Enhancement of homology-directed repair with chromatin donor templates in cells
Figures
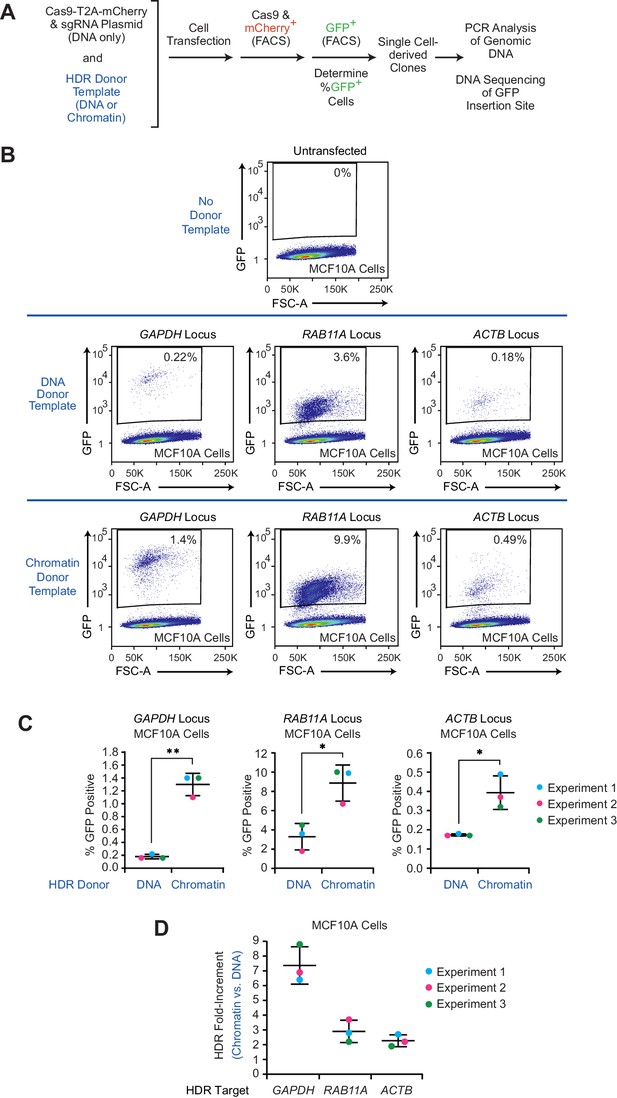
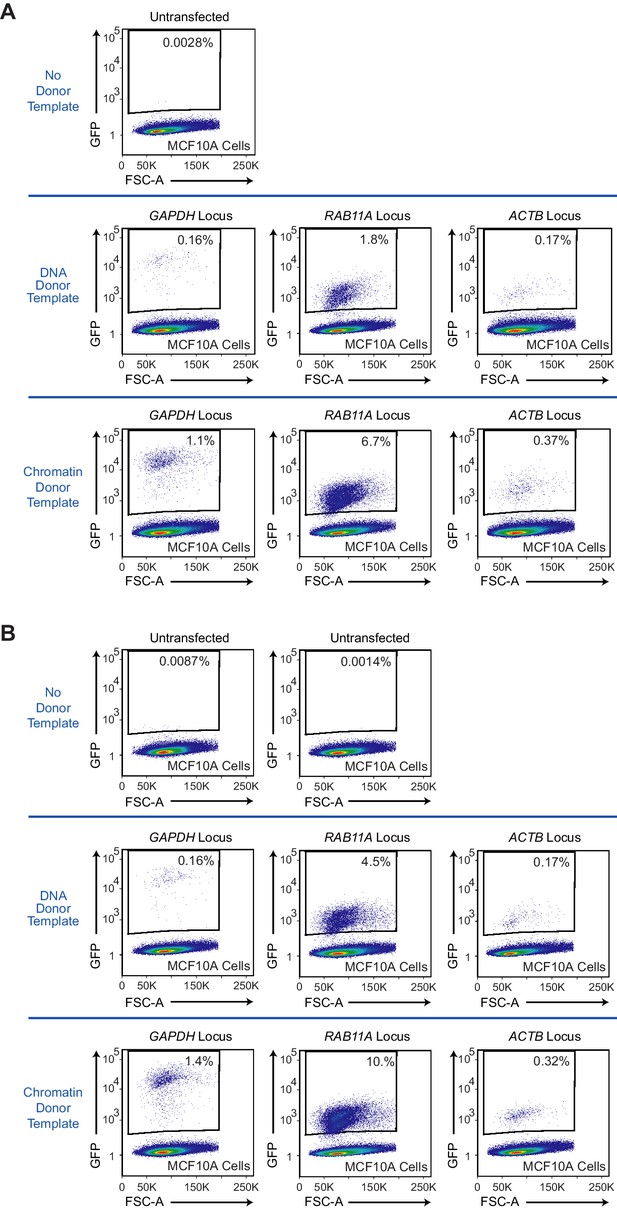
The efficiency of HDR-mediated gene editing with CRISPR-Cas9 is higher with chromatin donor templates than with DNA donor templates.
(A) Schematic outline of the workflow in the CRISPR-Cas9-mediated editing experiments with DNA or chromatin donor templates. The HDR-mediated insertion of the GFP sequence was directed to different loci as follows. Plasmid DNA containing the coding sequence for Cas9-T2A-mCherry and a target-specific sgRNA sequence was co-transfected into different human cell lines with the corresponding HDR donor template as either DNA or chromatin. At 24 hr post-transfection, mCherry-positive cells were enriched by FACS and cultured for an additional 10 days. The expression of GFP was then analyzed by flow cytometry, and individual GFP-positive cells were sorted by FACS to generate independent clones. To determine whether there was partial or complete conversion of the multiple chromosomes containing the target genes, genomic DNA samples from each of several independent GFP-positive clones were analyzed by PCR. In addition, the precise integration of the GFP sequence at the target sites in representative edited clones was confirmed by DNA sequencing. These experiments were performed under standard CRISPR-Cas9 genome-editing conditions, as in Ran et al., 2013. (B) Flow cytometry analysis reveals an increase in GFP-positive cells with chromatin relative to DNA donor templates. HDR experiments were performed, as outlined in A with MCF10A cells and GAPDH, RAB11A, or ACTB donor templates. The population of GFP-positive cells was gated based on control cells that show no GFP expression (no donor template; upper panel; see also Figure 1—figure supplement 3). Representative data from one out of three independent experiments are shown. The results of the other two biological replicates are in Figure 1—figure supplement 4. The percentage of GFP-positive cells is indicated in each plot. FSC-A: forward scatter area. (C) Individual results from three independent experiments with each of the target loci. The data points from each independent experiment are designated with the same colored dots. The mean and standard deviation are indicated for each set of experiments. The p-values were determined by using Welch's t test. **, p <0.01; *, p <0.05. The calculated p-values are as follows: p = 0.0062 for the GAPDH data set; p = 0.017 for the RAB11A data set; p = 0.048 for the ACTB data set. (D) The use of chromatin relative to naked DNA donor templates results in a 2.3- to 7.4-fold enhancement of GFP-positive cells. The data for each of three independent HDR experiments with each locus are shown. The bars represent mean and standard deviation for each locus.
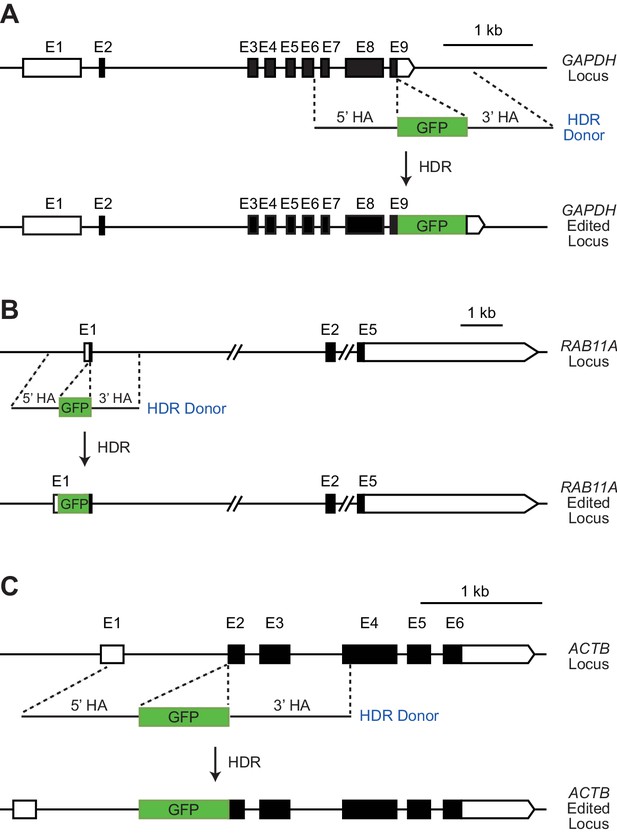
Schematic representations of the CRISPR-Cas9 target regions for HDR-mediated insertion of a GFP reporter sequence.
(A) GAPDH locus. A DNA sequence that encodes the T2A self-cleaving peptide fused to the GFP protein (T2A-GFP, indicated in the figure as ‘GFP’) is inserted in exon 9 (E9) of the GAPDH locus. This results in the production of a GAPDH-T2A-GFP polypeptide that is spontaneously cleaved into separate GAPDH and GFP proteins. (B) RAB11A locus. The GFP sequence is inserted in the first exon (E1) of the RAB11A locus. This in-frame HDR-mediated insertion yields a GFP-RAB11A fusion protein. (C) ACTB locus. The monomeric enhanced GFP sequence (mEGFP; indicated as ‘GFP’) is inserted into the second exon (E2) of the ACTB locus. This in-frame HDR-mediated insertion results in a mEGFP-ACTB fusion protein. All three donor repair templates contain the desired insert sequence flanked by two homology arms of about 1 kb each. The dashed lines indicate the regions of homology between the HDR donor templates and the CRISPR-Cas9 targeted loci. The black boxes represent coding regions, and white boxes represent untranslated regions. E, exon; HA, homology arm.
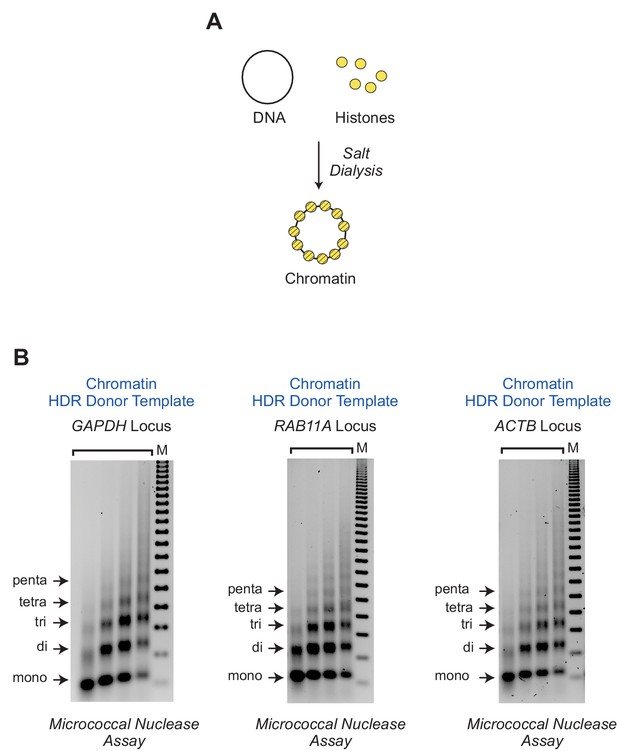
Reconstitution of plasmid DNA donor templates into chromatin.
(A) Salt dialysis reconstitution of chromatin. The HDR donor template plasmids were reconstituted into chromatin with purified core histones by the salt dialysis method. (B) Micrococcal nuclease digestion analysis of chromatin reconstituted with purified components. Preparations of chromatin that were reconstituted with each of the HDR donor template plasmids (which correspond to the GAPDH, RAB11A, and ACTB loci) were subjected to partial digestion with four different concentrations of micrococcal nuclease. The samples were deproteinized, and the resulting DNA fragments were resolved by agarose gel electrophoresis and visualized by staining with ethidium bromide. The arrows indicate the DNA bands that correspond to mono-, di-, tri-, tetra-, and pentanucleosomes. The DNA size markers (M) are the 123 bp ladder (Millipore Sigma).
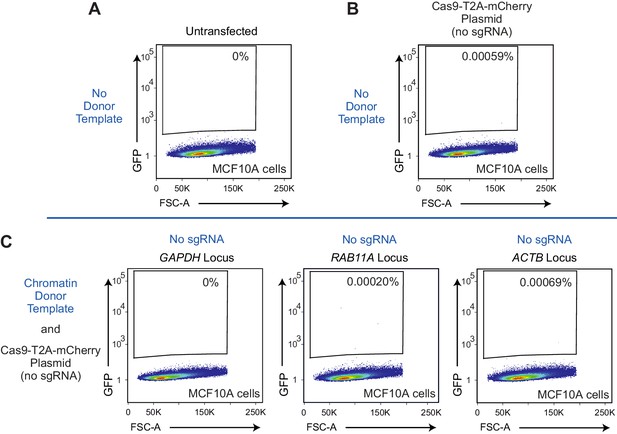
Flow cytometry analysis of MCF10A cells in control experimental conditions.
(A) Untransfected cells. (B) Cells were transfected with a Cas9-T2A-mCherry plasmid (lacking an sgRNA) in the absence of a donor template. (C) Cells were transfected with a Cas9-T2A-mCherry plasmid (lacking an sgRNA) in the presence of the indicated chromatin donor templates. GFP positive cells in B and C, were gated based on control cells that do not contain the GFP sequence (untransfected cells). The percentage of GFP-positive cells is indicated in each plot. Representative data from one out of three experiment is shown. FSC-A: forward scatter area.
Flow cytometry analyses of biological replicates of HDR-mediated gene integration experiments in MCF10A cells.
(A) Data from HDR experiment two with GAPDH, RAB11A, or ACTB donor templates. (B) Data from HDR experiment three with GAPDH, RAB11A, or ACTB donor templates. HDR experiments were performed as outlined in Figure 1A. GFP-positive cells were gated based on control cells that show no GFP expression (no donor template condition).
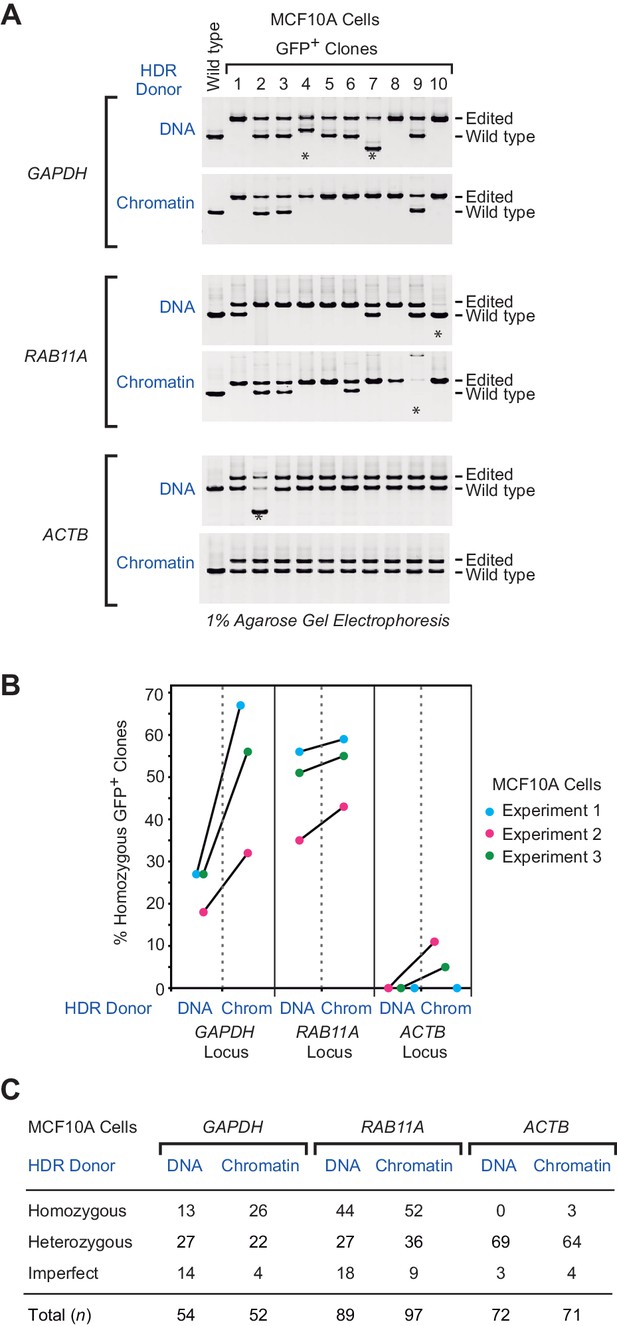
The use of chromatin donor templates increases the efficiency of HDR-mediated homozygous gene editing relative to that seen with DNA donor templates.
(A) PCR analysis of gDNA from MCF10A GFP-positive clones. Three independent HDR experiments were performed as shown in Figure 1A, and the gDNA from individual GFP-positive clones was analyzed by PCR. The positions of the PCR amplification products from edited and wild-type alleles are indicated. The PCR products derived from control wild-type cells are also included (left lane of each panel). The asterisks indicate imperfect clones that appear to contain at least one improperly edited chromosome, as indicated by either the absence of an edited chromosome or the presence of a PCR product whose size is not consistent with that of an edited or wild-type chromosome. The positions of the primer pairs (F1, R1) in the PCR analysis of each locus are shown in Figure 2—figure supplement 1. The results from a representative subset of the GFP-positive clones are shown. The complete set of PCR results are in Figure 2—figure supplements 2, 3 and 5. (B) The percentages of GFP-positive homozygous clones in three independent HDR experiments at each of the target loci. The results from each independent experiment (with DNA versus chromatin donor templates) are denoted with a connector line. The p-values were determined by using Welch's t-test. The calculated p-values are as follows: p = 0.062, p = 0.56, and p = 0.17 for the GAPDH, RAB11A and ACTB data sets, respectively. (C) Summary of the PCR analysis. MCF10A cells are diploid, and each clone was classified as homozygous (with two precisely edited chromosomes), heterozygous (with one precisely edited chromosome and one wild-type chromosome), or imperfect, as defined in A.
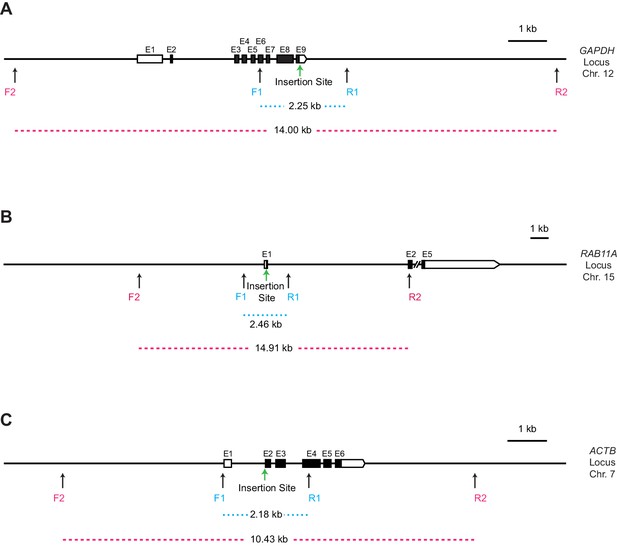
Diagrams of the positions of the primer sets for the PCR analysis of GFP-positive clones at the GAPDH, RAB11A, and ACTB loci.
(A) GAPDH locus. (B) RAB11A locus. (C) ACTB locus. The expected PCR product sizes with wild-type gDNA (dashed lines), the positions of the primers (F1, R1, F2, R2; black arrows), and the DNA insertion sites (green arrows) at each locus are indicated. Two primer pairs are shown for each locus: F1, forward primer 1; R1, reverse primer 1; F2, forward primer 2; R2, reverse primer 2. E, Exon. The HDR-mediated insertions increase the lengths of the PCR products by 771 bp, 732 bp, and 730 bp at the GAPDH, RAB11A, and ACTB loci, respectively.
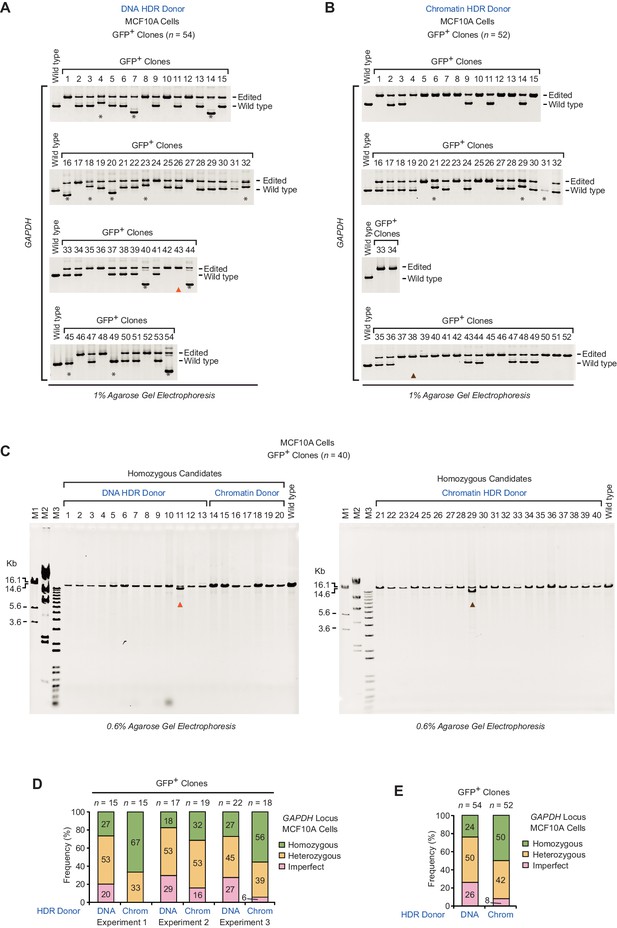
PCR analysis of gDNA from GFP-positive clones at the GAPDH locus in MCF10A cells.
(A) Clones (n = 54) collected from three independent HDR experiments with a DNA donor template. Lanes 1 to 15, 16 to 32, and 33 to 54 correspond to experiment 1, experiment 2, and experiment 3, respectively. (B) Clones (n = 52) collected from three independent HDR experiments with a chromatin donor template. Lanes 1 to 15, 16 to 34, and 35 to 52 correspond to experiment 1, experiment 2, and experiment 3, respectively. In panels A and B, the positions of the PCR amplification products from edited and wild-type alleles are indicated. Asterisks denote imperfect clones. Clones were classified as defined in the figure legend of Figure 2 of the main text. The triangles indicate imperfect clones (as assessed with long-range PCR analysis; see panel C, below) with an apparently homozygous genotype in the standard PCR analysis, as in panels A and B. (C) Long-range PCR analysis of homozygous candidate clones (n = 40). Clones collected from three independent HDR experiments with either a DNA donor template (lanes 1 to 13) or a chromatin donor template (lanes 14 to 40) were analyzed. These clones were preliminarily classified as homozygous based on the PCR analysis shown in A and B. Clones that have a deletion within a 14.0 kb region surrounding the target insertion site, as indicated by the presence of an additional PCR product that is smaller than that of the properly edited allele, are denoted with triangles. The PCR product (14.0 kb) from gDNA of wild-type cells is also shown. The positions of the primer pairs (F2, R2) for the PCR analyses (panels A–C) are depicted in Figure 2—figure supplement 1A. DNA size markers: M1 (1 kb Plus DNA Ladder, Invitrogen); M2 (λ DNA-HindIII Digest, NEB); M3 (bacteriophage T7 DNA digested with HindIII). (D) Frequency of occurrence of homozygous, heterozygous, and imperfect clones in three independent HDR experiments. n, number of clones analyzed. (E) Summary of the combined results at the GAPDH locus in MCF10A cells. The percentages were calculated based on the data for the GAPDH locus in Figure 2C.
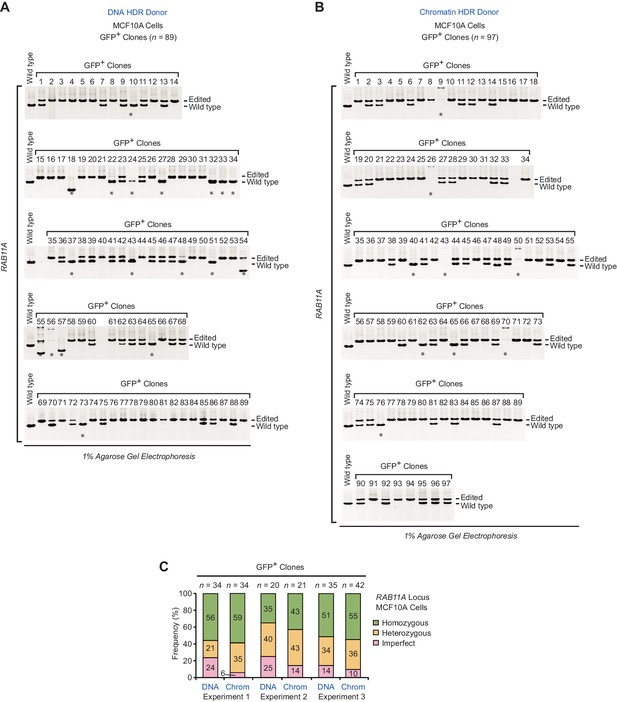
PCR analysis of gDNA from GFP-positive clones at the RAB11A locus in MCF10A cells.
(A) Clones (n = 89) collected from three independent HDR experiments with a DNA donor template. Lanes 1 to 34, 35 to 54, and 55 to 89 correspond to experiment 1, experiment 2, and experiment 3, respectively. (B) Clones (n = 97) collected from three independent HDR experiments with a chromatin donor template. Lanes 1 to 34, 35 to 55, and 56 to 97 correspond to experiment 1, experiment 2, and experiment 3, respectively. In A and B, the positions of the PCR amplification products from edited and wild-type alleles are indicated. Asterisks indicate imperfect clones, as defined in the figure legend of Figure 2. (C) Frequency of occurrence of homozygous, heterozygous, and imperfect clones in each of three independent HDR experiments. n, number of clones analyzed.
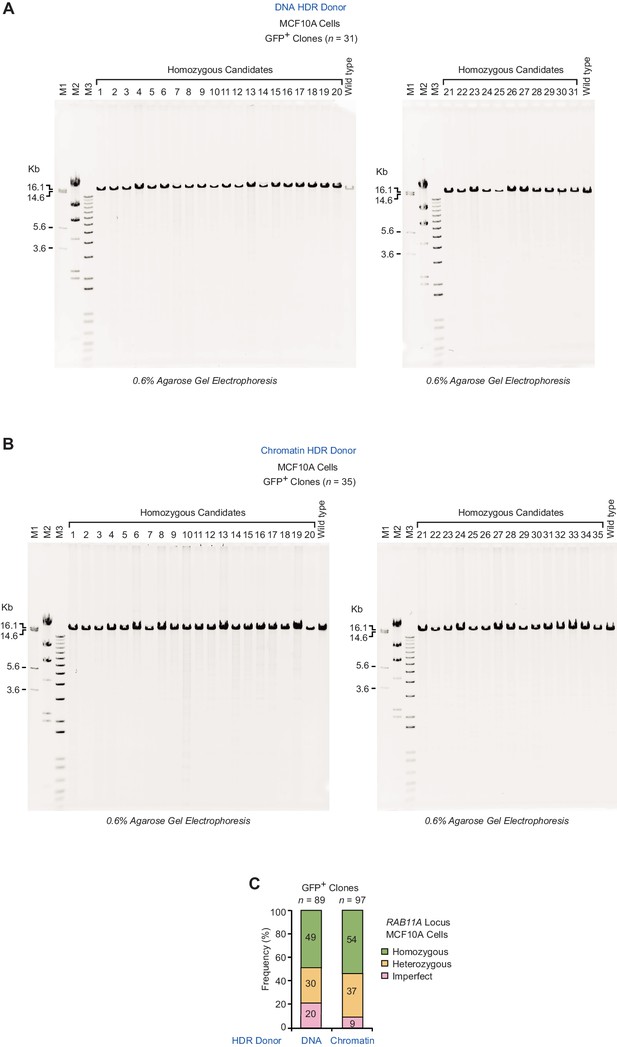
Long-range PCR analysis of gDNA from GFP-positive clones at the RAB11A locus in MCF10A cells.
(A) Analysis of homozygous candidates (n = 31) collected from three independent HDR experiments with a DNA donor template. (B) Analysis of homozygous candidates (n = 35) collected from three independent HDR experiments with a chromatin donor template. In panels A and B, the PCR product (14.91 kb) from gDNA of wild-type cells is also shown. The positions of the primers (F2, R2) in the PCR analysis are depicted in Figure 2—figure supplement 1B. DNA size markers: M1 (1 kb Plus DNA Ladder, Invitrogen); M2 (λ DNA-HindIII Digest, NEB); M3 (bacteriophage T7 DNA digested with HindIII). (C) Summary of the combined results at the RAB11A locus in MCF10A cells. The percentages were calculated based on the data for the RAB11A locus in Figure 2C.
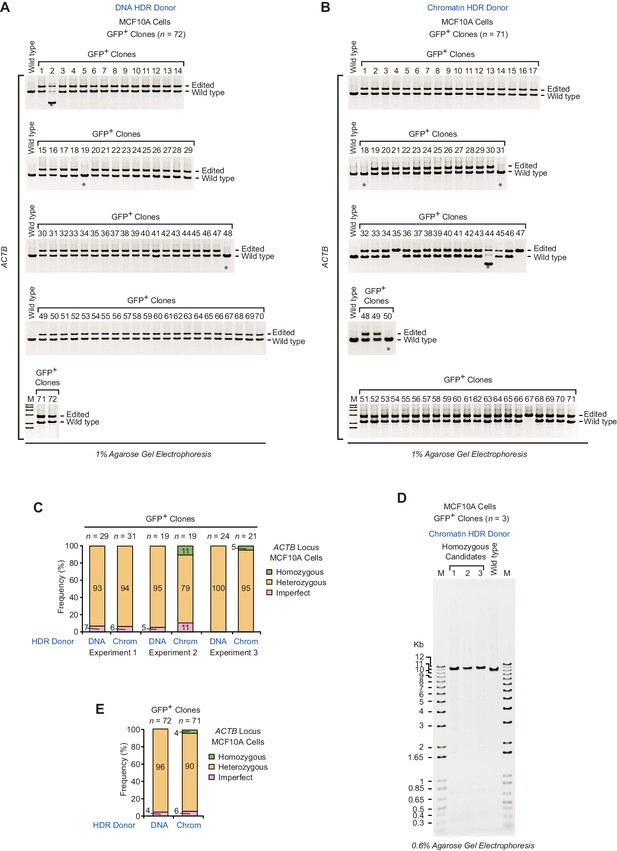
PCR analysis of gDNA from GFP-positive clones at the ACTB locus in MCF10A cells.
(A) Clones (n = 72) collected from three independent HDR experiments with a DNA donor template. Lanes 1 to 29, 30 to 48, and 49 to 72 correspond to experiment 1, experiment 2, and experiment 3, respectively. (B) Clones (n = 71) collected from three independent HDR experiments with a chromatin donor template. Lanes 1 to 31, 32 to 50, and 51 to 71 correspond to experiment 1, experiment 2, and experiment 3, respectively. In A and B, the positions of the PCR amplification products from edited and wild-type alleles are indicated. M, DNA size markers (1.65, 2, 3, 4, 5, 6 kb; 1 kb Plus DNA Ladder, Invitrogen). Asterisks denote imperfect clones as defined in Figure 2. (C) Frequency of occurrence of homozygous, heterozygous, and imperfect clones in three independent HDR experiments. n, number of clones analyzed. (D) Long-range PCR analysis of homozygous candidates collected from HDR experiments with a chromatin donor template. The PCR product (10.43 kb) from gDNA of wild-type cells is also shown. The positions of the primers (F2, R2) in the PCR analysis are depicted in Figure 2—figure supplement 1C. (E) Summary of the combined results at the ACTB locus in MCF10A cells. The percentages were calculated based on the data for the ACTB locus in Figure 2C.
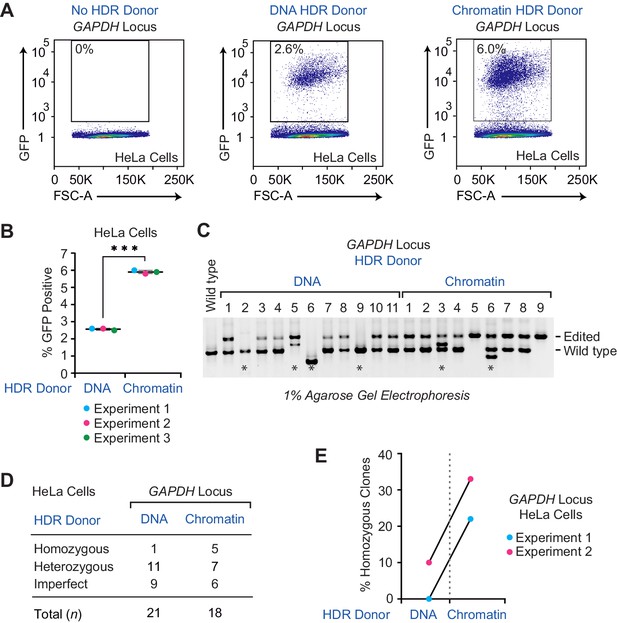
The efficiency of HDR-mediated gene editing with CRISPR-Cas9 is higher with a chromatin donor template than with a DNA donor template in HeLa cells.
(A) The use of a chromatin donor template relative to a naked DNA donor template results in an increase of GFP-positive cells. HDR experiments were performed as depicted in Figure 1A with HeLa cells and the GAPDH locus donor template. The population of GFP-positive cells was gated based on control cells that show no GFP expression (no HDR donor; left panel). Representative data from one out of three independent experiments are shown. The results of the other two biological replicates are in Figure 3—figure supplement 1. The percentage of GFP-positive cells is indicated in each plot. FSC-A: forward scatter area. (B) Individual results of flow cytometry analysis from three independent experiments with the GAPDH locus and HeLa cells. The data points from each independent experiment are designated with the same colored dots. The p-value was determined by using Welch's t-test. ***, p <0.0001. The mean and standard deviation are indicated. (C) The use of a chromatin HDR donor template results in an increase in the efficiency of homozygous edited clones relative to that seen with a DNA donor template. PCR analysis of edited genomic DNA was carried out as in Figure 2A. The positions of the PCR amplification products from edited and wild-type chromosomes are shown. The PCR products from control wild-type cells are also included (left lane). The results from a representative subset of the GFP-positive clones are shown. The results from the other GFP-positive clones that were analyzed are in Figure 3—figure supplement 2. (D) Summary of the PCR analysis of clones obtained in the HDR-mediated insertion of GFP sequences at the GAPDH locus in HeLa cells. The homozygous clones have four copies of the integrated GFP sequence, the heterozygous clones have one to three copies of the integrated GFP sequence, and the imperfect clones appear to contain improperly edited chromosomes, as indicated by either the absence of an edited chromosome or the presence of a PCR product whose size is not consistent with that of an edited or wild-type chromosome. (E) The percentages of GFP-positive homozygous clones in two independent HDR experiments. The results from each independent experiment (with DNA versus chromatin donor templates) are denoted with a connector line.
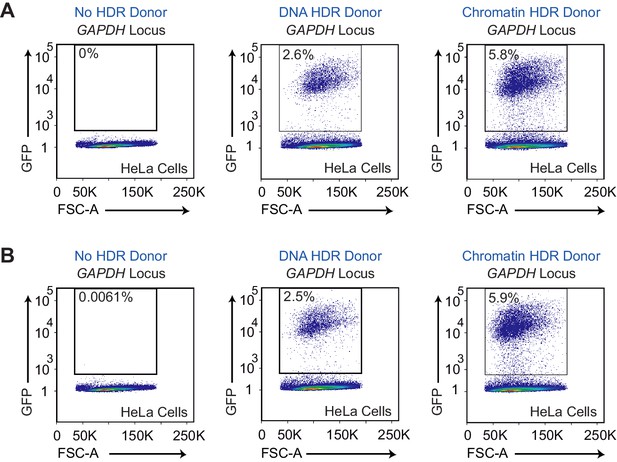
Flow cytometry analyses of biological replicates of HDR-mediated gene integration experiments in HeLa cells.
(A) Data from HDR experiment 2. (B) Data from HDR experiment 3. HDR experiments were performed as outlined in Figure 1A. GFP-positive cells was gated based on cells that show no GFP expression (no HDR donor; left panels).
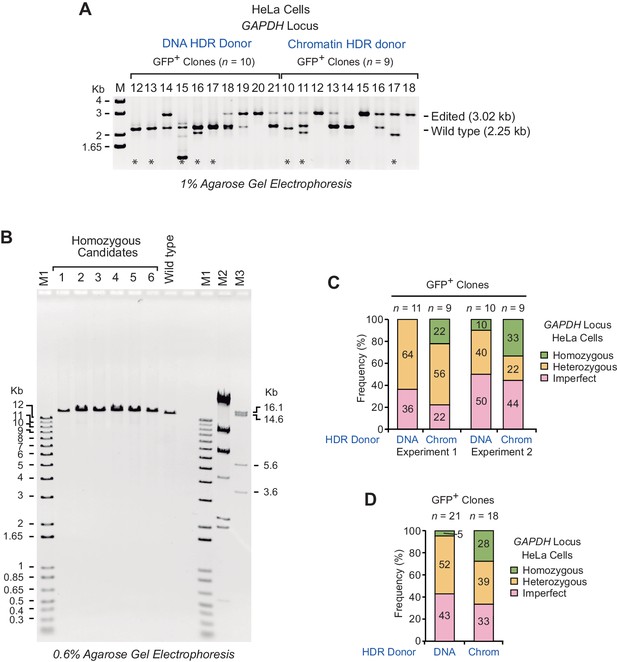
PCR analysis of gDNA from GFP-positive clones in HeLa cells.
(A) Clones collected from HDR experiments with a DNA donor template (clones 12 to 21) or a chromatin donor template (clones 10 to 18). The positions of the PCR products of the wild-type and HDR-edited alleles are indicated. The positions of the primer pairs (F1, R1) are depicted in Figure 2—figure supplement 1A. The asterisks denote imperfect clones, as specified in the figure legend of Figure 2. M, DNA size marker (1 kb DNA ladder, Invitrogen). (B) Long-range PCR analysis of six homozygous clones collected from two independent HDR experiments. The PCR product (14.0 kb) from gDNA of wild-type cells is also shown. The positions of the primer pairs (F2, R2) are depicted in Figure 2—figure supplement 1A. DNA size markers: M1 (1 kb Plus DNA Ladder, Invitrogen); M2 (λ DNA-HindIII Digest, NEB); M3 (bacteriophage T7 DNA digested with HindIII). (C) Frequency of occurrence of homozygous, heterozygous, and imperfect clones in two independent HDR experiments. n, number of clones analyzed. (D) Summary of the combined results at the GAPDH locus in HeLa cells. The percentages were calculated based on the data in Figure 3D. n, number of clones analyzed.
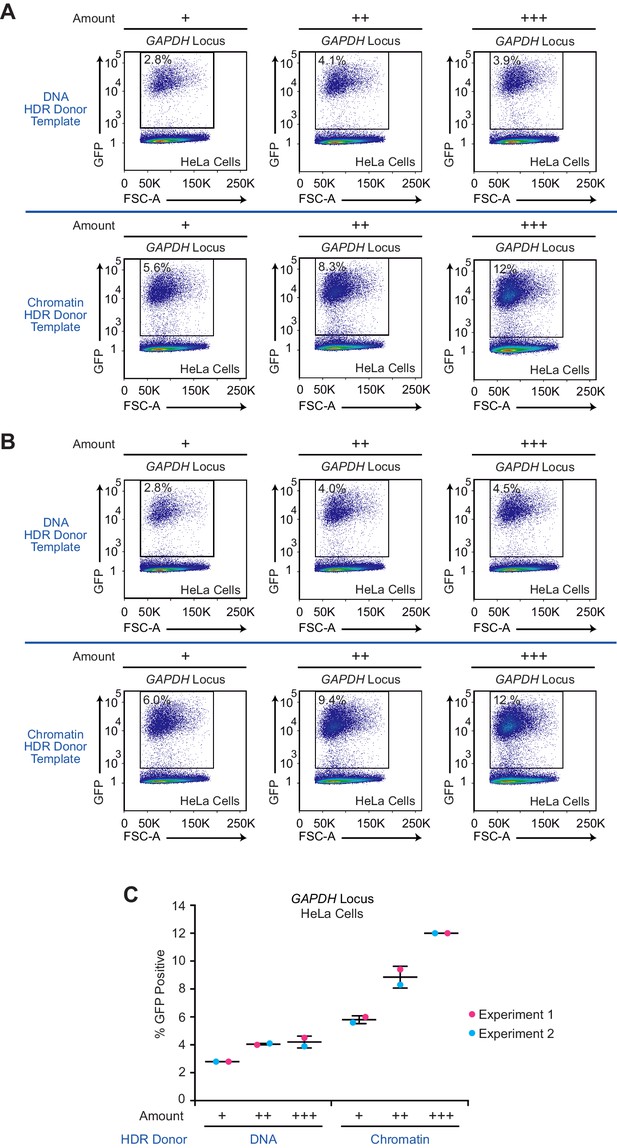
The efficiency of GFP insertion with different amounts of donor template in HeLa cells is higher with chromatin than with DNA.
(A) The results from HDR experiment 1. (B) The results from HDR experiment 2. In A and B, the experiments were performed as depicted in Figure 1A. HeLa cells were co-transfected with the Cas9-T2A-mCherry plasmid containing the sgRNA sequence targeting the GAPDH locus and 0.625 µg (+), 1.25 µg (++), or 1.88 µg (+++) of the corresponding HDR donor template as either DNA or chromatin. As a reference, we used 1.25 µg (++) of donor template as DNA or chromatin in our standard experiments, such as those shown in the main figures. At 24 hr post-transfection, mCherry-positive cells were enriched by FACS and cultured for an additional 10 days. The expression of GFP was then analyzed by flow cytometry. (C) Summary of the results from HDR experiments 1 and 2. The percentages of GFP-positive cells in each experiment are shown. The mean and standard deviation (horizontal bars) are depicted for each experimental condition (n = 2).
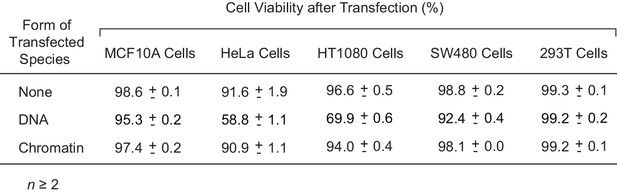
Chromatin templates are of comparable or lower toxicity to cells relative to naked DNA templates.
Cell viability after transfection with a 3 kb plasmid as either naked DNA or chromatin was determined along with the viability of mock-transfected (no DNA or chromatin) cells. The cell viability was assessed by flow cytometry in the presence of DAPI (4',6-diamidino-2-phenylindole). The analysis was performed 48 hr after transfection. The mean and standard deviation from at least two independent experiments with each cell line are shown.





