The wtf4 meiotic driver utilizes controlled protein aggregation to generate selective cell death
Figures
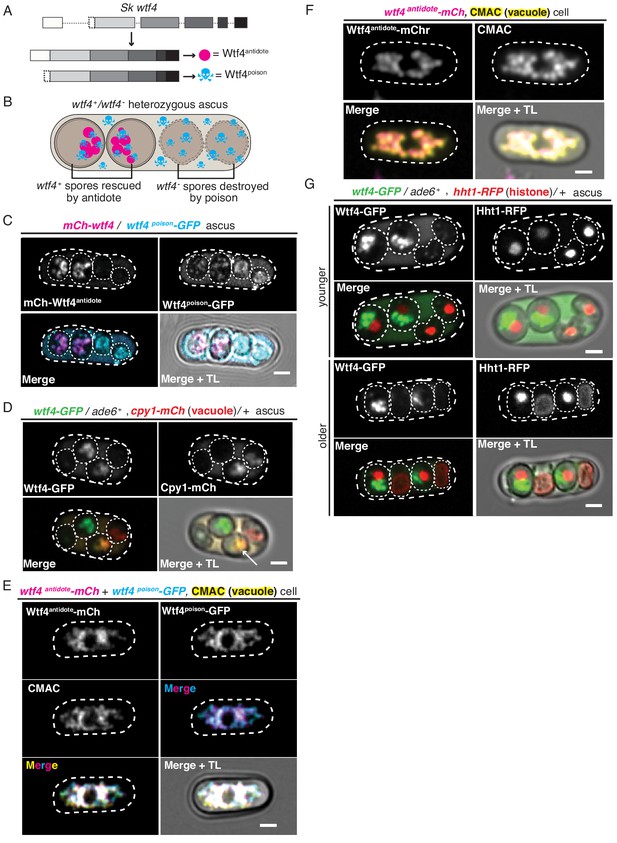
Wtf4poison and Wtf4antidote protein localization in both S. pombe meiosis and vegetative growth.
(A) The wtf4 gene utilizes alternate transcriptional start sites to encode for the Wtf4antidote and Wtf4poison proteins. (B) Model of a tetrad generated from a wtf4+/wtf4- diploid. wtf4+ spores are rescued by the spore-enriched antidote (magenta circles), while the poison (cyan skulls) spreads throughout the ascus. (C) An ascus generated by an mCherry-wtf4/wtf4poison-GFP diploid showing the localization of mCherry-Wtf4antidote (magenta in merged images) and Wtf4poison-GFP (cyan in merged images) (Nuckolls et al., 2017). (D) An ascus generated from a wtf4-GFP/ade6+, +/cpy1 mCherry diploid showing localization of Wtf4-GFP (green in merged images) and Cpy1-mCherry (red in merged images). The arrow highlights the spore that inherited both tagged alleles and thus contains both tagged proteins. (E) A vegetatively growing haploid cell expressing Wtf4poison-GFP (cyan in merged images) and Wtf4antidote-mCherry (magenta in merged images) using the β-estradiol-inducible system. CMAC is a vacuole lumen stain (yellow in merged images). Both Wtf4 proteins colocalize with the vacuole. Cells were imaged 4 hr after induction with 100 nM β-estradiol. (F) A vegetatively growing haploid cell expressing Wtf4antidote-mCherry (magenta in the merged images) using the β-estradiol system and stained with the CMAC vacuole stain (yellow in the merged images) shows Wtf4antidote-mCherry in the vacuole. Cells were induced in the same way as in (E). (G) Asci generated from a wtf4-GFP/ade6+, hht1-RFP/+ diploid. Hht1-RFP (red in merged images) is a histone marker. The nuclei in the spores that do not inherit wtf4-GFP (e.g. lacking GFP signal) can exhibit nuclear condensation and fragmentation. All scale bars represent 2 µm. TL = transmitted light.
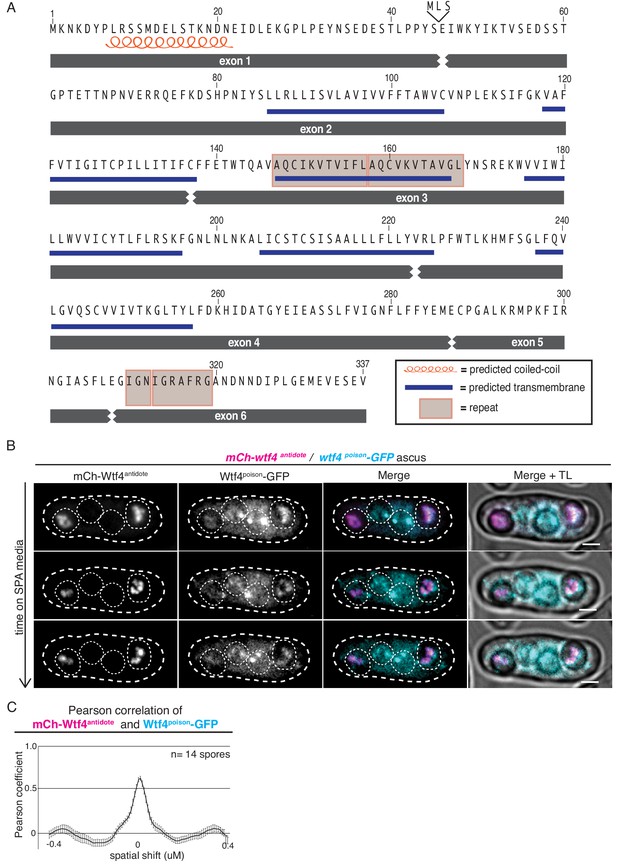
Wtf4poison and Wtf4antidote protein sequences and colocalization of the proteins in S. pombe asci.
(A) Wtf4 protein sequence. Wtf4antidote is encoded by all six exons. Wtf4poison is encoded by exons 2–6 and an additional three amino acids (MLS) encoded in intron 1. Exon 1 contains a predicted coiled-coil domain (depicted with an orange coil) (Lupas et al., 1991). There are six predicted transmembrane domains (depicted by blue lines) (TMHMM model, Krogh et al., 2001) found throughout the amino acid sequences shared by both proteins. There are also two regions of repetitive sequences: one found in exon 3 and one found in exon 6 (Eickbush et al., 2019). These are depicted with gray boxes. (B) Time-lapse microscopy of an ascus generated by an mCherry-wtf4/wtf4poison-GFP diploid. mCherry-Wtf4antidote is shown in magenta and Wtf4poison-GFP is shown in cyan in the merged images. All scale bars represent 2 µm. TL = transmitted light. (C) Pearson correlation of mCherry-Wtf4antidote and Wtf4poison-GFP from spores within asci generated from mCherry-wtf4/wtf4poison-GFP diploids is shown. Error bars represent standard deviation.
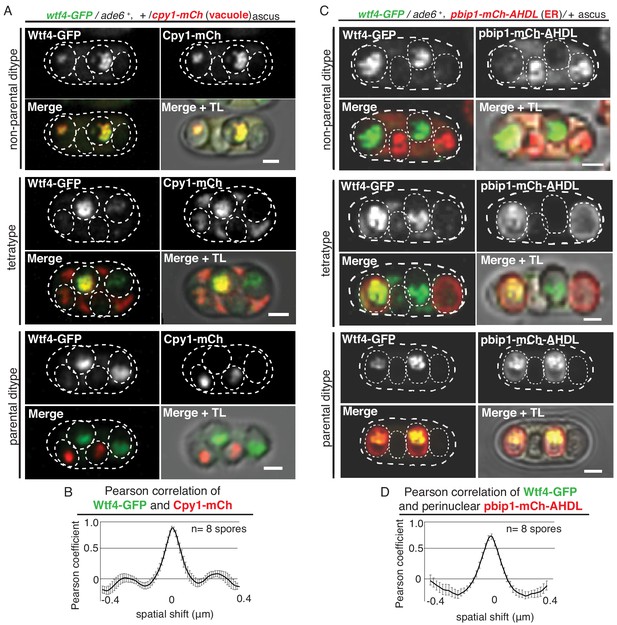
Wtf4poison and Wtf4antidote colocalize in the vacuole and endoplasmic reticulum (ER) in spores.
(A) Non-parental ditype, tetratype, and parental ditype asci generated by wtf4-GFP/ade6+, +/cpy1-mCherry diploids. Cpy1-mCherry (red in merged images) is a vacuole luminal marker and Wtf4-GFP is shown in green in merged images. Scale bars represent 2 µm. TL = transmitted light. (B) Pearson correlation of Wtf4-GFP and Cpy1-mCherry in spores that inherited both alleles generated by wtf4-GFP/ade6+, +/cpy1-mCherry diploids shows a coefficient of approximately 0.89. (C) Non-parental ditype, tetratype, and parental ditype asci generated by wtf4-GFP/ade6+, pbip1-mCherry-AHDL/+ diploids. pbip1-mCherry-AHDL (red in merged images) is an ER marker and Wtf4-GFP is shown in green in merged images. (D) Pearson correlation of Wtf4-GFP and pbip1-mCherry-AHDL from spores that inherited both alleles generated by wtf4-GFP/ade6+, pbip1-mCherry-AHDL/+ diploids shows a coefficient of approximately 0.74. Error bars represent standard error. TL = transmitted light.

Wtf4poison and Wtf4antidote colocalize and are functional in vegetative S. pombe cells.
(A) Results of allele transmission assays from two diploids. Diploid 1 has an empty vector (EV) with a hygromycin resistance gene (hphMX) integrated at lys4 and another EV with a G418 resistance gene (kanMX) integrated at ade6. Diploid 2 has a vector with a wtf4antidote-mCherry allele (under the control of the β-estradiol promoter) and hygromycin resistance integrated at lys4. It also has a vector with a wtf4poison-GFP allele (under the control of the β-estradiol promoter) and G418 resistance integrated at ura4. The ura4 and lys4 loci are unlinked, and there are four possible spore genotypes in relation to drug resistance (R) or sensitivity (S). We sporulated diploids on SPA media (without β-estradiol) and performed random spore analysis (Smith, 2009). The expected percentage of each spore genotype and the observed percentages are shown, as well as the raw data (*=p < 0.01, G-test; NS = not significant). We were unable to recover strains carrying only the β-estradiol-inducible wtf4poison-GFP allele, suggesting it is toxic in the absence of the Wtf4antidote. (B) A vegetatively growing haploid cell expressing a β-estradiol inducible wtf4antidote-mCherry allele (magenta in merged images). This strain also contains a tagged sec63-YFP allele to mark the endoplasmic reticulum (ER) (yellow in merged images) and is stained with CMAC, a vacuole stain (green in merged images). Cells were imaged 4 hr after induction with 100 nM β-estradiol. (C) Pearson correlation of Wtf4antidote-mCherry and CMAC (vacuole, black line) and sec63-YFP (ER, blue line) from cells carrying a β-estradiol inducible wtf4antidote-mCherry allele, integrated sec63-YFP allele, and stained with CMAC, imaged f4 hours after induction with 100 nM β-estradiol. Wtf4antidote-mCherry and CMAC shows a Pearson coefficient of approximately 0.69, while Wtf4antidote-mCherry and Sec63-YFP shows −0.1. (D) A vegetatively growing haploid cell expressing a β-estradiol inducible wtf4antidote-mCherry allele (magenta in merged images) and a β-estradiol inducible wtf4poison-GFP allele (cyan in merged images) imaged 4 hr after induction with 100 nM β-estradiol. All scale bars represent 2 µm. TL = transmitted light. (E) Pearson correlation of Wtf4antidote-mCherry and Wtf4poison-GFP from cells carrying a β-estradiol inducible wtf4antidote-mCherry allele and a β-estradiol inducible wtf4poison-GFP allele 4 hr after induction with 100 nM β-estradiol shows a Pearson coefficient of approximately 0.64.
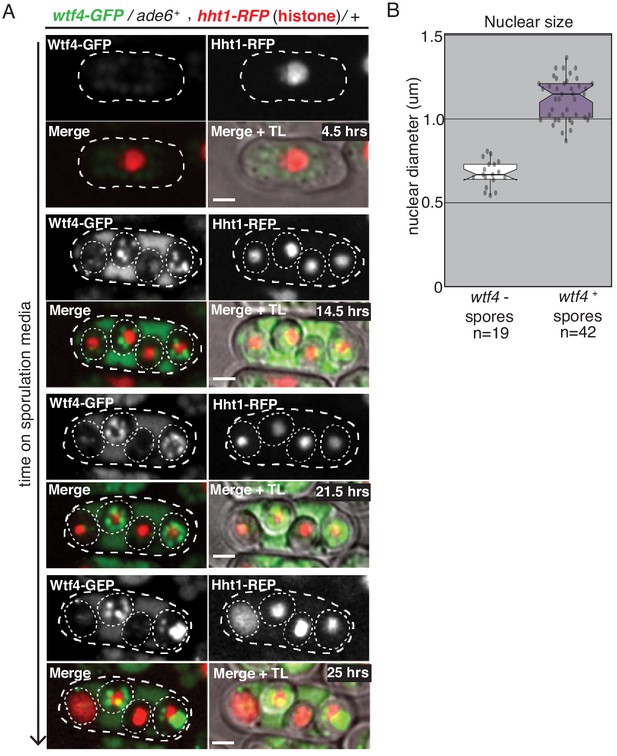
Spores destroyed by Wtf4poison exhibit nuclear condensation followed by fragmentation.
(A) A time course showing the nuclear phenotypes representative of gametogenesis starting from a wtf4-GFP/ade6+, hht1-RFP/+ diploid. The nucleus was visualized using the histone Hht1-RFP marker (red in merged images). The spores that inherit the wtf4-GFP allele can be distinguished from those that do not, because the wtf4+ spores have enriched GFP signal (green in merged images) from the accumulation of the Wtf4antidote. During spore development, the nuclei of the wtf4- spores appear to condense (14.5 hr) and, in some spores, fragment (25 hr). All scale bars represent 2 µm. (B) Quantification of nuclear diameter (µm) in wtf4- and wtf4+ spores in asci produced by wtf4-GFP/ade6+, hht1-RFP/+ diploids, imaged two days after placement on SPA media. We excluded any nuclei that appeared to have already exploded (p<0.01, t-test). TL = transmitted light.
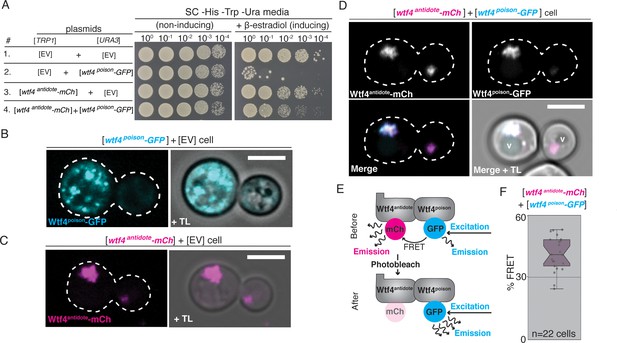
Wtf4poison and Wtf4antidote proteins physically interact and are functional in vegetative S. cerevisiae cells.
(A) Spot assay of serial dilutions on non-inducing (SC -His -Trp -Ura) and inducing (SC -His -Trp -Ura + 500 nM β-estradiol) media. Each strain contains [TRP1] and [URA3] ARS CEN plasmids that are either empty (EV) or carry the indicated β-estradiol-inducible wtf4 alleles. (B) A cell carrying an empty [TRP1] vector and a [URA3] vector with a β-estradiol inducible wtf4poison-GFP allele (cyan). (C) A haploid cell carrying an empty [URA3] vector and a [TRP1] plasmid with a β-estradiol inducible wtf4antidote-mCherry allele (magenta). (D) A haploid cell carrying a [URA3] plasmid with a β-estradiol inducible wtf4poison-GFP allele (cyan in merged images) and a [TRP1] plasmid with a β-estradiol inducible wtf4antidote-mCherry allele (magenta in merged images). The vacuole is marked with ‘v’. (E) Cartoon of acceptor photobleaching Fluorescence Resonance Energy Transfer (FRET). If the two proteins interact, Wtf4poison-GFP (the donor) transfers energy to Wtf4antidote-mCherry (the acceptor). After photobleaching of the acceptor, the donor emission will increase. (F) Quantification of FRET values measured in cells carrying β-estradiol inducible Wtf4antidote-mCherry and β-estradiol inducible Wtf4poison-GFP. The cells showed an average of 40% FRET. These data are also presented in Figure 5C. In all experiments, the cells were imaged approximately 4 hr after induction in 500 nM β-estradiol. All scale bars represent 4 µm. TL = transmitted light.
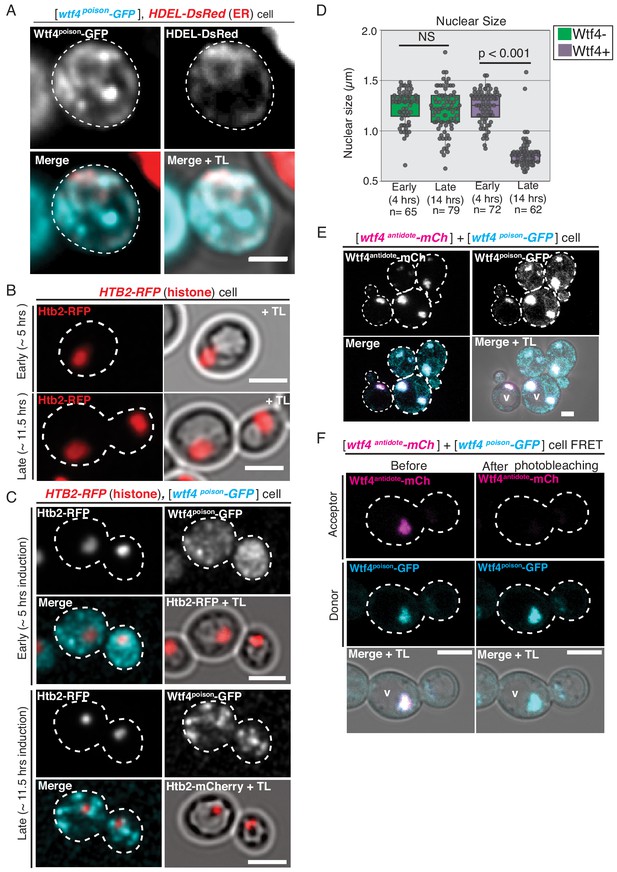
Similarities in Wtf4 protein localization and Wtf4poison-induced cell death between S. pombe and S. cerevisiae.
(A) A cell carrying a [TRP1] ARS CEN plasmid with a HDEL-DsRed allele serving as an ER marker (red in merged images) and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele (cyan in merged images) imaged 4 hr after being placed in 500 nM β-estradiol media. (B) A cell containing an integrated HTB2-RFP histone marker (red) ~5 hr and ~11.5 after switching to SC galactose (inducing) media. (C) A cell containing the HTB2-RFP histone marker (red in merged images) and carrying a [URA3] ARS CEN plasmid with a galactose inducible wtf4poison-GFP allele (cyan in merged images) ~5 hr and ~11.5 after switching to SC galactose (inducing) media. (D) Quantification of nuclear size (μm) in wild-type cells and in cells carrying the [URA3] ARS CEN plasmid with the galactose inducible wtf4poison-GFP allele 4 and 14 hr after being placed in SC galactose (inducing) media (p<0.01, t-test, NS = not significant). (E) A cell carrying both a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote-mCherry allele and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele imaged 4 hr after being placed in 500 nM β-estradiol media. Both Wtf4 proteins colocalize in a large puncta next to the vacuole (v), but there is a faint circle of Wtf4poison-GFP (cyan in merged images) that is devoid of Wtf4antidote-mCherry (magenta in merged images, arrow). (F) Representative image of Acceptor Photobleaching Fluorescence Resonance Energy Transfer (FRET) of Wtf4poison-GFP (the donor, cyan) and Wtf4antidote-mCherry (the acceptor, magenta) before and after photobleaching of Wtf4antidote-mCherry. The GFP signal is brighter following photobleaching of mCherry. FRET was assayed 4 hr after induction in 500 nM β-estradiol media. All scale bars represent 4 µm. TL = transmitted light.
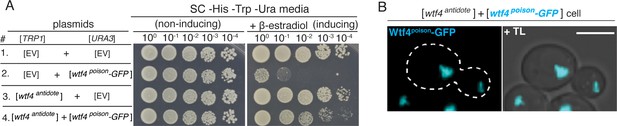
Untagged Wtf4antidote causes altered localization of Wtf4poison-GFP and neutralizes Wtf4poison-GFP toxicity in S. cerevisiae.
(A) Spot assay of serial dilutions on non-inducing (SC -His -Trp -Ura) and inducing (SC -His -Trp -Ura + 500 nM β-estradiol) media. Each strain contains [TRP1] and [URA3] ARS CEN plasmids that are either empty (EV) or carry the indicated β-estradiol inducible wtf4 alleles. (B) A haploid cell carrying a [TRP1] ARS CEN plasmid with an β-estradiol inducible wtf4antidote allele and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele (cyan) imaged 4 hr after induction in 500 nM β-estradiol. Scale bar represents 4 µm. TL = transmitted light.
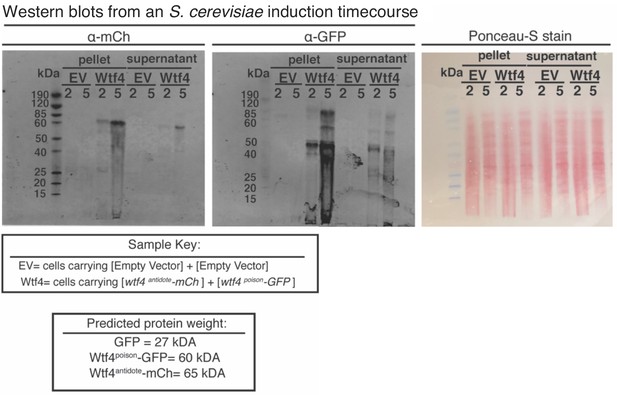
Potential Wtf4antidote-mCherry and Wtf4poison-GFP degradation in S. cerevisiae cells.
Western blot analysis of Wtf4antidote-mCherry and Wtf4poison-GFP in the supernatant and pellet of whole-cell lysates of S. cerevisiae cells carrying either empty [URA3] and [TRP1] vectors or with the same vectors carrying β-estradiol inducible tagged wtf4 alleles. Whole-cell lysates were made at 2- and 5 hr post-induction with 500 nM β-estradiol. The right image is of Ponceau-S staining on the membranes to show protein levels, acting as a loading control.

Wtf4poison and Wtf4antidote proteins assemble into aggregates at all detectable concentrations.
(A) Model of Distributed Amphifluoric FRET (DAmFRET) assay. (B) Spot assay of serial dilutions on non-inducing (SC -His -Trp -Ura) and inducing (SC -His -Trp -Ura + 500 nM β-estradiol) media. Each strain contains [TRP1] and [URA3] ARS CEN plasmids that are either empty (EV) or carry the indicated β-estradiol inducible wtf4 alleles. (C) DAmFRET plots of flow cytometry data of cells carrying a [URA3] ARS CEN plasmid carrying galactose inducible mEos3.1 (control for a monomeric protein population), cells carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4poison-mEos3.1 allele and an empty [URA3] vector, cells carrying a [URA3] plasmid with a β-estradiol inducible wtf4antidote-mEos3.1 allele and an empty [TRP1] vector, and cells carrying a [URA3] plasmid with a β-estradiol inducible wtf4antidote-mEos3.1 allele and a [TRP1] plasmid with a β-estradiol inducible wtf4poison-mEos3.1. Cells carrying vectors with the β-estradiol inducible Wtf4-mEos3.1 protein were assayed four hours after induction in 500 nM β-estradiol. Cells carrying the galactose-inducible mEos3.1 control were assayed 16 hr after induction in SC galactose media.

Overexpression of various chaperones does not neutralize Wtf4poison-GFP toxicity.
Spot assays of serial dilutions on non-inducing (SC -His -Trp -Ura -Leu), Wtf4poison inducing (SC -His -Trp -Ura + 50 nM β-estradiol in (A) and 100 nM β-estradiol in (B)), chaperone inducing (SC galactose -His -Trp -Ura -Leu), and Wtf4poison+chaperone double inducing (SC galactose -His -Trp -Ura -Leu + 50 nM β-estradiol) media. Each strain (except for the Hsp104 experiments) contains a [TRP1] ARS CEN plasmid that is either empty (EV) or carries a β-estradiol inducible wtf4 poison-mEos3.1 allele and a [URA3] ARS CEN plasmid that is either empty (EV) or carries the indicated galactose inducible chaperone. The Hsp104 strains contain a [URA3] ARS CEN plasmid that is either empty (EV) or carries a β-estradiol inducible wtf4poison-GFP allele and a [LEU2] ARS CEN plasmid that is either empty (EV) or carries the galactose inducible HSP104 allele.

Homotypic interactions facilitate Wtf4poison-Wtf4antidote co-assembly and Wtf4antidote function.
(A) DNA sequence encoding 18 amino acids (marked with an *) were added to the repeat sequences within exon 6 of the wtf4 allele to create wtf4* alleles. (B) Spot assay of serial dilutions on non-inducing (SC -His -Trp -Ura) and inducing (SC -His -Trp -Ura + 500 nM β-estradiol) media. Each strain contains [TRP1] and [URA3] ARS CEN plasmids that are either empty (EV) or carry the indicated β-estradiol inducible wtf4 alleles. (C) Representative image of a haploid cell carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote*-mCherry allele (magenta in merged images) and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison*-GFP (cyan in merged images). (D) Representative image of a haploid cell carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote*-mCherry allele (magenta in merged images) and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele (cyan in merged images). (E) Representative image of a haploid cell carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote-mCherry allele (magenta in merged images) and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison*-GFP allele (cyan in merged images). In all experiments, the cells were imaged ~4 hr after induction in 500 nM β-estradiol. All scale bars represent 4 µm. TL = transmitted light.
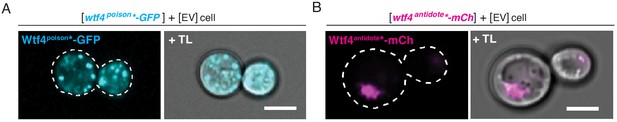
Wtf4poison* and Wtf4antidote* have the same localization as the wild-type Wtf4 proteins in S. cerevisiae.
(A) Representative image of a haploid cell carrying a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison*-GFP allele (cyan) and an empty [TRP1] vector. (B) Representative image of a haploid cell with carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote*-mCherry allele (magenta) and an empty [URA3] vector. In both experiments, the cells were imaged ~4 hr after induction in 500 nM β-estradiol. All scale bars represent 4 µm. TL = transmitted light.

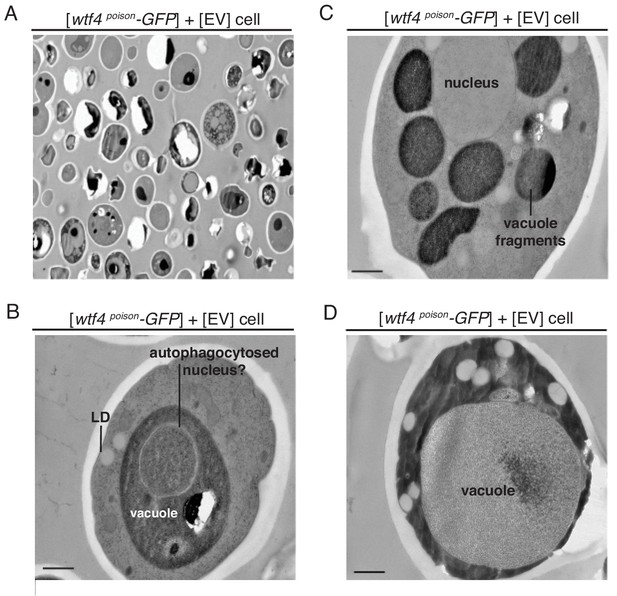
Cells expressing Wtf4poison and Wtf4antidote or Wtf4antidote have large vesicles that cluster within the Wtf4 aggregate.
(A) Representative tomograph of immuno-gold Transmission Electron Microscopy (TEM) of a haploid cell carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote allele and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele. A monoclonal antibody against GFP was used. Immunogold particles (black dots) are enriched in a cluster near light staining organelles. (B) Representative TEM tomograph of cells of the same genotype as in (A). Arrows point to vesicle structures. (C) Representative TEM tomograph of a cell carrying an empty [TRP1] vector and an empty [URA3] vector. (D) Quantification of the number of lipid droplets (left) or vesicles (right) per cell of two samples: 1. Cells carrying empty [TRP1] and [URA3] vectors (EV, white, n = 203 cells) and 2. Cells carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote allele and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele (purple, n = 133 cells), (p<0.01, t-test). All samples were processed ~4 hr after induction in 500 nM β-estradiol. All scale bars represent 0.5 µm. Lipid droplet = LD.
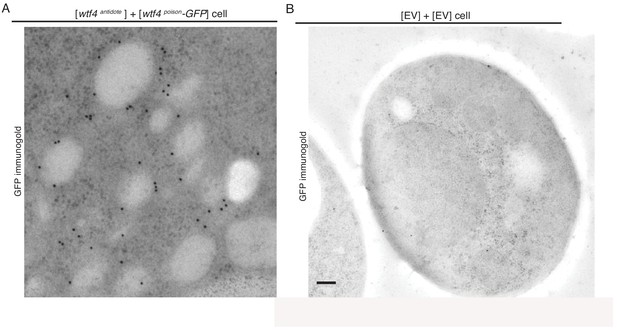
Cells expressing Wtf4poison and Wtf4antidote or Wtf4antidote have large vesicles that cluster within the Wtf4 aggregate.
(A) A cropped, magnified immuno-gold Transmission Electron Microscopy (TEM) tomograph of a region of a haploid cell carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote allele and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele. A monoclonal antibody against GFP was used. Immunogold particles (black dots) are enriched in a cluster near light staining organelles. (B) Representative Immuno-gold TEM tomograph of a cell carrying an empty [TRP1] vector and an empty [URA3] vector. The same antibody was used as in (A), but very few particles were observed, suggesting little background staining is present. Both samples were processed after ~4 hr of induction in 500 nM β-estradiol. Scale bar represents 0.2 µm.

Cells expressing Wtf4poison+Wtf4antidote or Wtf4antidote have large vesicles that cluster within the Wtf4 aggregate.
(A) Transmission Electron Microscopy (TEM) tomograph of a haploid cell carrying an empty [TRP1] vector and an empty [URA3] vector. (B) TEM tomograph of a haploid cell carrying a [TRP1] vector with a wtf4antidote allele and a [URA3] vector with a wtf4poison-GFP allele. (C) Models generated using array tomography, a process that utilizes serial slices and Scanning Electron Microscopy (SEM) to generate a 3D reconstruction of the cell. Top is the reconstruction of a haploid cell of the same genotype as that in (A). The bottom is the 3D reconstruction of a haploid cell of the same genotype as that in (B). The key shows the colors representative of the cellular structures in the images. (D) TEM tomograph of a haploid cell carrying a [TRP1] vector with a wtf4antidote-mCherry allele and an empty [URA3] vector. (E) TEM tomograph of a haploid cell carrying a [TRP1] ARS CEN plasmid with a RNQ1-mCardinal allele and an empty [URA3] vector. (F) Quantification of the size of lipid droplets (left) or vesicles (right) in two samples: wild-type cells like those in (A) (EV, white, n = 203) and cells like those in (B) expressing both Wtf proteins (purple, n = 133 cells). (G) Quantification of the average volume of mitochondria per cell (left) or number of mitochondria per cell (right) in wild-type (EV, white, n = 15 cells) and cells expressing Wtf proteins (purple, n = 12 cells), as in (F) (ns = not significant) (p<0.01, t-test). All samples were processed after ~4 hr of induction in 500 nM β-estradiol. Scale bar represents 0.5 µm.
Cells producing only Wtf4poison have variable phenotypes.
(A–D) are representative Transmission Electron Microscopy (TEM) tomographs of cells carrying a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP and an empty [TRP1] vector. Cells were processed 4 hr after induction with 500 nM β-estradiol. (A) TEM micrograph at a lower magnification, showing a field of cells. Many of the cells appear dead and have lost integrity through the TEM process. There are few cells surviving long enough to have recognizable phenotypes. We grouped the remaining cells into phenotype classes listed in B-D. (B) One phenotype of the remaining cells is increased autophagy. The cell shown has an organelle, presumed to be the nucleus, inside the darker staining vacuole. (C) Some cells show numerous, fragmented vacuoles. (D) Others show enlarged vacuoles, with minimal, darker staining cytoplasm. All scale bars represent 0.5 µm. LD = lipid droplet.
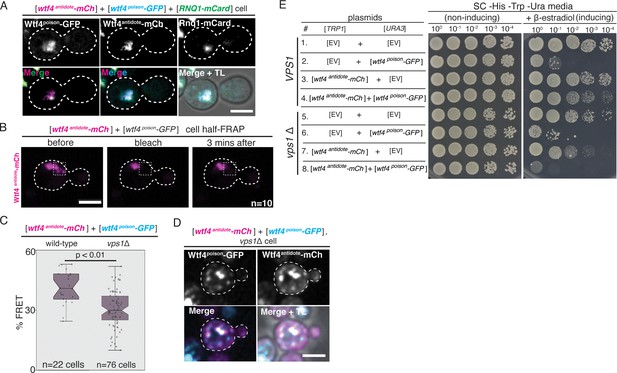
The Wtf4 proteins colocalize at the Insoluble Protein Deposit (IPOD).
(A) Representative image of a vegetatively growing haploid cell carrying a [TRP1] vector with a β-estradiol inducible wtf4antidote-mCherry (magenta in merged images) allele, a [URA3] vector with a β-estradiol inducible wtf4poison-GFP (cyan) allele, and a [LEU2] vector with a β-estradiol inducible RNQ1-mCardinal allele, acting as an IPOD marker (green in merged images). (B) half-Fluorescence Recovery After Photobleaching (half-FRAP) of the Wtf4antidote-mCherry aggregate in cells carrying a [TRP1] vector with a β-estradiol inducible wtf4antidote-mCherry (magenta) allele and a [URA3] vector with a β-estradiol inducible wtf4poison-GFP allele. Cells were imaged for 3 min after bleaching and no recovery of mCherry fluorescence was observed. (C) Quantification of FRET values of Wtf4antidote-mCherry and Wtf4poison-GFP measured in wild-type (same data as Figure 2F) and vps1Δ cells carrying vectors with β-estradiol inducible wtf4antidote-mCherry allele and β-estradiol inducible wtf4poison-GFP allele (p>0.01, t-test). (D) Representative image of a vegetatively growing, haploid vps1Δ cell carrying a [URA3] vector with a β-estradiol inducible wtf4poison-GFP allele (cyan in merged images) and a [TRP1] vector with a β-estradiol inducible wtf4antidote-mCherry (magenta in merged images) allele. All fluorescence microscopy images acquired after ~4 hr in 500 nM β-estradiol media. All scale bars represent 4 µm. TL = transmitted light. (E) Spot assay of serial dilutions on non-inducing inducing (SC -His -Trp -Ura) and inducing (SC -His -Trp -Ura + 500 nM β-estradiol) media of both wild-type (top, samples 1–4) and vps1Δ (bottom, samples 5–8) cells. Each strain contains [TRP1] and [URA3] ARS CEN plasmids that are either empty (EV) or carry the indicated β-estradiol-inducible wtf4 alleles.
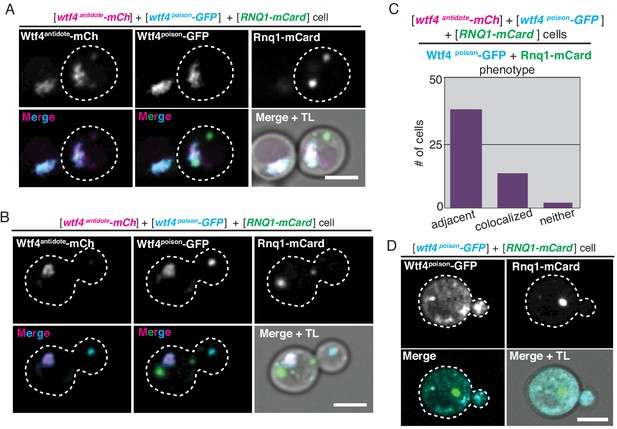
The Wtf4 proteins colocalize at the Insoluble Protein Deposit (IPOD).
(A) A cell carrying a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele (cyan in merged images), a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote-mCherry allele (magenta in merged images), and a [LEU2] ARS CEN plasmid with a β-estradiol inducible RNQ1-mCardinal allele (green in merged images) acting as an IPOD marker. The image shown is representative of the colocalization phenotype, where both Wtf4 proteins colocalize with the Rnq1-mCardinal IPOD maker (n = 11). (B) Cells of the same genotype as in (A) showing the adjacent phenotype, where both Wtf4 proteins colocalize directly adjacent to the Rnq1-mCardinal IPOD maker (n = 30). Scale bars represent 4 µm. (C) We quantified the localization phenotypes of Wtf4poison-GFP and Rnq1-mCardinal in 45 cells. Eleven cells showed colocalization of Wtf4poison-GFP and Rnq1-mCardinal, while 30 showed Wtf4poison-GFP localizing adjacent to Rnq1-mCardinal. Four cells showed neither colocalization nor adjacency of Wtf4poison-GFP and Rnq1-mCardinal. (D) Representative image of a cell carrying a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP allele (cyan in merged images), an empty [TRP1] vector, and a [LEU2] ARS CEN plasmid with a β-estradiol inducible RNQ1-mCardinal acting as an IPOD marker (green in merged images). Cells were images ~4 hr in 500 nM β-estradiol media. Scale bar represents 4 µm. TL = transmitted light.

A screen of the S. cerevisiae deletion collection to identify genes necessary for survival upon Wtf4poison and Wtf4antidote expression.
(A) Cartoon of the screen designed to identify genes necessary for survival of cells after co-induction of the Wtf4 poison and antidote proteins. (B) Representative images of a plate from the screen. The control is a wild-type cell carrying a [LEU2] ARS CEN plasmid with a galactose inducible wtf4antidote-mCherry allele and a [URA3] ARS CEN plasmid with a galactose inducible wtf4poison-GFP allele. Cells that show reduced growth in comparison to the control on SC galactose (inducing) media were deemed potential ‘hits’. This identified 250 strains. We then transformed empty [LEU2] and [URA3] vectors into these 250 strains and screened for survival on SC galactose media to remove false hits that cannot grow on galactose. At the end, we identified 106 hits that grew poorly upon Wtf4 protein expression. (C)-(G) are representative images of vegetatively growing, haploid cells carrying a [LEU2] ARS CEN plasmid with a galactose inducible wtf4antidote-mCherry (magenta in merged images) and a [URA3] ARS CEN plasmid with a galactose inducible wtf4poison-GFP allele (cyan in merged images). Cells were imaged 4 hr in galactose (inducing) media. (C) ynl170wΔ and (D) phd1Δ are representative of multiple samples (81/106) that showed the phenotype of the Wtf4 proteins localizing as dispersed puncta throughout the cell. (E) mne1Δ, and (F) qri5Δ, are representative of multiple samples (5/106) that showed the phenotype of the Wtf4antidote-mCherry protein localizing as one aggregate, while the Wtf4poison-GFP protein looked more dispersed. (G) abz2Δ, is representative of multiple samples (20/106) that showed the Wtf4antidote-mCherry and Wtf4poison-GFP proteins localizing as a soluble haze throughout the cell. All scale bars represent 4 µm. TL = transmitted light.
-
Figure 5—figure supplement 2—source data 1
Results of a genetic screen of the S. cerevisiae deletion collection to identify genes necessary for survival upon Wtf4 protein expression.
Tab 1 is a table of contents describing the rest of the file. The file includes all the genes assayed in the screen and the hits before and after the secondary screen (described in Figure 5—figure supplement 2A). Additional information about the localization of the Wtf4 proteins in the screen hits, the annotated functions of the screen hits and their S. pombe homologs are also provided.
- https://cdn.elifesciences.org/articles/55694/elife-55694-fig5-figsupp2-data1-v1.xls
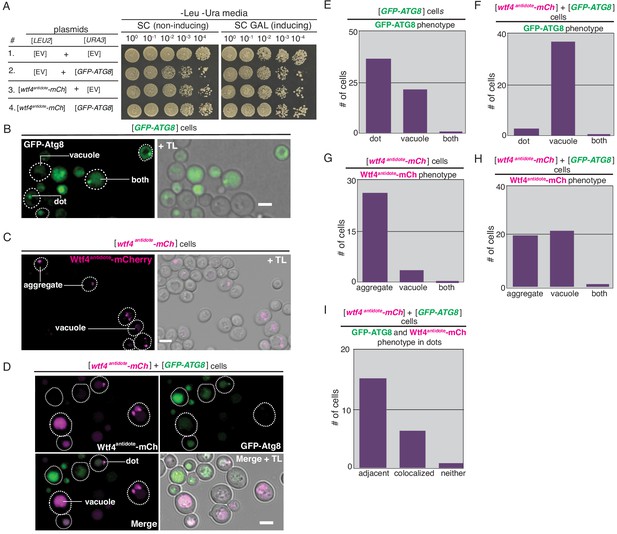
Overexpression of ATG8 causes vacuolar localization of Wtf4antidote-mCherry.
(A) Spot assay of serial dilutions on non-inducing inducing (SC -Leu -Ura) and inducing (SC Galactose -Leu -Ura) media. Each strain contains [LEU2] and [URA3] ARS CEN plasmids that are either empty (EV), carry the galactose-inducible wtf4antidote-mcherry allele, or the GFP-ATG8 allele under the endogenous promoter. (B) Representative image of a cell carrying a [URA3] ARS CEN plasmid with the GFP-ATG8 under its endogenous promoter acting as a pre-autophagosomal site (PAS) marker. (C) Representative image of a cell carrying a [LEU2] ARS CEN plasmid with a galactose inducible wtf4antidote-mCherry (magenta). (D) Representative image of a cell carrying a [LEU2] ARS CEN plasmid with a galactose inducible wtf4antidote-mCherry (magenta in merged images) and a [URA3] ARS CEN plasmid with a GFP-ATG8 allele (green in merged images). Arrow points to a rare cell that showed GFP-Atg8 and Wtf4antidote-mCherry localizing as a vacuole-associated dot and aggregate, respectively. These were the cells quantified in (I). All images acquired after ~4 hr in SC galactose media. All scale bars represent 4 µm. TL = transmitted light. (E) Quantification of the localization phenotypes of GFP-Atg8 in 26 cells carrying a [URA3] ARS CEN plasmid containing the GFP-ATG8 allele. (F) Quantification of the localization phenotypes of GFP-Atg8 in 40 cells carrying both a [URA3] ARS CEN plasmid containing the GFP-ATG8 allele and a [LEU2] ARS CEN plasmid carrying the wtf4antidote-mCherry allele. (G) Quantification of the localization phenotypes of Wtf4antidote-mCherry in 33 cells carrying a [LEU2] ARS CEN plasmid containing the wtf4antidote-mCherry allele. (H) Quantification of the localization phenotypes of Wtf4antidote-mCherry in 40 cells carrying both a [URA3] ARS CEN plasmid containing the GFP-ATG8 allele and a [LEU2] ARS CEN plasmid containing the wtf4antidote-mCherry allele. (I) Quantification of the localization phenotypes of Wtf4antidote-mCherry and GFP-Atg8 in 23 cells where the proteins remained outside of the vacuole.
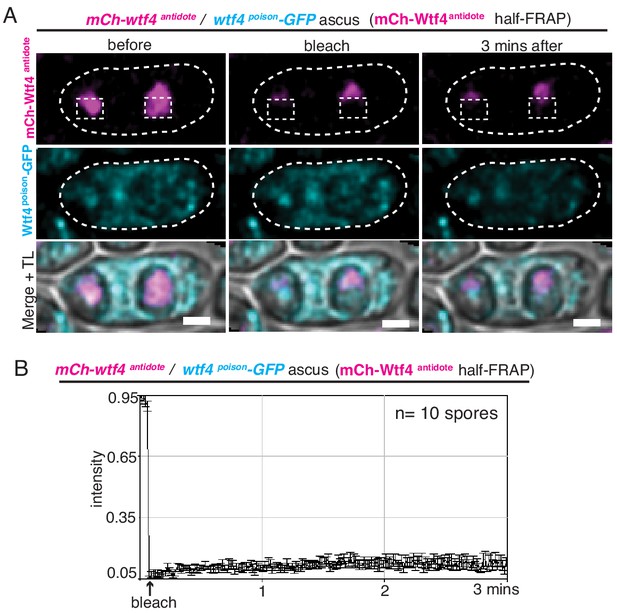
Wtf4antidote-mCherry is stable in S. pombe spores.
(A) Representative image of half-Fluorescence Recovery After Photobleaching (half-FRAP) of the Wtf4antidote-mCherry (magenta) aggregate in asci that were generated from heterozygous mCherry-wtf4/wtf4poison-GFP diploids. (B) Quantification of the FRAP data shown in A. Cells were imaged for 3 min after bleaching and very little recovery was observed. TL = transmitted light.
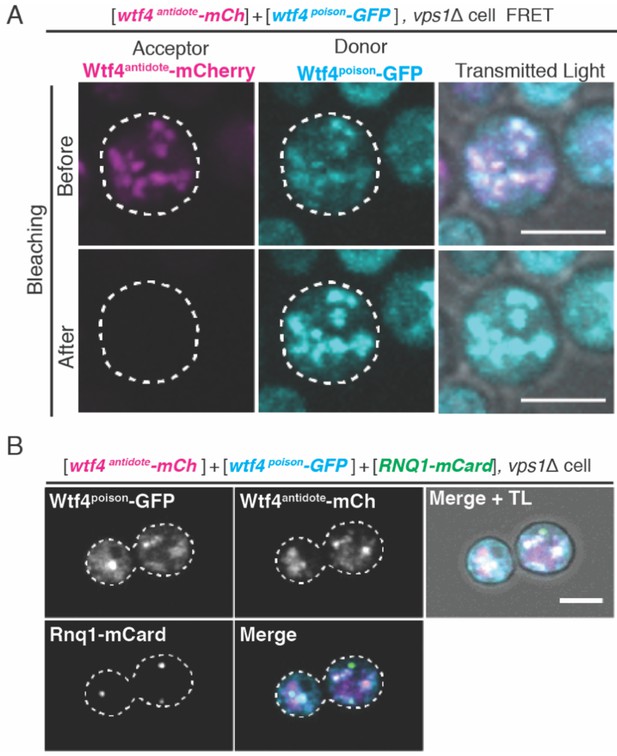
Vps1 is necessary for the recruitment of Wtf4 proteins to the Insoluble Protein Deposit (IPOD).
(A) Representative image of acceptor photobleaching Fluorescence Resonance Energy Transfer (FRET) of Wtf4antidote-mCherry and Wtf4poison-GFP in vps1Δ cells carrying a carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote-mCherry (magenta in merged images) allele and a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP (cyan in merged images) allele. After photobleaching of Wtf4antidote-mCherry (the acceptor), Wtf4poison-GFP (the donor) signal increases. (B) Representative image of a vegetatively growing haploid vps1Δ cell carrying a [TRP1] ARS CEN plasmid with a β-estradiol inducible wtf4antidote-mCherry (magenta in merged images) allele, a [URA3] ARS CEN plasmid with a β-estradiol inducible wtf4poison-GFP (cyan in merged images) allele, and a [LEU2] ARS CEN plasmid with the β-estradiol inducible IPOD marker RNQ1-mCardinal (green in merged images). All cells were imaged 4 hr after induction in 500 nM β-estradiol media. All scale bars represent 4 µm. TL = transmitted light.
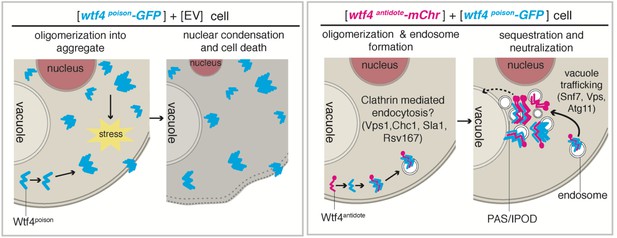
Model of Wtf4poison and Wtf4antidote mechanism in S. cerevisiae.
Wtf4poison assembles into toxic aggregates that spread throughout the cell potentially causing stress. This stress could lead to nuclear condensation and ultimately cell death. Wtf4antidote co-assembles with Wtf4poison, at least partially driven by shared sequences within the C-terminus. These assemblies are recruited to a compartment next to the vacuole and are also vesicle-associated. This sequestration neutralizes the Wtf4poison toxicity, rescuing cell viability.
Additional files
-
Supplementary file 1
Yeast strains.
Column 1 is the name of strain used, while column 2 refers to the species of the yeast (Sp = S. pombe; Sc = S. cerevisiae). Columns 3 lists the reference for the yeast strain. If it was made in this study, we also detail how the strain was made in column 5. Column 6 lists which figures the yeast strain was used in.
- https://cdn.elifesciences.org/articles/55694/elife-55694-supp1-v1.xlsx
-
Supplementary file 2
Plasmids used in this study.
Column 1 is the name of plasmid used, while column 2 describes the content of the plasmid. Columns 3 lists the reference for each plasmid.
- https://cdn.elifesciences.org/articles/55694/elife-55694-supp2-v1.xlsx
-
Supplementary file 3
Oligos used in this study.
Column 1 lists the oligo numbers and column 2 provides the sequence of the oligo. Columns 3 gives a short description of each oligo.
- https://cdn.elifesciences.org/articles/55694/elife-55694-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55694/elife-55694-transrepform-v1.pdf





