The peroxisome counteracts oxidative stresses by suppressing catalase import via Pex14 phosphorylation
Figures
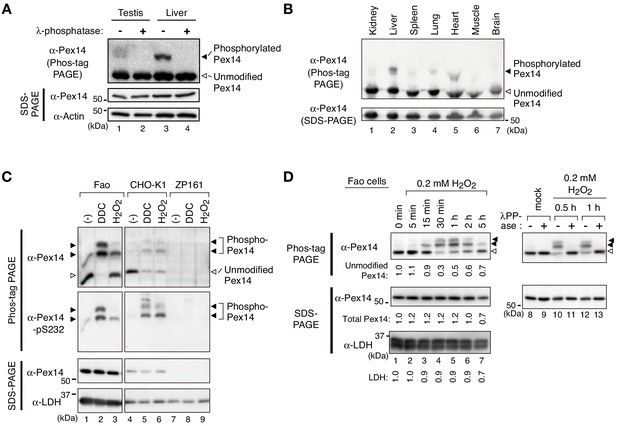
Pex14 is phosphorylated in vivo.
(A) Lysates of testis and liver (20 µg each) from an 8-week-old male mouse were incubated with vehicle (-, lanes 1 and 3) and 400 unit λ-protein phosphatase (+, lanes 2 and 4). Samples were separated by Phos-tag PAGE (top panel) and SDS-PAGE (middle and bottom panels) and analyzed by immunoblotting with antibodies to Pex14 and actin, a loading control. Solid and open arrowheads indicate phosphorylated and unmodified Pex14, respectively. (B) Lysates of various mouse tissues (15 µg each) indicated at the top were analyzed by Phos-tag PAGE (upper panel), SDS-PAGE (lower panel), and immunoblotting with anti-Pex14 antibody. (C) Phosphorylation of Pex14 upon treatment with oxidative agents. Fao, CHO-K1, and a PEX14-deficient (pex14) CHO mutant ZP161 cells (4 × 105 cells each) were treated for 30 min with vehicle (-), 100 μM diethyldithiocarbamate (DDC), and 1 mM hydrogen peroxide (H2O2). Cell lysates were analyzed as in A with antibodies to Pex14, phosphorylated Pex14 at Ser232 (Pex14-pS232), and lactate dehydrogenase (LDH). Open and solid arrowheads indicate unmodified and phosphorylated Pex14, respectively. Note that antibody to phsopho-Pex14 at Ser232 specifically recognized slower-migrating bands of Pex14 in Phos-tag PAGE. (D) Left, Time course of Pex14 phosphorylation upon H2O2 treatment. Fao cells were treated with 0.2 mM H2O2 as in B for indicated time periods. Unmodified Pex14 in Phos-tag PAGE and total Pex14 and LDH in SDS-PAGE were quantified and represented at the bottom of respective bands by taking as 1 those at 0 min. Right, λ-protein phosphatase treatment of phosphorylated Pex14. After the treatment with mock or 0.2 mM H2O2 for 0.5 hr and 1 hr, Fao cells were incubated with vehicle (-) and λ-phosphatase (+) as in A. The cell lysates were analyzed by Phos-tag PAGE (upper panels), SDS-PAGE (lower panels), and immunoblotting with anti-Pex14 antibody.
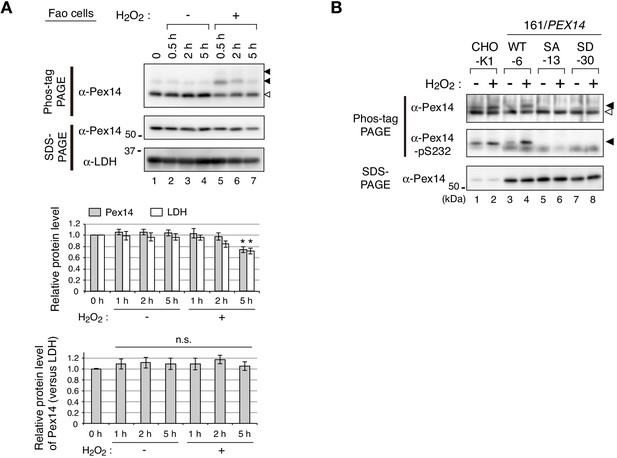
Little effect of H2O2 treatment on Pex14 level and evaluation of Pex14 variants with anti-Pex14-pS232 antibody.
(A) Fao cells (4 × 106 cells each) treated with vehicle (-) or 0.2 mM H2O2 (+) for indicated time periods were solubilized in an equal volume of the buffer and analyzed by SDS-PAGE and Phos-tag PAGE as in Figure 1D. Upper panels, Representative images of immunoblotting were shown. Solid and open arrowheads indicate phosphorylated and unmodified Pex14, respectively. Middle and lower graphs, Data of upper panels was quantified and the protein levels of Pex14 (solid bars) and LDH (open bars; middle graph) or relative protein level of Pex14 to LDH (lower graph) were represented by taking as 1 those at 0 min. Data represents means ± SEM of four independent experiments. Statistical analysis was performed by one-way ANOVA with Dunnett’s post hoc test as compared with 0 min. *p<0.05; n.s., not significant. (B) CHO-K1 and pex14 ZP161 stable cell lines each expressing wild-type His-Pex14 (WT-6), Pex14-S232A (SA-13), and Pex14-S232D (SD-30) were treated for 30 min with vehicle (-) and 0.8 mM H2O2. Cell lysates were analyzed by SDS-PAGE (upper panel), Phos-tag PAGE (lower panels), and immunoblotting with indicated antibodies. Open and solid arrowheads are as in A.
-
Figure 1—figure supplement 1—source data 1
Data for graphs depicted in Figure supplement 1A.
- https://cdn.elifesciences.org/articles/55896/elife-55896-fig1-figsupp1-data1-v2.xlsx
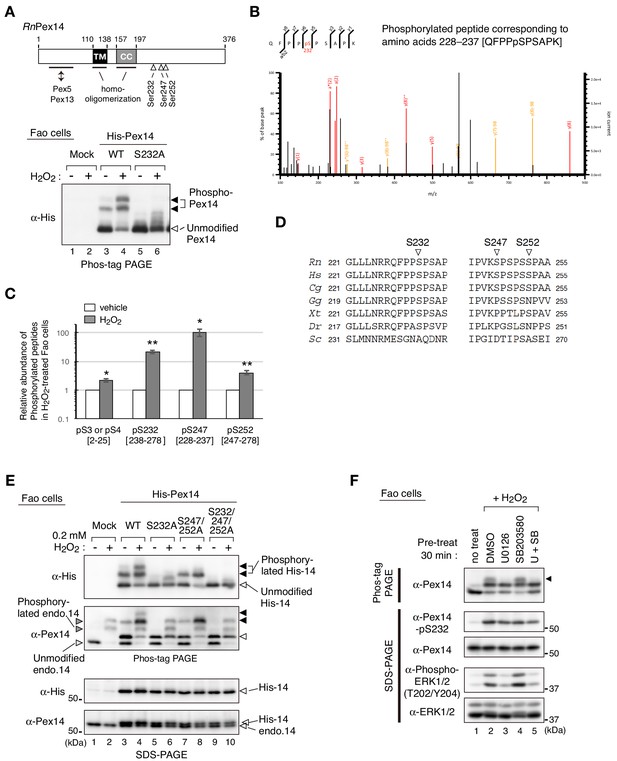
Hydrogen peroxide induces phosphorylation of Pex14 at three distinct serine residues in Fao cells.
(A) Upper, a schematic view of domain structure of rat Pex14. Solid box, putative transmembrane (TM) domain; gray box, coiled-coil (CC) domain. Lower, Fao cells (4 × 105 cells) were transiently transfected with plasmids encoding wild-type His-RnPex14 (WT) and S232A mutant harboring a substitution at Ser232 to Ala, and a mock plasmid (mock). At 24 hr after transfection, cells were treated for 30 min with vehicle (-) and 1 mM H2O2 (+) and the cell lysates were analyzed by Phos-tag PAGE and immunoblotting with anti-His antibody. Open and solid arrowheads indicate unmodified and phosphorylated forms of His-Pex14, respectively. (B) Mass spectrometric analysis of phosphorylated Pex14 induced by H2O2-treatment. Endogenous Pex14 in Fao cells (8 × 106 cells) treated for 30 min with vehicle or 0.2 mM H2O2 was immunoprecipitated with anti-Pex14 antibody and subjected to LC-MS/MS analysis. Fragment spectrum of a phosphorylated peptide corresponding to amino acids 228–237 [QFPPpSPSAPK] showed phosphorylation of endogenous Pex14 at Ser232 (pS232) upon H2O2 treatment. (C) Quantification of phosphorylated Pex14 upon H2O2-treatment. Phosphorylated peptides were identified in Pex14 isolated from Fao cells that had been treated with vehicle or H2O2 as described in B. The levels of respective phosphopeptides in H2O2-treated cells (solid bars) were quantified with label-free precursor ion quantification and represented by taking as 1.0 that in vehicle-treated cells (open bars). Error bars represent means ± SEM of eight measurements in three independent experiments. *, p<0.05; **, p<0.01; unpaired Student’s t test versus vehicle treated cells. (D) Multiple amino-acid sequence alignment of Pex14 neighboring Ser232, Ser247, and Ser252 of rat Pex14. (E) Fao cells transiently expressing wild-type His-Pex14 (WT) and the variants with indicated mutations were treated with 0.2 mM H2O2 for 30 min as in A and analyzed as in Figure 1A with antibodies to His and Pex14. Open and solid arrowheads were as in A. (F) Fao cells (4 × 105 cells) pre-incubated for 30 min with vehicle (DMSO), 10 μM U0126, 10 μM SB203580, and 10 μM U0126 plus SB203580 were further treated with 0.2 mM H2O2 for 30 min. Cell lysates were analyzed as in Figure 1A by immunoblotting with indicated antibodies.
-
Figure 2—source data 1
Data for the phosphorylated peptides of Pex14 shown in Figure 2C.
- https://cdn.elifesciences.org/articles/55896/elife-55896-fig2-data1-v2.xlsx
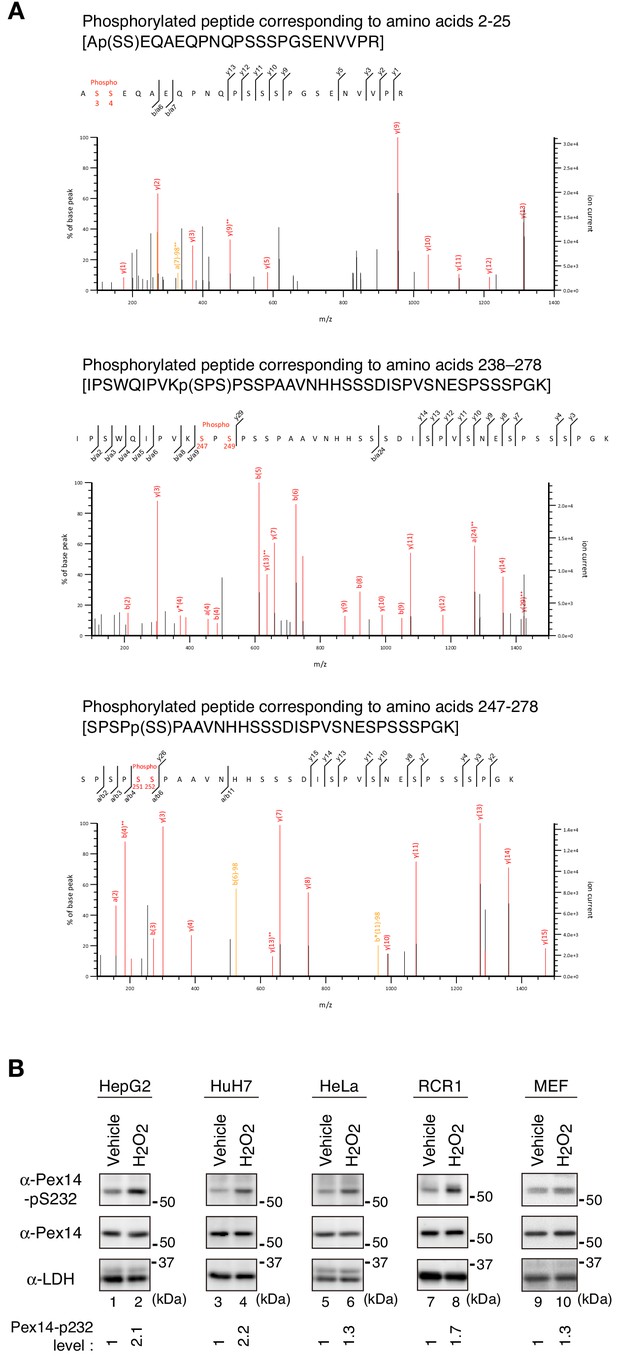
H2O2-induced phosphorylation of Pex14: identification the phosphorylation sites by LC-MS/MS analysis and detection of Pex14-pS232 in various cultured cell lines.
(A) LC-MS/MS analysis for phosphorylation sites of Pex14 upon H2O2 treatment was performed as in Figure 2B. Fragment spectra of phosphorylated peptides corresponding to amino acids 2–25 containing pS3 or pS4 (upper panel), amino acids 238–278 containing pS247 (middle panel), and amino acids 247–278 containing pS252 (lower panel) are shown. (B) Phosphorylation of Pex14 at Ser232 was induced in various cultured cells upon H2O2-treatment. Several mammalian cell lines (4 × 105 cells each) were treated with vehicle and 0.5 mM H2O2 for 0.5 hr (HepG2, HuH7, and MEF) and 1 hr (HeLa and RCR1), similarly to Figure 1B, and were analyzed by SDS-PAGE and immunoblotting with indicated antibodies. The levels of Pex14 phosphorylation at Ser232 upon H2O2-treatment were quantified, normalized by that of total Pex14, and represented at the bottom by taking as one those in vehicle-treated cells.
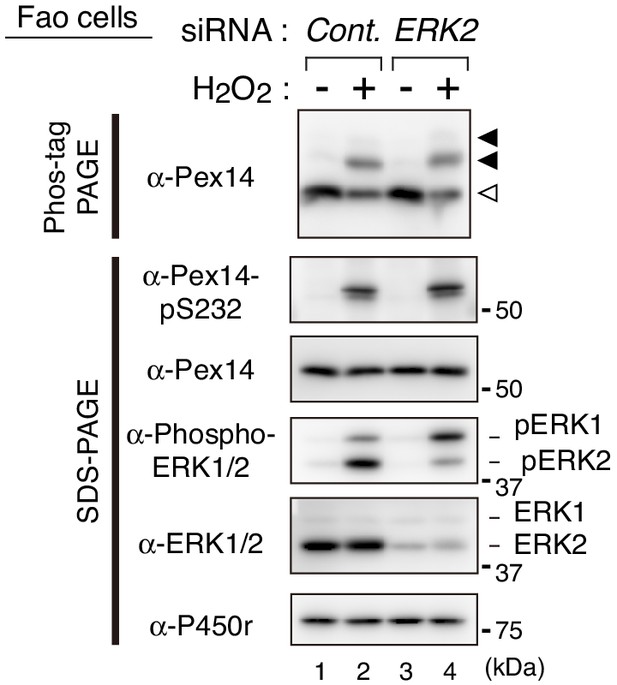
ERK2 siRNA shows no apparent effect on the H2O2-induced phosphorylation of Pex14.
Fao cells transfected for 48 hr with a control siRNA (lanes 1 and 2) or ERK2 siRNA (lanes 3 and 4) were treated for 30 min with vehicle (-) and 0.2 mM H2O2 (+). Cell lysates were analyzed as in B with indicated antibodies. Open and solid arrowheads were as in B.
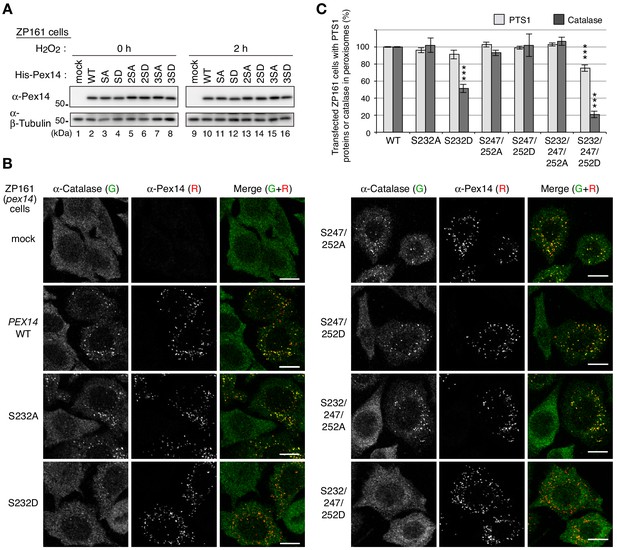
Phosphorylation of Pex14 suppresses peroxisomal import of catalase.
(A) pex14 ZP161 cells were transiently transfected for 36 hr with an empty vector (mock), wild-type (WT), and respective Ser mutants of His-PEX14. Cell lysates were analyzed by SDS-PAGE and immunoblotting with antibodies indicated on the left. (B) pex14 ZP161 cells was transiently transfected with PEX14 variants as in A. Cells were immunostained with antibodies to catalase (green) and Pex14 (red). Bar, 10 μm. (C) Quantification of the data in B and those for PTS1 proteins in Figure 3—figure supplement 1B. Percentages of the cells where PTS1 proteins (light gray) and catalase (dark gray) were mostly localized in peroxisomes in Pex14-expressing cells were represented as the means ± SEM by taking those as 100% in Pex14-WT-expressing cells. Transfected cells (n > 50) were counted in three independent experiments. ***p<0.001; one-way ANOVA with Dunnett’s post hoc test versus cells expressing Pex14-WT.
-
Figure 3—source data 1
Data for the import of catalase and PTS1 proteins shown in Figure 3C.
- https://cdn.elifesciences.org/articles/55896/elife-55896-fig3-data1-v2.xlsx
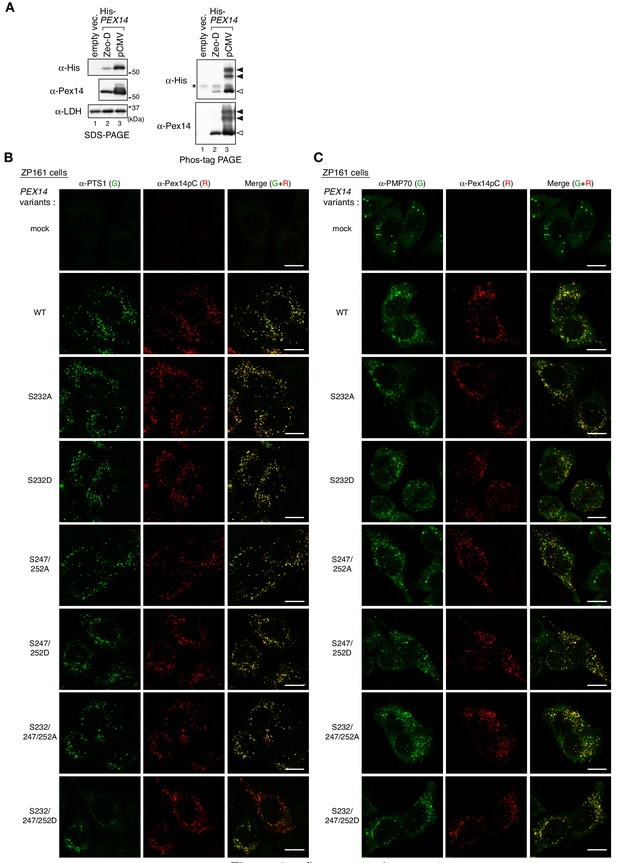
Phosphomimetic Pex14-S232/247/252D mutation affects PTS1 protein import, but not peroxisomal localization of Pex14.
(A) Protein level of exogenously expressed Pex14. pex14 ZP161 cells were transfected with a pCMVSPORT1 vector harboring no insert (mock, lane 1) or that encoding His-Pex14 (pCMV, lane 3), and pcDNAZeo-D vector encoding His-Pex14 (Zeo-D, lane 2) At 24 hr after transfection, cells were lysed and analyzed by SDS-PAGE (left panels) and Phos-tag PAGE (right panels), and immunoblotting with indicated antibodies. Open and solid arrowheads indicate unmodified and phosphorylated forms of His-Pex14, respectively. *, a nonspecific band. (B and C) Peroxisomal localization of Pex14 variants and the restoring activity in PTS1 protein import. pex14 ZP161 was transiently transfected with wild-type (WT) and respective PEX14 variants as in Figure 3B. At 36 hr after transfection, cells were immunostained with antibodies to PTS1 peptides (B, green), PMP70 (C, green), and Pex14 (B and C, red). Representative images are shown with the merged views. Bar, 10 μm.
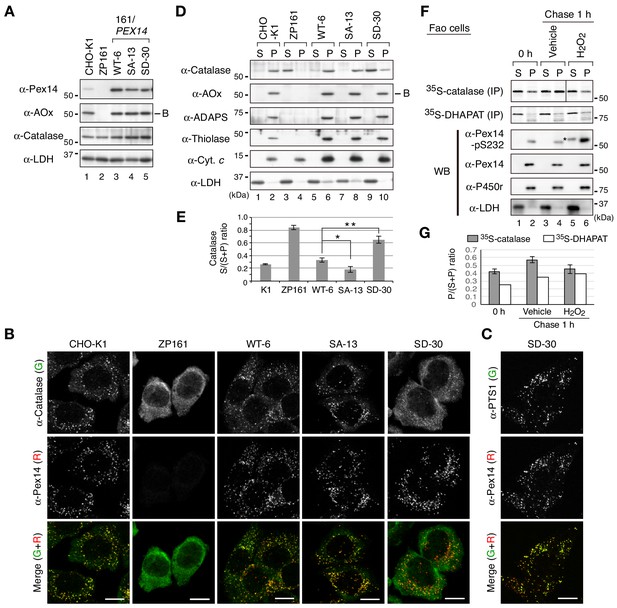
Phosphomimetic Pex14 mutant, Pex14-S232D, reduces catalase import into peroxisomes.
(A) Cell lysates of CHO-K1, pex14 ZP161, and stable cell lines of ZP161 each expressing wild-type His-Pex14 (WT-6) and its mutants, phosphorylation-deficient Pex14-S232A (SA-13) and phosphomimetic Pex14-S232D (SD-30) (4 × 105 cells each) were analyzed by SDS-PAGE and immunoblotting with antibodies indicated on the left. Only the B-chain of acyl-CoA oxidase (AOx) that is generated by intraperoxisomal proteolytic processing of full-length AOx is shown. (B) Catalase was less imported into peroxisomes in Pex14-S232D-expressing cells. CHO-K1, pex14 ZP161, and stable cell lines each expressing Pex14 variants were immunostained with antibodies to catalase (green) and Pex14 (red). Merged images were also shown. Bar, 10 μm. (C) Stable lines of pex14 ZP161 expressing phosphomimetic Pex14-S232D (SD-30) were likewise immunostained with antibodies to PTS1 (green) and Pex14 (red). Bar, 10 μm. (D) Catalase in cytosolic fraction was increased in Pex14-S232D-expressing cells. Cells indicated at the top (8 × 105 cells each) were separated into cytosolic (S) and organelle (P) fractions by permeabilization with 25 μg/mL digitonin and subsequent ultracentrifugation. Equal aliquots of respective fractions were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. AOx, a typical PTS1 protein; alkyl-dihydroxyacetonephosphate synthase (ADAPS) and 3-ketoacyl-CoA thiolase (thiolase), PTS2 proteins; Cyt. c, cytochrome c. LDH is a marker for cytosolic fraction. (E) Catalase level in the cytosolic and organelle fractions assessed in D was quantified and shown as a ratio of cytosol (S) to total (S plus P). Data represent means ± SEM of three independent experiments. Statistical analysis was performed by one-way ANOVA with Dunnett’s post hoc test as compared with the S/(S+P) ratio of catalase in WT-6 cells. *p<0.05 and **p<0.01. (F) Pulse-chase experiment of catalase translocation. Fao cells were labeled with 35S-methionine and 35S-cysteine for 1 hr and were chased for 1 hr in the presence of vehicle or 0.2 mM H2O2. Cells were fractionated into the cytosol (S) and organelle (P) fractions as described in Material and methods. Equal aliquots of respective fractions were solubilized and subjected to immunoprecipitation with antibodies to catalase and DHAPAT. 35S-labeled catalase was analyzed by SDS-PAGE and detected by autoradiography (two upper panels). Equal aliquots of the cytosol and organelle fractions were analyzed by SDS-PAGE and immunoblotting using indicated antibodies. P450r, an ER membrane protein, cytochrome P450 reductase; LDH, a cytosolic protein. *, a putative nonspecific band. (G) 35S-labelled bands in F were quantified and 35S-catalase and 35S-DHAPAT in peroxisomes were shown as the ratio of respective bands in organelle (P) to total (S plus P). The P/(S+P) ratios of 35S-catalase and 35S-DHAPAT were represented as an average of two independent experiments and a single experiment, respectively.
-
Figure 4—source data 1
Data for the level of cytosolic catalase shown in Figure 4E and those of 35S-catalase and 35S-DHAPAT in the organelle fractions shown in Figure 4G.
- https://cdn.elifesciences.org/articles/55896/elife-55896-fig4-data1-v2.xlsx
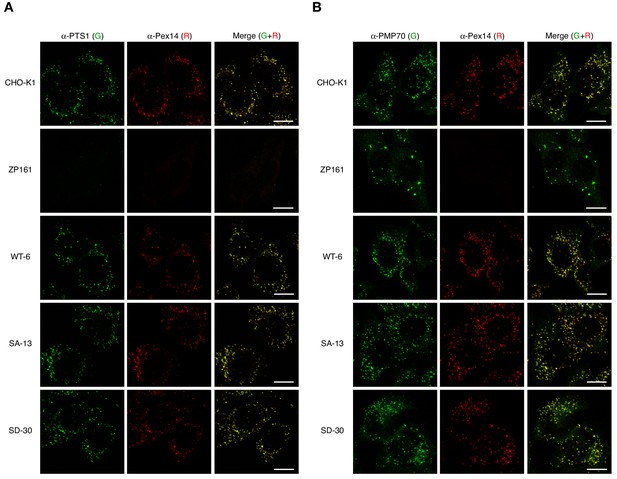
Phosphomimetic Pex14-S232D has no apparent effect on import of PTS1 protein and peroxisomal localization of Pex14.
(A and B) CHO-K1, pex14 ZP161, and stable cell lines each expressing Pex14 variants were immunostained as in Figure 4B with antibodies to PTS1 peptides (A, green), PMP70 (B, green), and Pex14 (A and B). Representative images are shown with the merged views. Bar, 10 μm.
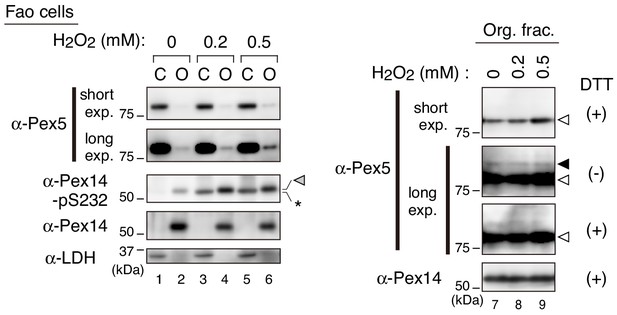
Pex5 recycling upon H2O2-treatment.
Fao cells treated with H2O2 at indicated concentration for 1 hr were homogenized in the presence 5 mM NEM. PNS fractions were separated into the cytosolic (C) and organelle (O) fractions. Equal aliquots of the cytosolic and organelle fractions in Laemmli sample buffer with 0.1 M DTT (left panels) and the organelle fractions in the presence (+) or absence (-) of DTT (right panels) were analyzed by SDS-PAGE and immunoblotting with indicated antibodies. Gray, solid, and open arrowheads indicate phosphorylated Pex14 at S232, mono-ubiquitinated Pex5 at Cys11 (Okumoto et al., 2011), and unmodified Pex5, respectively. *, a nonspecific band.
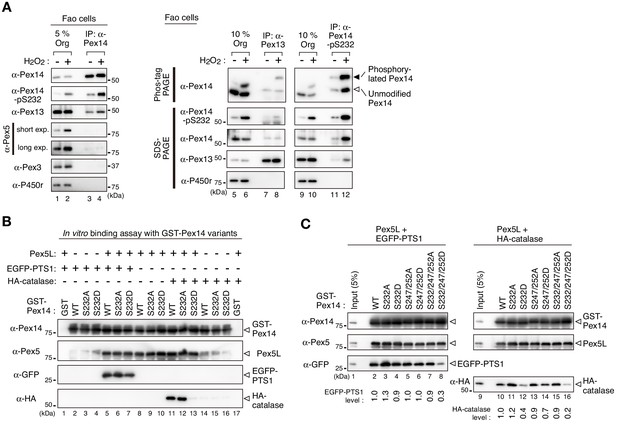
Phosphorylation of Pex14 exclusively affects Pex5-mediated complex formation with catalase.
(A) Phosphorylated Pex14 forms a complex with Pex13. Organelle fractions of Fao cells (4 × 106 cells each) treated for 30 min with vehicle (-) or 0.2 mM H2O2 were solubilized and subjected to immunoprecipitation with antibodies to Pex14 (lanes 3 and 4), Pex13 (lanes 7 and 8), and Pex14-pS232 (lanes 11 and 12). Equal-volume aliquots of immunoprecipitates (IP) and the input of organelle fractions (Org. input; 5% for IP of Pex14, 10% for IP of Pex13 and phosphorylated Pex14) were analyzed by SDS-PAGE and immunoblotting with antibodies indicated on the left. (B) In vitro binding assays were performed using recombinant proteins, that is GST-Pex14 variants, Pex5L, EGFP-PTS1, and HA-catalase. Components added to the assay mixtures, including GST in place of GST-Pex14 variants, are indicated at the top. Pex5L, EGFP-PTS1, and HA-catalase in the fractions bound to GST-Pex14-conjugated glutathione-Sepharose beads were analyzed by immunoblotting with antibodies indicated on the left. (C) In vitro binding assays were likewise performed using GST-Pex14 variants with mutations in three distinct Ser residues as in B. Five percent input of the reaction used was also loaded. Levels of the recovered EGFP-PTS1 and HA-catalase were quantified, normalized by that of GST-Pex14, and represented at the bottom by taking as one those pulled-down by GST-Pex14 WT.
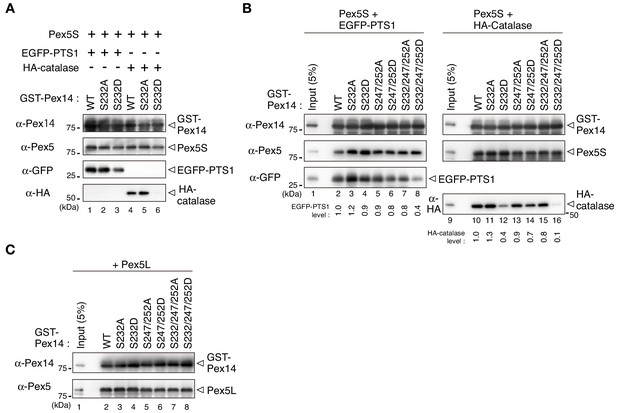
S232D mutation in Pex14 affects Pex5-catalase interaction.
(A) Pex14-S232D also shows lower affinity to catalase in the complex with Pex5S. In vitro binding assay was performed using GST-Pex14 variants, EGFP-PTS1, HA-catalase, and Pex5S as in Figure 5B. (B) Pex5S shows a similar interaction profile as Pex5L in forming a ternary complex with Pex14 and the cargos. In vitro binding assay using Pex5S was performed with a pair of GST-Pex14 variants and Pex5S. Levels of the recovered EGFP-PTS1 and HA-catalase were quantified and represented as in Figure 5C. (C) Direct interaction of Pex14 variants with Pex5L. GST pull-down assay was performed with Pex5L as in Figure 5B.
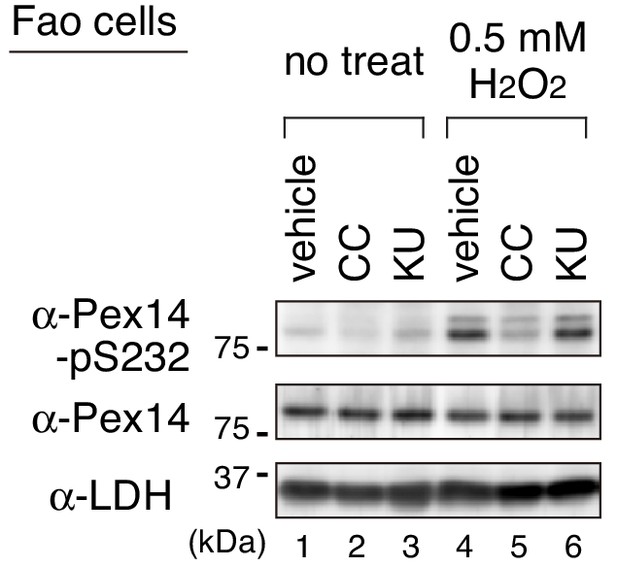
An ATM inhibitor KU-55933 shows no effect on the H2O2-induced phosphorylation of Pex14.
Fao cells pretreated for 30 min with DMSO (vehicle), 10 μM Compound C (CC), or 10 μM KU-55933 (KU) were cultured for 1 hr in the absence (no treat) and presence of 0.5 mM H2O2. Cell lysates were analyzed as in Figure 1B by immunoblotting with the indicated antibodies.
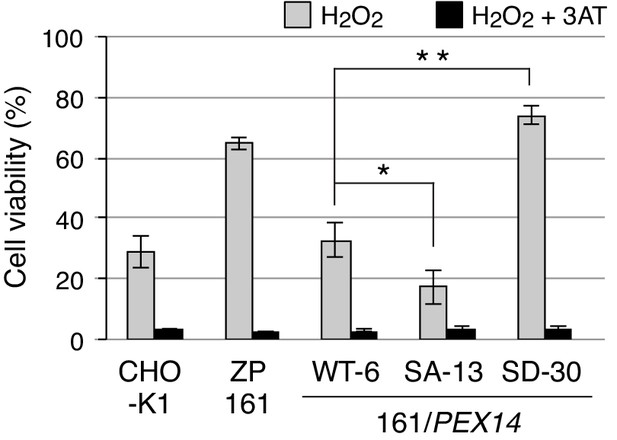
Phosphorylation at Ser232 of Pex14 is important for cell resistance to exogenous hydrogen peroxide.
CHO-K1, pex14 ZP161, and its stable cell lines expressing Pex14 variants (1 × 104 cells each) were treated with 0.8 mM H2O2 in the absence (gray bars) and presence (solid bars) of 20 mM 3-aminotriazole (3AT), a catalase inhibitor. Cell viability was determined by MTS assay at 16 hr after H2O2 treatment and represented as percentages relative to that of each mock-treated, H2O2-untreated cells. Data represent means ± SEM of three independent experiments. *p<0.05 and **p<0.01; one-way ANOVA with Dunnett’s post hoc test versus a stable cell line of ZP161 expressing Pex14-WT.
-
Figure 6—source data 1
Data for the cell viability upon H2O2-treatment shown in Figure 6.
- https://cdn.elifesciences.org/articles/55896/elife-55896-fig6-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit polyclonal anti-FLAG | Sigma | F7425 RRID:AB_439687 | (1:1000) |
| Antibody | Rabbit polyclonal anti-phospho-Erk1/2 | Cell signaling | 9101S RRID:AB_331646 | (1:2000) |
| Antibody | Mouse monoclonal anti-Erk1/2 | Cell signaling | 4694S | (1:2000) |
| Antibody | Mouse monoclonal anti-FLAG (M2) | Sigma | F1804 RRID:AB_262044 | (1:1000) |
| Antibody | Mouse monoclonal anti-HA (16B12) | Covance | MMS-101R-200 RRID:AB_291263 | (1:2000) |
| Antibody | Mouse monoclonal anti-hexa-histidine tag | Qiagen | 34650 RRID:AB_2687898 | (1:500) |
| Antibody | Mouse monoclonal anti-GFP (B-2) | Santa Cruz Biotechnology | Sc-9996 | (1:1000) |
| Antibody | Mouse monoclonal anti-Tom20 (F-10) | Santa Cruz Biotechnology | sc-17764 RRID:AB_628381 | (1:1000) |
| Antibody | Mouse monoclonal anti-Cytochrome P450 reductase | Santa Cruz Biotechnology | sc-25270 RRID:AB_627391 | (1:2000) |
| Antibody | Mouse monoclonal anti-Cytochrome c | BD Pharmingen | 556433 RRID:AB_396417 | (1:1000) |
| Antibody | Mouse monoclonal anti-ß-actin | MBL | M177-3 | (1:2000) |
| Antibody | Goat polyclonal anti-lactate dehydrogenase | Rockland | 110–1173 | (1:1000) |
| Antibody | Donley anti-Rabbit IgG, HRP-linked F(ab')2 fragment | GE Healthcare | NA9340 RRID:AB_772191 | (1:4000) |
| Antibody | Sheep anti-Mouse IgG, HRP-linked whole Antibody | GE Healthcare | NA931 RRID:AB_772210 | (1:4000) |
| Antibody | Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 488 | Invitrogen | A11034 RRID:AB_2576217 | (1:10000) |
| Antibody | Goat anti-Guinea Pig IgG (H+L) Secondary Antibody, Alexa Fluor 568 | Invitrogen | A11075 RRID:AB_141954 | (1:10000) |
| Cell line (C. griseus) | CHO-K1 | Tsukamoto et al., 1990 | ||
| Cell line (C. griseus) | pex14 ZP161 | Shimizu et al., 1999 | A PEX14-deficient CHO mutant | |
| Cell line (C. griseus) | ZP161 stably expressing His-RnPEX14 WT (WT-6) | This paper | A stable cell line of ZP161 expressing Pex14-WT | |
| Cell line (C. griseus) | ZP161 stably expressing His-RnPEX14 S232A (SA-13) | This paper | A stable cell line of ZP161 expressing Pex14-S232A | |
| Cell line (C. griseus) | ZP161 stably expressing His-RnPEX14 S232D (SD-30) | This paper | A stable cell line of ZP161 expressing Pex14-S232D | |
| Cell line (R. norvegicus) | Fao | Motojima et al., 1994 | ||
| Cell line (R. norvegicus) | RCR-1 | Abe et al., 2020 | ||
| Cell line (H. sapiens) | HuH-7 | RIKEN | RCB1366 | |
| Cell line (H. sapiens) | HeLa | Yagita et al., 2013 | ||
| Cell line (H. sapiens) | HepG2 | Honsho et al., 2017 | ||
| Cell line (M. musculus) | MEF | Itoyama et al., 2013 | ||
| Transfected construct (R. norvegicus) | siRNA to ERK2 | Sigma-Aldrich | SASI_Rn01_00107866 | GUAUAUACAUUCAGCUAAU |
| Recombinant DNA reagent | MISSION siRNA Universal Negative Control #1 | Sigma-Aldrich | SIC001 | |
| Recombinant DNA reagent | pCMVSPORT/His-RnPEX14 WT (plasmid) | Itoh and Fujiki, 2006 | His-Pex14 WT | |
| Recombinant DNA reagent | pCMVSPORT/His-RnPEX14 SA or SD variants (plasmid) | This paper | His-Pex14 S232A, S232D, S247/252A, S247/252D, S232/247/252A, S232/247/252D | |
| Recombinant DNA reagent | pcDNAZeo-D (plasmid) | This paper | A mammalian expression vector with low transcription | |
| Recombinant DNA reagent | pcDNAZeo-D/His-RnPEX14 WT (plasmid) | This paper | Wild-type His-Pex14 | |
| Recombinant DNA reagent | pcDNAZeo-D/His-RnPEX14 SA or SD variants (plasmid) | This paper | His-Pex14 S232A, S232D, S247/252A, S247/252D, S232/247/252A, S232/247/252D | |
| Recombinant DNA reagent | pGEX/RnPEX14 WT (plasmid) | Itoh and Fujiki, 2006 | GST-Pex14 WT | |
| Recombinant DNA reagent | pGEX/His-RnPEX14 SA or SD variants (plasmid) | This paper | GST-Pex14 S232A, S232D, S247/252A, S247/252D, S232/247/252A, S232/247/252D | |
| Recombinant DNA reagent | pGEX/HA-HsCatalase (plasmid) | This paper | for recombinant GST-HA-Catalase | |
| Recombinant DNA reagent | pGEX/ClPEX5S (plasmid) | Otera et al., 2002 | ||
| Recombinant DNA reagent | pGEX/ClPEX5L (plasmid) | Otera et al., 2002 | ||
| Recombinant DNA reagent | pGEX/EGFP-PTS1 (plasmid) | Okumoto et al., 2011 | ||
| Recombinant DNA reagent | pGEX6P-1 (plasmid) | GE Healthcare | 28954648 | |
| Sequence-based reagent | Truncated CMV.Fw | This paper | PCR primer | ATGGGCGGTAGGCGTGTACG |
| Sequence-based reagent | Truncated CMV.Rv: | This paper | PCR primer | CGCGAAGCAGCGCAAAACG |
| Sequence-based reagent | RnPEX14-S232A.InvFw: | This paper | PCR primer | GCCCCGTCAGCCCCGAAGATCCCCTCCT- |
| Sequence-based reagent | RnPEX14-S232D.InvFw: | This paper | PCR primer | GACCCGTCAGCCCCGAAGATCCCCTCCT |
| Sequence-based reagent | RnPEX14-S232A/D.InvRv: | This paper | PCR primer | GGGAGGGAACTGTCTCCGATTC |
| Sequence-based reagent | RnPEX14-S252A.InvFw: | This paper | PCR primer | GCCCCCGCGGCCGTGAACCACCACAGC |
| Sequence-based reagent | RnPEX14-S252D.InvFw: | This paper | PCR primer | GACCCCGCGGCCGTGAACCACCACAGC |
| Sequence-based reagent | RnPEX14-S247_252A.InvRv: | This paper | PCR primer | GGAGGGTGACGGAGCCTTCACTGGG |
| Sequence-based reagent | RnPEX14-S247_252D.InvRv: | This paper | PCR primer | GGAGGGTGACGGGTCCTTCACTGGG |
| Sequence-based reagent | GST-HA-HsCatalase.BglFw: | This paper | PCR primer | GCGCAGATCTATGGCTTATCCATACGAC |
| Sequence-based reagent | pUcD3.Rv: | Otera and Fujiki, 2012 | PCR primer | TTTCCACACCTGGTTGC |
| Chemical compound, drug | U0126 | Cell signaling | 9903S | |
| Chemical compound, drug | SB203580 | Cell signaling | 5633S | |
| Chemical compound, drug | KU-55933 | Abcam | ab120637 | |
| Chemical compound, drug | Compound C | Merck | 171260 | |
| Chemical compound, drug | Complete protease inhibitor cocktail | Roche | 11836170001 | |
| Chemical compound, drug | PhosStop phosphatase inhibitor cocktail | Sigma | 4906845001 | |
| Commercial assay or kit | CellTiter 96 AQueous One Solution Cell Proliferation Assay | Promega | G3580 | |
| Software, algorithm | R | R-project | http://www.r-project.org | |
| Software, algorithm | Image J | NIH | https://imagej.nih.gov/ij/ |





