc#16 | DASC | Variable | ER | 1.25 | −2.9528 | 0.0993 | 0.0000 | Highly conserved |
| dasc#17 | DASC | Variable | ER | 1.08 | −3.0772 | 0.0854 | 0.0000 | Highly conserved |
| dasc#2 | DASC | Variable | ER | 2.50 | 0.8850 | 0.6697 | 0.8989 | Variable |
| dasc#3 | DASC | Variable | ER | 2.66 | 1.5025 | 0.4077 | 0.7440 | Variable |
| dasc#4 | DASC | Variable | ER | 18.90 | 0.1325 | 0.8670 | 0.0644 | Somewhat conserved (Brownian?) |
| dasc#5 | DASC | Variable | ARD | 6.83 | −1.6165 | 0.2487 | 0.0032 | Somewhat conserved (Brownian?) |
| dasc#6 | DASC | Variable | ER | 37.88 | 0.8552 | 0.2487 | 0.7392 | Variable |
| dasc#9 | DASC | Variable | ARD | 17.90 | 0.5267 | 0.6116 | 0.3702 | Variable |
| npar#1 | NPAR | Variable | ER | 1.17 | −2.2388 | 0.0854 | 0.0000 | Highly conserved |
| npar#2 | NPAR | Variable | ER | 1.19 | −2.2318 | 0.0854 | 0.0000 | Highly conserved |
| npar#3 | NPAR | Variable | ARD | 8.11 | −0.5835 | 0.6463 | 0.0644 | Somewhat conserved (Brownian?) |
| pasa#9 | PASC | Variable | ARD | 7.16 | −0.7273 | 0.6116 | 0.0254 | Somewhat conserved (Brownian?) |
| pasc#1 | PASC | Variable | ARD | 7.89 | −0.1643 | 0.8601 | 0.0644 | Somewhat conserved (Brownian?) |
| pasc#12 | PASC | Variable | ER | 2.52 | −1.6914 | 0.0513 | 0.0000 | Highly conserved |
| pasc#9 | PASC | Variable | ER | 2.52 | −1.6928 | 0.0513 | 0.0000 | Highly conserved |
| paso#9 | PASC | Variable | ARD | 4.19 | −1.7483 | 0.1108 | 0.0000 | Somewhat conserved (Brownian?) |
| ubas#1 | UBAS | Variable | ER | 5.94 | −1.0425 | 0.2825 | 0.0006 | Somewhat conserved (Brownian?) |
| ubas#12 | UBAS | Variable | ER | 5.99 | 0.6570 | 0.6116 | 0.6005 | Variable |
| ubas#2 | UBAS | Variable | ARD | 11.46 | −0.0160 | 0.9172 | 0.1368 | Variable |
| ubas#26 | UBAS | Variable | ARD | 6.37 | 2.2410 | 0.0513 | 0.0993 | Over-dispersed |
| ubas#27 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#28 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#29 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#3 | UBAS | Variable | ARD | 11.67 | −0.1931 | 0.8262 | 0.0255 | Somewhat conserved (Brownian?) |
| ubas#30 | UBAS | Variable | ER | 2.66 | 1.6939 | 0.2825 | 0.6054 | Variable |
| ubas#32 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#33 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#34 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#35 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#36 | UBAS | Singleton | – | – | – | – | – | Young |
| ubas#4 | UBAS | Variable | ER | 6.01 | 0.6606 | 0.6116 | 0.6005 | Variable |
| ubas#5 | UBAS | Variable | ER | 9.22 | 0.6413 | 0.6007 | 0.5546 | Variable |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Caenorhabditis elegans) | N2 | Caenorhabditis Genetics Center | ||
| Strain, strain background (Pristionchus pacificus) | RS2333 | RJ Sommer Lab | ||
| Strain, strain background (P. pacificus) | RS5410 (Ex[eud-1]) | RJ Sommer Lab | ||
| Strain, strain background (P. exspectatus) | RS5522 | RJ Sommer Lab | ||
| Strain, strain background (P. occultus) | RS5811 | RJ Sommer Lab | ||
| Strain, strain background (P. sikae) | RS5901 | RJ Sommer Lab | ||
| Strain, strain background (P. arcanus) | RS5527 | RJ Sommer Lab | ||
| Strain, strain background (P. kurosawai) | RS5527 | RJ Sommer Lab | ||
| Strain, strain background (P. taiwanensis) | RS5797 | RJ Sommer Lab | ||
| Strain, strain background (P. laevicollis) | RS5939 | RJ Sommer Lab | ||
| Strain, strain background (P. maxplancki) | RS5594 | RJ Sommer Lab | ||
| Strain, strain background (P. japonicus) | RS5238 | RJ Sommer Lab | ||
| Strain, strain background (P. riukiariae) | RS5937 | RJ Sommer Lab | ||
| Strain, strain background (P. neolucani) | RS5949 | RJ Sommer Lab | ||
| Strain, strain background (P. degawai) | RS5938 | RJ Sommer Lab | ||
| Strain, strain background (P. hongkongensis) | RS5957 | RJ Sommer Lab | ||
| Strain, strain background (P. quartusdecimus) | RS5230 | RJ Sommer Lab | ||
| Strain, strain background (P. pseudoaerivorus) | RS5139 | RJ Sommer Lab | ||
| Strain, strain background (P. aerivorus) | RS5106 | RJ Sommer Lab | ||
| Strain, strain background (P. marianneae) | RS5108 | RJ Sommer Lab | ||
| Strain, strain background (P. maupasi) | RS0143 | RJ Sommer Lab | ||
| Strain, strain background (P. boliviae) | RS5518 | RJ Sommer Lab | ||
| Strain, strain background (P. mayeri) | RS5460 | RJ Sommer Lab | ||
| Strain, strain background (P. americanus) | RS5140 | RJ Sommer Lab | ||
| Strain, strain background (P. atlanticus) | CZ3975 | RJ Sommer Lab | ||
| Strain, strain background (P. brevicauda) | RS5231 | RJ Sommer Lab | ||
| Strain, strain background (P. bulgaricus) | RS5283 | RJ Sommer Lab | ||
| Strain, strain background (P. uniformis) | RS0141 | RJ Sommer Lab | ||
| Strain, strain background (P. entomophagus) | RS0144 | RJ Sommer Lab | ||
| Strain, strain background (P. fukushimae) | RS5595 | RJ Sommer Lab | ||
| Strain, strain background (P. hoplostomus) | JU1090 | RJ Sommer Lab | ||
| Strain, strain background (P. triformis) | RS5233 | RJ Sommer Lab | ||
| Strain, strain background (P. yamagatae) | RS5964 | RJ Sommer Lab | ||
| Strain, strain background (P. bucculentus) | RS5596 | RJ Sommer Lab | ||
| Strain, strain background (P. paulseni) | RS5918 | RJ Sommer Lab | ||
| Strain, strain background (P. fissidentatus) | RS5133 | RJ Sommer Lab | ||
| Strain, strain background (Micoletzkya japonicus) | RS5524B | RJ Sommer Lab | ||
| Strain, strain background (Parapritionchus giblindavisi) | RS5555B | RJ Sommer Lab | ||
| Strain, strain background (Diplogasteroides magnus) | RS5740 | RJ Sommer Lab | ||
| Strain, strain background (Diplogasteroides magnus) | RS1987 | RJ Sommer Lab | ||
| Strain, strain background (Allodiplogaster sudhausi) | SB413 | RJ Sommer Lab | ||
| Strain, strain background (Fuchsnema sp.) | RS5592 | RJ Sommer Lab | ||
| Strain, strain background (Acrostichus sp.) | RS5722 | RJ Sommer Lab |
Additional files
-
Supplementary file 1
NMR spectra of isolated or enriched NDMMs from Pristionchus species.
a. NMR spectra of simple ASCR. b. NMR spectra of DASC chemicals. c. NMR spectra of PASC chemicals. d. NMR spectra of UBAS chemicals. e. NMR spectra of UPAS chemicals.
- https://cdn.elifesciences.org/articles/55687/elife-55687-supp1-v2.pdf
-
Supplementary file 2
MS/MS spectral data of NDMMs detected from Pristionchus species.
a. MS/MS spectral data of DASC chemicals. b. MS/MS spectral data of PASC chemicals. c. MS/MS spectral data of UBAS chemicals. d. MS/MS spectral data of UPAS chemicals.
- https://cdn.elifesciences.org/articles/55687/elife-55687-supp2-v2.pdf
-
Supplementary file 3
Detected NDMMs from Pristionchus species and NMR data for isolated or enriched NDMMs from Pristionchus species.
- https://cdn.elifesciences.org/articles/55687/elife-55687-supp3-v2.pdf
-
Supplementary file 4
Ancestral reconstruction for individual compounds.
- https://cdn.elifesciences.org/articles/55687/elife-55687-supp4-v2.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55687/elife-55687-transrepform-v2.pdf