Roles of the ClC chloride channel CLH-1 in food-associated salt chemotaxis behavior of C. elegans
Figures
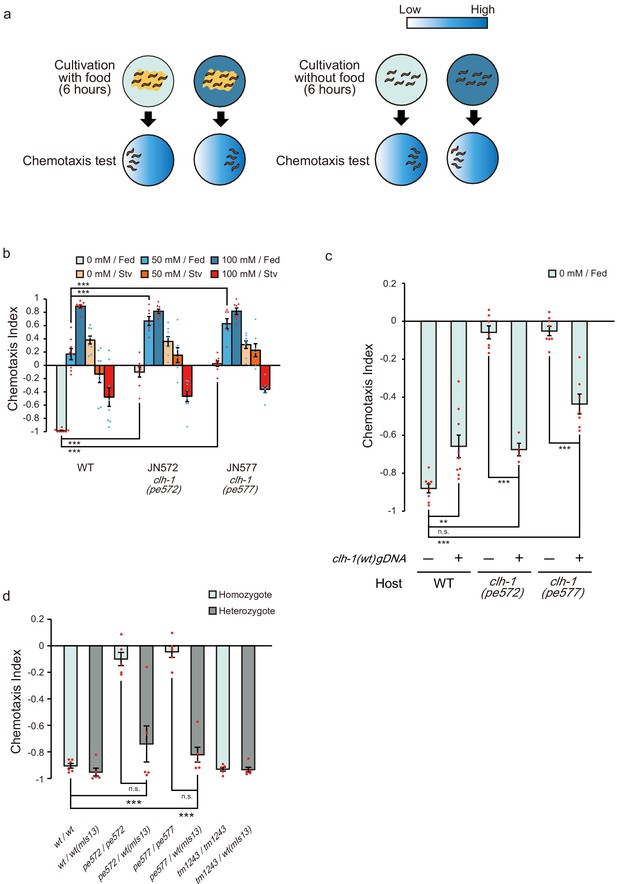
Two missense mutations in clh-1 give rise to food-associated salt chemotaxis disorder.
(a) Salt concentration chemotaxis of wild type. Adult hermaphrodites were cultivated at 0 mM or 100 mM of NaCl with or without food for 6 hr and placed on a chemotaxis assay plate on which NaCl gradient was created. Distribution of animals was quantified by calculating a chemotaxis index. See Materials and methods for details. (b) Chemotaxis of wild-type animals and two mutants obtained from screening, JN572: clh-1(pe572) and JN577: clh-1(pe577). Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n = 8 assays, Dunnett’s test, ***p<0.001. (c) Rescue of clh-1(pe572) and clh-1(pe577) mutants by a clh-1 genomic DNA fragment. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 4, Tukey’s test, ***p<0.001, **p<0.01, n.s. not significant. (d) Chemotaxis of clh-1 heterozygotes. clh-1 homozygotes were crossed with a clh-1(wt) reporter strain that express GFP in pharyngeal muscle (mIs13). Resulted F1 animals were used for assay. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 4, Tukey’s test, ***p<0.001, n.s. not significant.
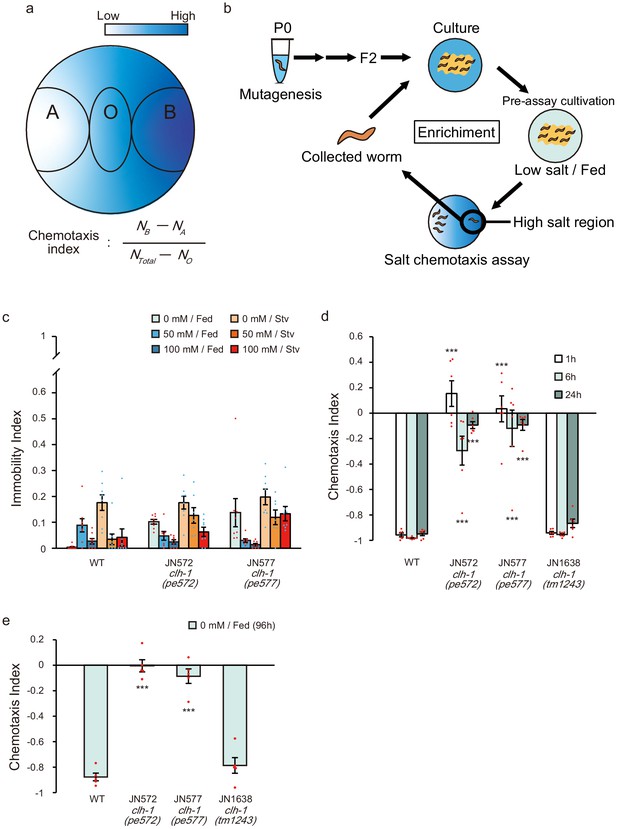
Isolation and characterization of salt chemotaxis mutants JN572 and JN577.
(a) A schematic diagram of salt chemotaxis assay plate and calculation of the chemotaxis index. See Materials and methods for details. (b) Procedure for forward genetic screening to obtain food-associated salt chemotaxis mutants. Wild-type animals were mutagenized with ethyl methanesulfonate (EMS), and F2 animals were applied for salt chemotaxis assay. Animals that showed defective chemotaxis were isolated and propagated for another round of test to enrich the ratio of mutants. (c) Immobility index of wild type, JN572 and JN577 on chemotaxis assay plate. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n = 8 assays. (d and e) Chemotaxis of wild type and clh-1 mutants after different duration of pre-assay cultivation. Animals were fed at 0 mM NaCl for 1, 6 or 24 hr (d) or 96 hr (from birth until just prior to assay, e) and tested for salt chemotaxis assay. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 6, compared with wild type, Dunnett’s test, ***p<0.001.
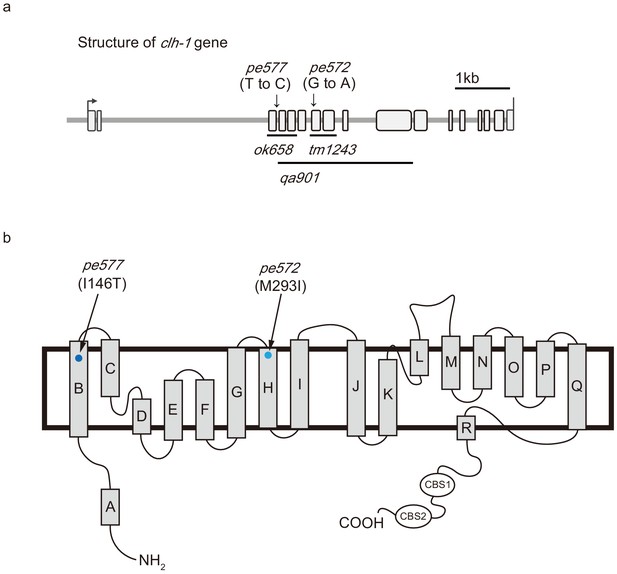
Missense mutations in clh-1 responsible for salt chemotaxis defect.
(a) Gene structure of clh-1 and the positions of clh-1 mutations. The positions of pe572 (T to C) and pe577 (G to A) are noted by arrows. ok658 removes exons 3– 5, tm1243 removes exons 6 and 7, qa901 removes exons 4–10. (b) A schematic diagram of protein structure of CLH-1, which is based on structural studies for ClCs (Duran et al., 2009; Dutzler et al., 2003; Dutzler et al., 2002; Feng et al., 2010; Jentsch, 2008; Miyazaki et al., 2012; Wang et al., 2019). Gray rectangles represent α-helices. The positions of pe572 (I146T) and pe577 (M293I) are noted by arrows. CBS1 and CBS2 indicate cystathionine beta synthase domains, which are known as protein-binding domains.
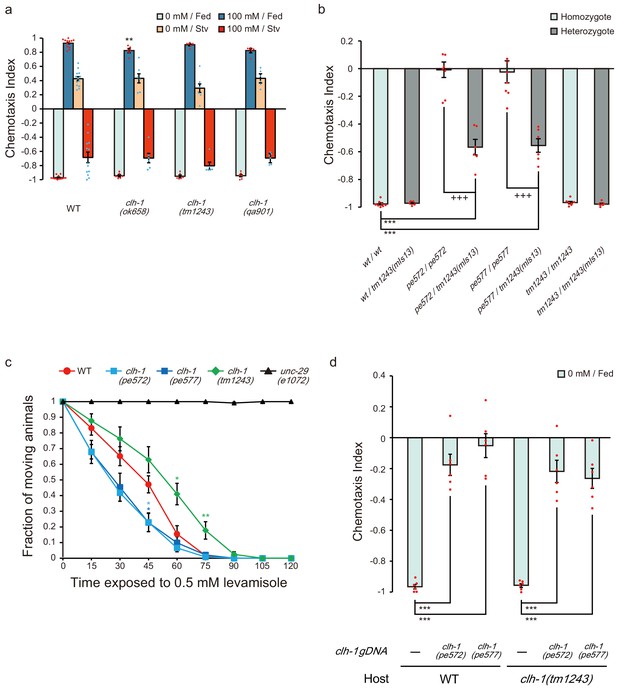
Characterization of clh-1 mutants.
(a) Chemotaxis of clh-1 deletion mutants. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 6 assays, compared with wild type, Dunnett’s test, **p<0.01. (b) pe572 and pe577 show haploinsufficiency. For generating heterozygotes, hermaphrodites of either wild type, clh-1(pe572), clh-1(pe577) or clh-1(tm1243) were crossed with clh-1(tm1243) males which express GFP in pharyngeal muscle (mIs13), and F1 animals were used for the assay. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 5, Tukey’s test, ***p<0.001, compared with wild-type homozygote. +++p<0.001, compared with each original missense allele homozygote. (c) Levamisole resistance test. The graph shows fraction of non-paralyzed (moving) animals at the indicated time points on 0.5 mM levamisole. Mean +/- s.e.m., n = 6 assays, compared with wild type, Dunnett’s test, **p<0.01, *p<0.05. Asterisks are colored according to the strain (light blue for clh-1(pe572), deep blue for clh-1(pe577), and green for clh-1(tm1243)). (d) Excess genomic DNA fragments of clh-1(pe572) or clh-1(pe577) recapitulate salt chemotaxis defect in wild type and clh-1(tm1243). Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n = 6 assays, Tukey’s test, ***p<0.001.
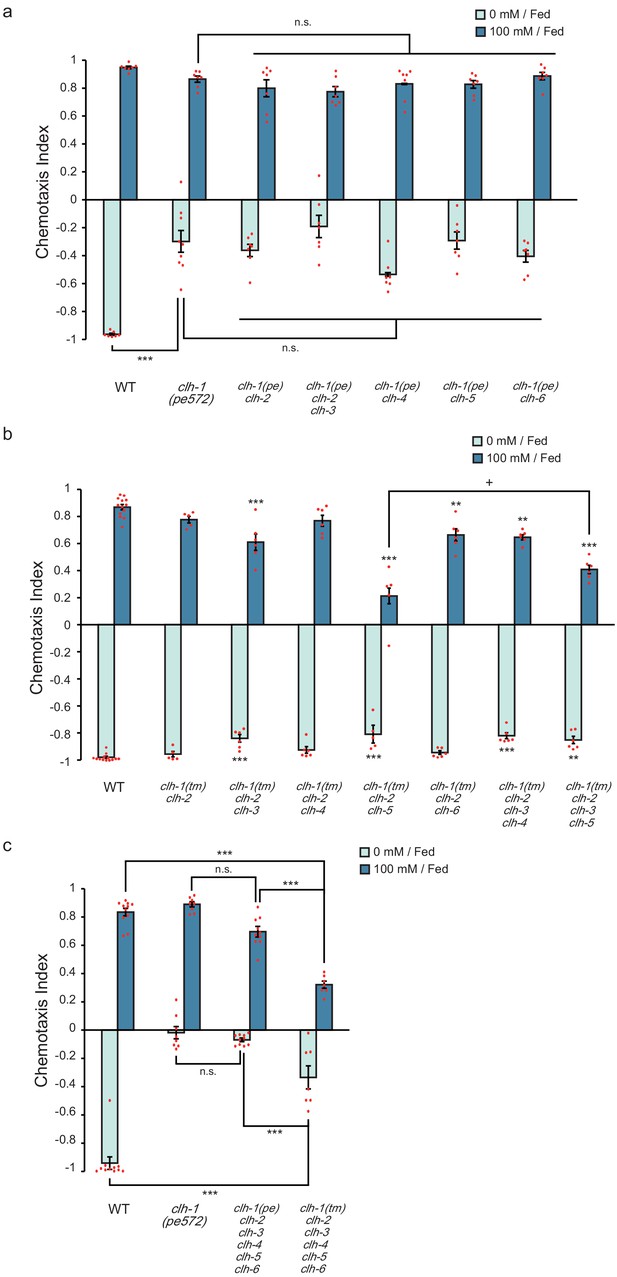
ClC genes redundantly function in salt chemotaxis.
(a) Chemotaxis of clh multiple mutants that carry clh-1(pe572) mutation with a deletion in other clh genes. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 6 assays, Tukey’s test, ***p<0.001, n.s. not significant. (b) Chemotaxis of clh multiple mutants that carry clh-1(tm1243) mutation and a deletion in other clh genes. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 5, Tukey’s test, ***p<0.001, **p<0.01, compared with wild type. +p<0.05, compared with indicated mutants. (c) Chemotaxis of clh hexatruple mutants. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 7, Tukey’s test, ***p<0.001, n.s. not significant.
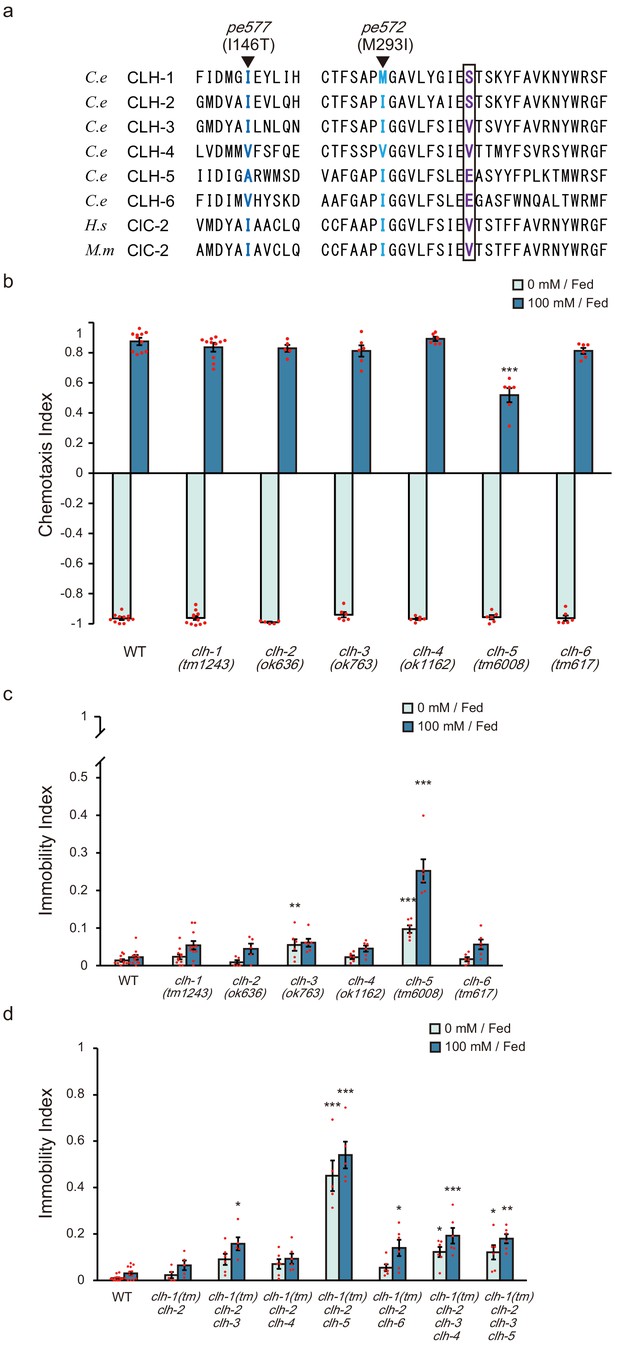
Effect of loss of ClC genes on salt chemotaxis.
(a) Comparison of C. elegans (C.e) CLHs, human (H.s), and murine (M.m) ClC-2. Amino acids altered in clh-1(pe) alleles (deep and light blue) and their flanking regions are shown. The ‘Proton glutamate’ residue is boxed, which predicts whether the corresponding ClC protein is an anion channel or a H+/Cl- antiporter (Accardi et al., 2005). Members that carry glutamate (E) at the position is proposed as a H+/Cl- antiporter, whereas anion channel if it is valine (V). Although CLH-1 and CLH-2 carry serine (S) residue at this position, they are presumed to be anion channels because they share high similarity with mammalian ClC-2 (Schriever et al., 1999; Nehrke et al., 2000). (b) Chemotaxis of the single deletion mutants of each ClC gene (clh-1 through clh-6). All mutant strains were outcrossed with wild type more than four times. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 5 assays, compared with wild type, Dunnett’s test, ***p<0.001. (c) Immobility index of the clh deletion mutants. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 5, compared with wild type, Dunnett’s test, ***p<0.001, **p<0.01. (d) Immobility index of the clh multiple mutants. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 5, Tukey’s test, ***p<0.001, **p<0.01, *p<0.05, compared with wild type.
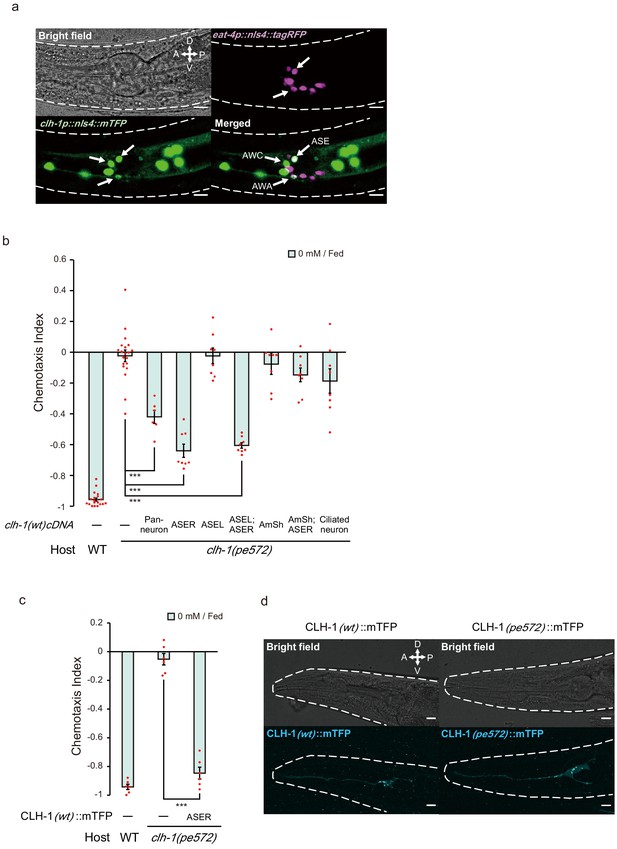
clh-1 acts in the salt-sensing neuron ASER.
(a) Expression pattern of clh-1p::nls4::mTFP (green, bottom left) in an adult animal. At least three pairs of sensory neurons, AWA, AWC and ASE expressed the marker. eat-4p::nls4::tagRFP and DiI, that marks glutamatergic neurons (magenta, top right) and six pairs of head sensory neurons (not shown), respectively, were used as position markers for cell identification. Scale bar = 10 µm. (b) Rescue of clh-1(pe572) mutants by cell-specific expression of clh-1(wt) cDNA. Promoters used in this experiment are as follows; rimb-1p for all neurons, gcy-5p for ASER, gcy-7p for ASEL, vap-1p for amphid sheath cells, dyf-11p for ciliated neurons. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n ≧ 6 assays, Tukey’s test. ***p<0.001. (c) Chemotaxis of clh-1(pe572) mutants that express clh-1(wt)cDNA::mTFP in ASER. mTFP-tagged CLH-1 is functional. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n = 6, Tukey’s test. ***p<0.001. (d) Subcellular localization of CLH-1 in ASER. Panels show gcy-5p::clh-1(wt)cDNA::mTFP in wild type (left) and gcy-5p::clh-1(pe572)cDNA::mTFP in clh-1(pe572) (right). Both CLH-1(wt)::mTFP and CLH-1(pe572)::mTFP localized to the membrane of dendrite, soma, axon, and cell organelles. Scale bar = 10 µm.
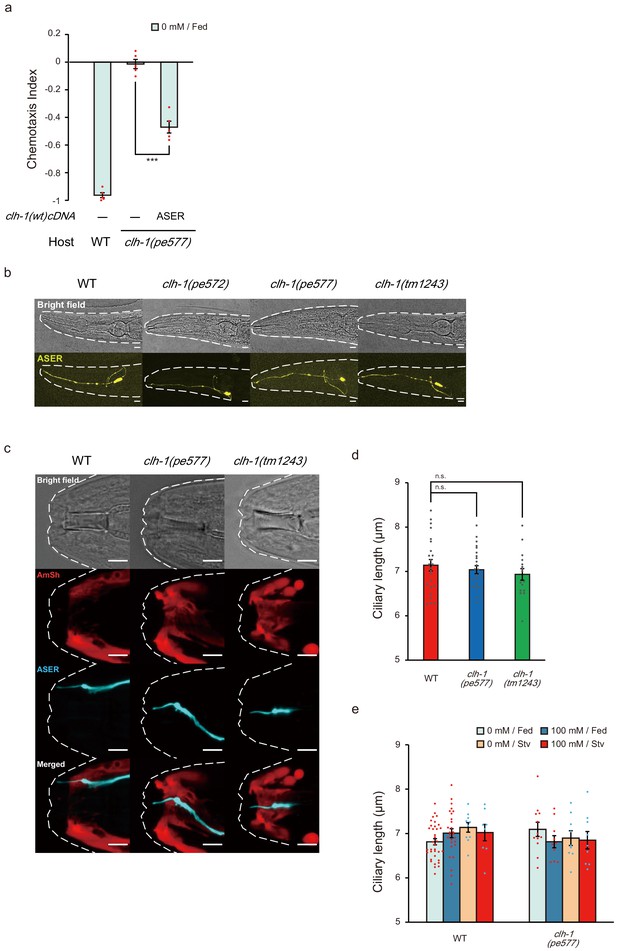
clh-1 acts in ASER in salt chemotaxis, and mutations in clh-1 do not affect morphology of ASER.
(a) Rescue of clh-1(pe577) mutants by ASER-specific expression of clh-1(wt) cDNA. Dots represent individual trials. Bars and the error bars represent mean +/- s.e.m., n = 5 assays, Tukey’s test. ***p<0.001. (b) Morphology of ASER in wild-type and clh-1 adult animals visualized by gcy-5p::GFP. No gross morphological defect was observed in the mutants. Animals were anesthetized by 10 mM NaN3. Bright-field (top) and stacked confocal fluorescence images (bottom). Scale bar = 10 µm. (c) Confocal images of ASER sensory cilium and amphid sheath glial cells in adult animals. Top; bright field, second top; AmSh (vap-1p::mCherry, red), second bottom; ASER (gcy-5p::mTFP, cyan), bottom; merged. Animals were anesthetized by 100 μM levamisole. Scale bar = 5 µm. (d) Length of ASER sensory cilium in adult animals. Animals were anesthetized by 100 μM levamisole and ciliary length was measured from confocal images. Mean +/- s.e.m., dots represent individual trials. n ≧ 16 animals, Dunnett’s test, n.s. not significant. (e) Length of ASER sensory cilium in adult animals after cultivation at the indicated conditions. Animals were anesthetized by 100 μM levamisole. Ciliary length was measured from confocal images. Mean +/- s.e.m., dots represent individual trials. n ≧ 8 animals.
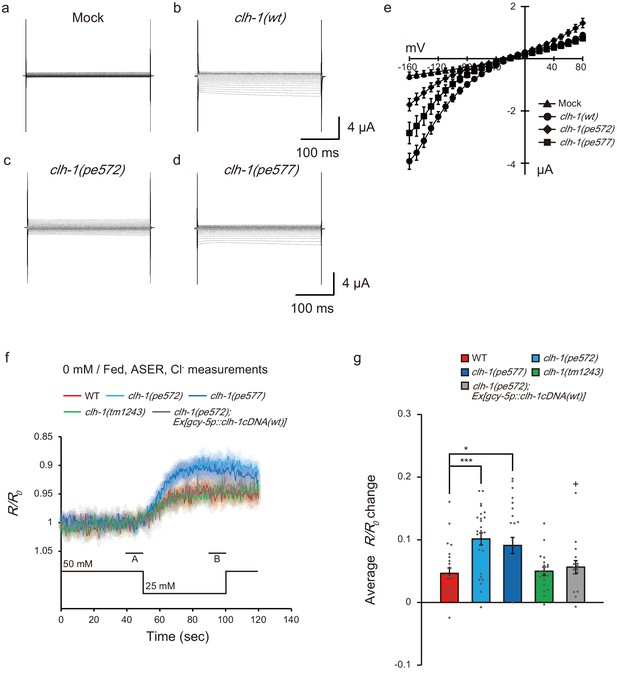
Mutations in clh-1 affect chloride dynamics of ASER in response to salt down-step stimulus.
(a–e) Representative current traces from Xenopus oocytes that expressed cRNA for mock (a), clh-1(wt) (b), clh-1(pe572) (c), and clh-1(pe577) (d) clamped at voltages ranging from −160 mV to 80 mV and perfused at pH 7 with Cl- as the dominant extracellular anion. (e) The current-voltage relationships of mock (triangle, n = 7), clh-1(wt) (circle, n = 20), clh-1(pe572) (diamond, n = 13) and clh-1(pe577) (square, n = 16). The error bars represent s.e.m. (f) Responses of SuperClomeleon expressed in ASER after cultivation at 0 mM NaCl in the presence of food. External NaCl concentration was shifted from 50 mM to 25 mM at time 50 s. Note that the scale of the vertical axis is inverted so that increase in chloride concentration is displayed as up-shift of traces. A and B indicate the time points for calculating R/R0 changes. The shaded region represents s.e.m., n ≧ 17 animals. (g) R/R0 changes upon salt decrease. Mean +/- s.e.m., dots represent individual trials. n ≧ 17 animals, Tukey’s test, ***p<0.001, *p<0.05, compared with wild type. +p<0.05, compared with clh-1(pe572) mutant.
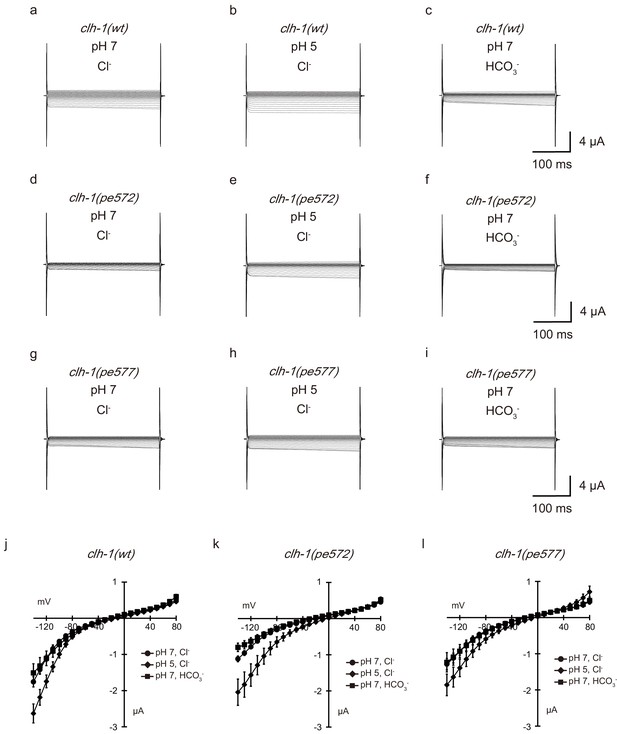
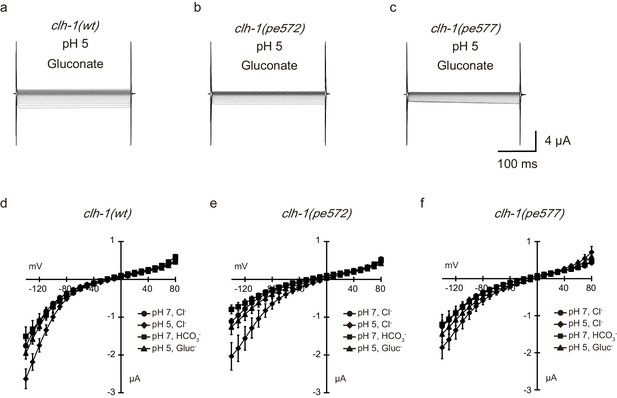
Extracellular pH-sensitivity and bicarbonate permeability of CLH-1.
(a–i) Representative current traces from Xenopus oocytes that expressed cRNA for clh-1(wt) (a–c), clh-1(pe572) (d–f), or clh-1(pe577) (g–i). Oocytes were perfused at pH 7 with Cl- as the dominant extracellular anion (a, c, d), pH 5 with Cl- (b, e, h), or pH 7 with HCO3- (c, f, i). See Materials and methods for details. The oocytes were clamped at voltages ranging from −140 mV to 80 mV. (j–l) The averaged current-voltage relationships of clh-1(wt) (j, n ≧ 16), clh-1(pe572) (k, n ≧ 16), clh-1(pe577) (l, n = 8), at pH 7 with Cl- (circle), at pH 5 with Cl- (diamond), and at pH 7 with HCO3- (square). The error bars represent s.e.m. Trials of the different conditions were performed using the same oocyte.
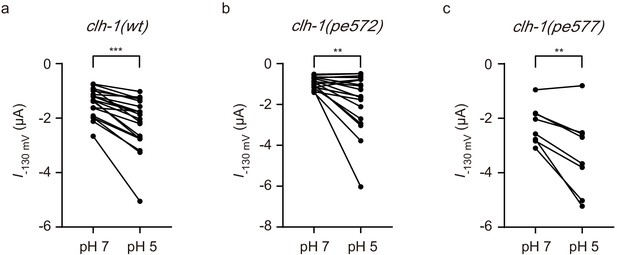
Mutant CLH-1 channels retained pH sensitivity and bicarbonate permeability.
(a–c) Comparison of current amplitudes at −130 mV between pH 7 and pH 5. clh-1(wt) (a, n = 20), clh-1(pe572) (b, n = 18), and clh-1(pe577) (c, n = 8). Two-tailed paired t-test, ***p<0.001, **p<0.01.
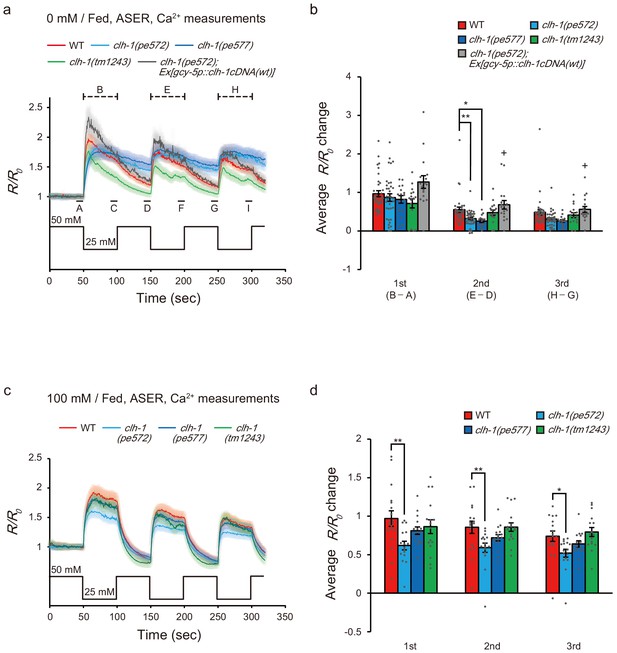
Calcium dynamics of ASER in response to repeated salt stimuli is altered in clh-1 mutants.
(a and c) Calcium responses of ASER stimulated by NaCl concentration changes between 50 mM and 25 mM after cultivation at 0 mM NaCl (a) or 100 mM NaCl (c) in the presence of food for 6 hr. A to I indicate the time points for calculation of R/R0 changes. A, D, and G are the time points for pre-stimulus R/R0, B, E, and H are the time points for peak R/R0 during stimulation, C, F, and I are the time points for decayed R/R0 during stimulation. The shaded region represents s.e.m., n ≧ 16 animals. (b and d) R/R0 changes at each NaCl down-step stimulus (B - A, E - D, and H - G for the 1st, 2nd, and 3rd stimulus, respectively). 0 mM NaCl cultivated (b) or 100 mM cultivated (d). See Materials and methods for details. Bars and the error bars represent mean +/- s.e.m., dots represent individual trials. n ≧ 16 animals, Tukey’s test, **p<0.01, *p<0.05. +p<0.05, compared with clh-1(pe572) mutant.
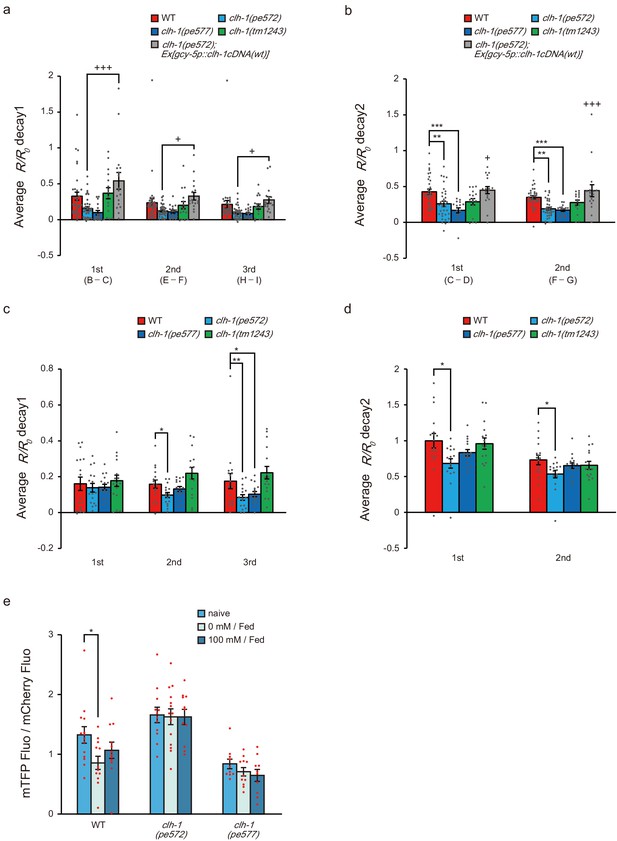
clh-1 missense mutations affect calcium dynamics of ASER.
(a and b) Decay of ASER calcium response during 25 mM NaCl (a) and 50 mM NaCl (b) derived from Figure 5a. Differences of R/R0 between the time points illustrated in Figure 5a (B to I) were calculated along with time as follows. Calculation of ‘decay1’ during 25 mM NaCl (a) was as follows; B - C for 1st, E - F for 2nd, H - I for 3rd. Calculation of ‘decay2’ during 50 mM NaCl (b) was as follows; C - D for 1st, F – G for 2nd. See Materials and methods for details. Bars and the error bars represent mean +/- s.e.m., dots represent individual trials. n ≧ 16 animals, Tukey’s test, ***p<0.001, **p<0.01, compared with wild type. +++p<0.001, +p<0.05, compared with clh-1(pe572) mutant. (c and d) Decays derived from Figure 5c as calculated above. Bars and the error bars represent mean +/- s.e.m., dots represent individual trials. n ≧ 16 animals, Dunnett’s test, **p<0.01, *p<0.05. (e) Expression level of the clh-1 promoter. Fluorescence intensity of the clh-1 promoter-driven mTFP relative to that of gcy-5 promoter-driven mCherry in ASER. Naive animals were directly measured after cultivation for 4 days at 50 mM NaCl in the presence of food. Otherwise, animals were further cultivated for 6 hr at either 0 mM or 100 mM NaCl in the presence of food. Bars and the error bars represent mean +/- s.e.m., dots represent individual trials. n ≧ 10 animals, Tukey’s test, *p<0.05.
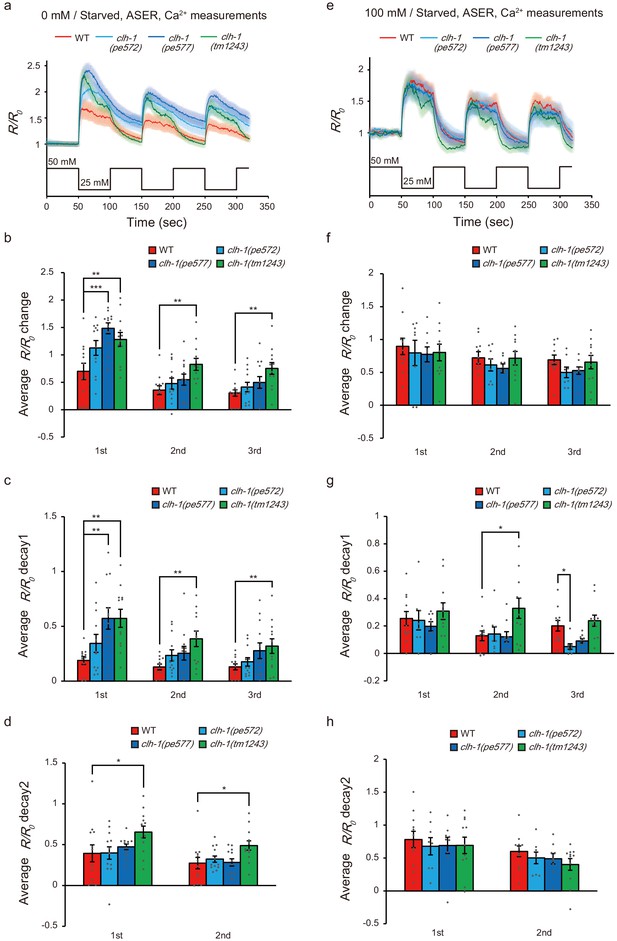
Effect of clh-1(pe) mutations on calcium dynamics of ASER in starved animals.
(a and e) Calcium responses of ASER stimulated by NaCl concentration changes between 50 mM and 25 mM after cultivation at 0 mM NaCl (a) or 100 mM NaCl (e) for 6 hr in the absence of food. The shaded region represents s.e.m., n ≧ 8 animals. (b–d and f–h) R/R0 changes at each NaCl down-step stimulus (b, f), decay of ASER calcium response during 25 mM NaCl (c, g) or 50 mM NaCl (d, h). 0 mM NaCl-cultivated (b–d) or 100 mM NaCl-cultivated (f–h). Calculation of R/R0 changes and decays was performed as described in Figure 5a and Figure 5—figure supplement 1. See Materials and methods for details. Bars and the error bars represent mean +/- s.e.m., dots represent individual trials. n ≧ 8 animals, Dunnett’s test, ***p<0.001, **p<0.01, *p<0.05.
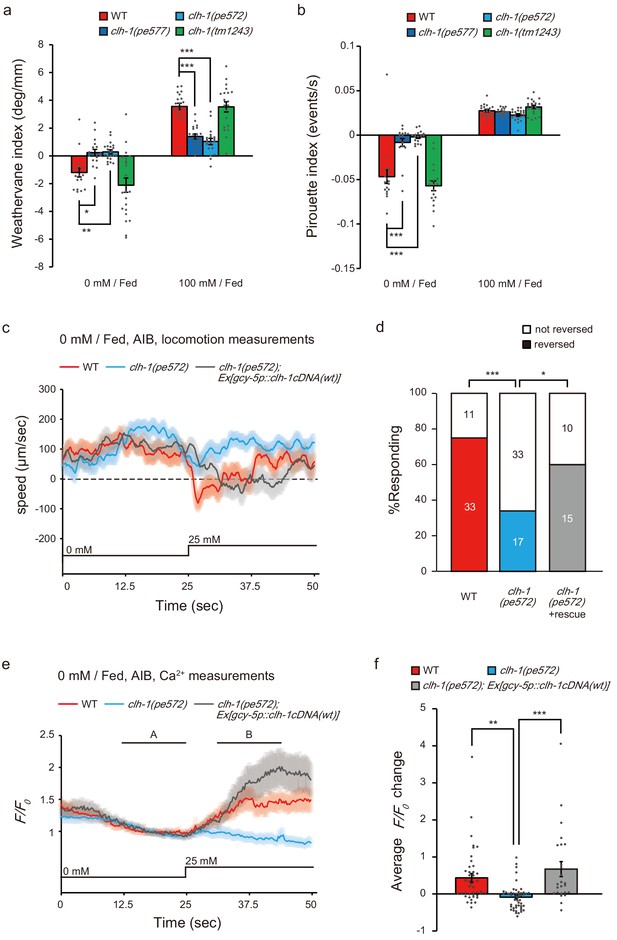
Missense mutations in clh-1 attenuate both klinotaxis and klinokinesis, as well as AIB response and reversal in response to salt increase.
(a and b) clh-1(pe) mutants show defects in migration bias in salt chemotaxis. Bias of klinotaxis (a) and klinokinesis (b), represented by weathervane index and pirouette index, respectively. In both mechanisms, positive and negative values indicate migration bias toward higher and lower salt concentrations, respectively. Bars and the error bars represent mean +/- s.e.m., dots represent individual trials. n ≧ 18 assays, Dunnett’s test, ***p<0.001, **p<0.01, *p<0.05. (c and e) Locomotion speed of animals (c) and calcium responses of AIB (e) after cultivation at 0 mM NaCl in the presence of food. In panel (e), A and B indicate the time points for calculation of F/F0 changes. NaCl concentration change from 0 mM to 25 mM at 25 s. The shaded region represents s.e.m., n ≧ 25 animals. (d) Proportion of animals that showed reversal after salt stimulus. Reversal was defined as follows; backward locomotion, whose velocity less than −100 µm/sec was continued for more than 1 s (35 frames). The error bars represent s.e.m., n ≧ 25 animals, Fisher’s exact test. ***p<0.001, *p<0.05. (f) F/F0 change upon salt stimulus (B - A, see Materials and methods for details). Bars and the error bars represent mean +/- s.e.m., dots represent individual trials. n ≧ 25 animals, Tukey’s test, ***p<0.001, **p<0.01.
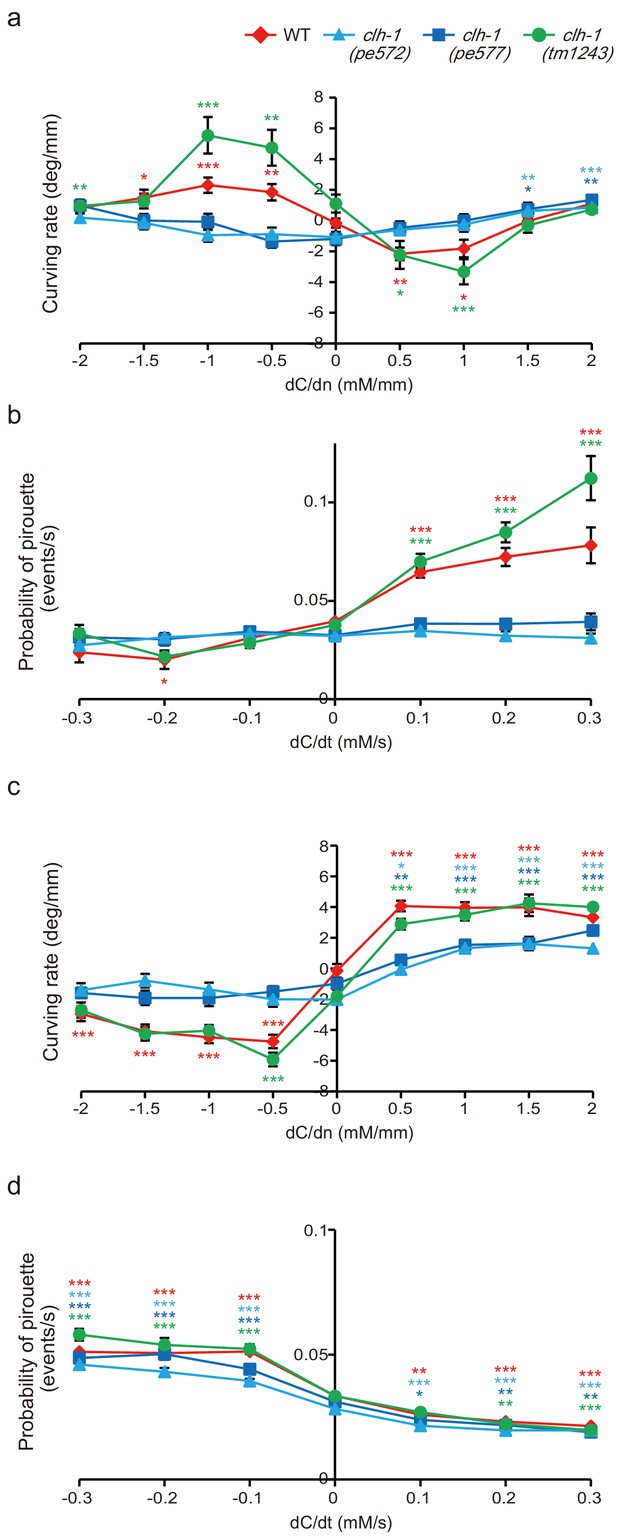
Quantification of the navigation behaviors.
(a and b) Chemotaxis after cultivation at 0 mM NaCl in the presence of food. Klinotaxis bias represented by curving rate (a) and klinokinesis bias represented by probability of pirouette (b). dC/dn represents spatial gradient of NaCl concentration perpendicular to the direction of centroid movement of the animal. Note that migration biases in both klinotaxis and klinokinesis toward low salt are weak in the clh-1(pe) mutants. The error bars represent s.e.m., n ≧ 18 animals, compared with the basal weathervane index (dC/dn = 0) or the basal pirouette index (dC/dt = 0), Dunnett’s test, ***p<0.001, **p<0.01, *p<0.05. Asterisks are colored according to the strain (red for wild type, light blue for clh-1(pe572), deep blue for clh-1(pe577), green for clh-1(tm1243)). (c and d) Chemotaxis after cultivation at 100 mM NaCl in the presence of food. Migration bias of klinotaxis (c) and klinokinesis (d). Klinotaxis, but not klinokinesis toward high salt is weak in the clh-1(pe) mutants. The error bars represent s.e.m., n ≧ 19 animals, compared with the basal weathervane index (dC/dn = 0) or the basal pirouette index (dC/dt = 0), Dunnett’s test, ***p<0.001, **p<0.01, *p<0.05. Asterisks are colored according to the strain.
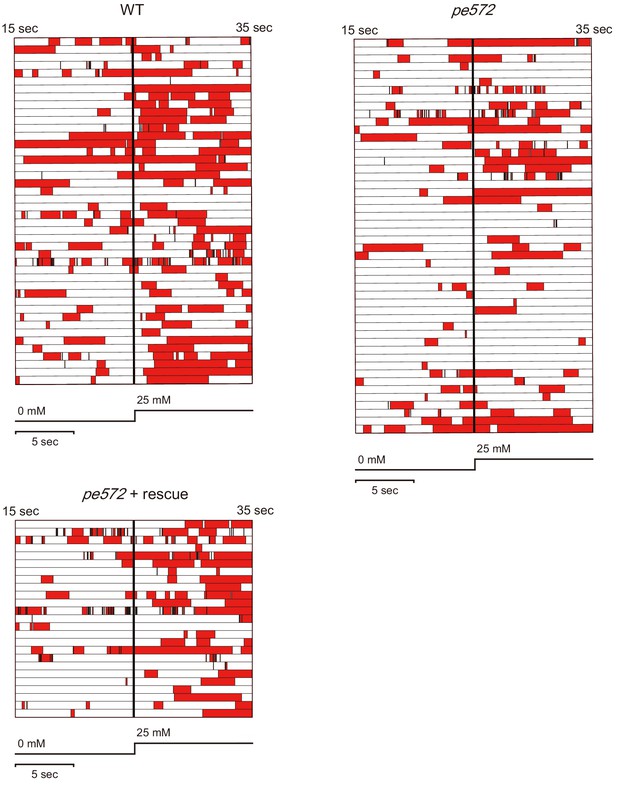
Quantification of reversal behaviors in microfluidics arena.
Locomotion of wild type, clh-1(pe572), and clh-1(pe572) that express clh-1(wt) cDNA in ASER (pe572 + rescue). Locomotion of animals was categorized as either forward (white) or reversal (red). Twenty second recordings, consisting of each 10 s of before and after salt up-step stimulus (bold bar), were aligned. Each row represents each individual. n ≧ 25 animals.
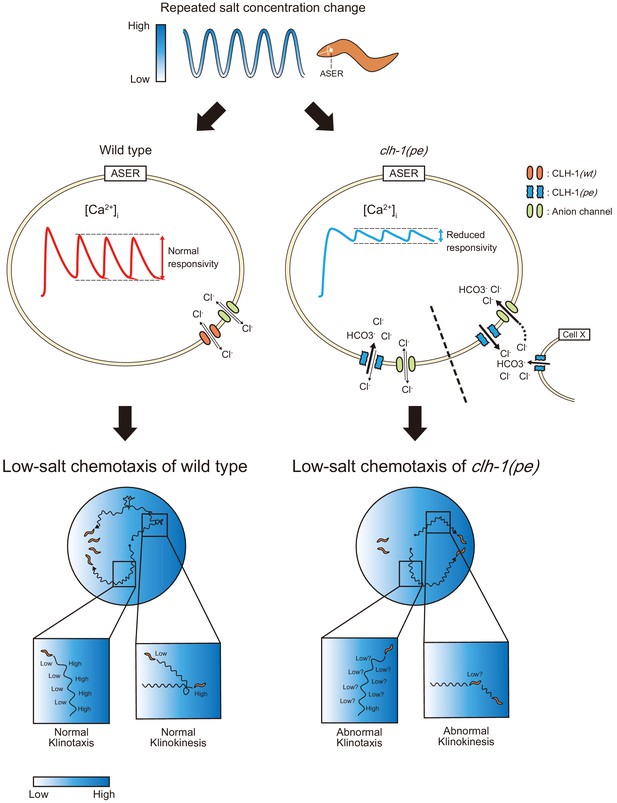
A model for the mechanism of impaired low salt chemotaxis of clh-1(pe) mutants.
Animals experience salt concentration fluctuation along with migration on salt gradient, which is depicted by blue waves (top). Salt information is sensed by the ASER neuron and translated into intracellular calcium transients (middle). In the clh-1(pe) mutants, calcium responses of ASER to repeated salt stimuli is diminished after cultivation at low-salt concentration. This is probably due to increased influx of chloride ions into the cell upon depolarization. Permeation of bicarbonate ions through CLH-1 may also be involved. Although the mechanism for increased chloride influx is currently unknown, we suggest two possible scenarios; increased anion influx through CLH-1(pe) channels (left of the dashed line) and elevated extracellular chloride level (right of the dashed line). Reduced responsivity of ASER then results in hampered migration bias toward low salt both in klinokinesis and klinotaxis (bottom).
Tables
The reversal potential of wild-type and mutant CLH-1 channels for Cl- and HCO3- at pH7.
Number of samples in parentheses, unit: mV.
| Cl- | HCO3- | |
|---|---|---|
| CLH-1(wt) | -20.89 ± 5.03 (20) | -26.07 ± 14.08 (19) |
| CLH-1(pe572) | -19.30 ± 4.44 (18) | -29.50 ± 16.36 (18) |
| CLH-1(pe577) | -14.20 ± 11.46 (8) | -19.69 ± 9.92 (8) |
Additional files
-
Supplementary file 1
Strain list.
- https://cdn.elifesciences.org/articles/55701/elife-55701-supp1-v1.docx
-
Supplementary file 2
Genotyping primer.
- https://cdn.elifesciences.org/articles/55701/elife-55701-supp2-v1.docx
-
Supplementary file 3
Plasmid list and use.
- https://cdn.elifesciences.org/articles/55701/elife-55701-supp3-v1.docx
-
Supplementary file 4
Plasmid sequences.
- https://cdn.elifesciences.org/articles/55701/elife-55701-supp4-v1.zip
-
Supplementary file 5
Statistics.
- https://cdn.elifesciences.org/articles/55701/elife-55701-supp5-v1.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55701/elife-55701-transrepform-v1.pdf