A single power stroke by ATP binding drives substrate translocation in a heterodimeric ABC transporter
Figures
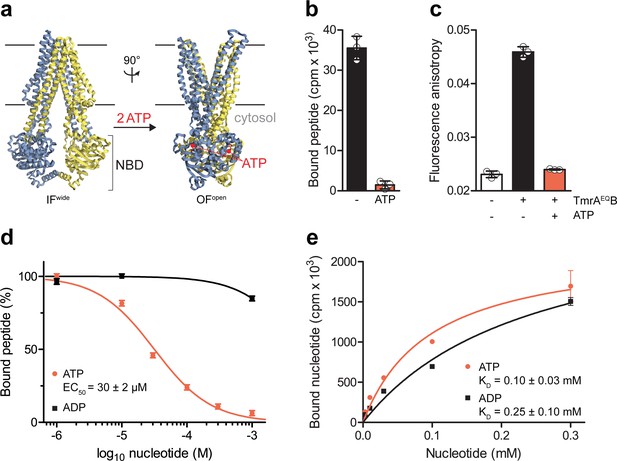
The IF-to-OF transition by ATP binding precludes substrate binding.
(a) Scheme of the IF-to-OF transition by comparing the IFwide and OFopen conformation of TmrAB. TmrA, TmrB, and ATP are shown in cyan, yellow, and red, respectively. (b) Loss in peptide binding after the ATP-induced IF-to-OF transition. TmrAEQB (0.2 µM) in Nds was incubated for 5 min at 45 °C with the R9LQK peptide (10 µM, supplemented with 3H-R9L) in the presence and absence of Mg2+-ATP (1 mM). Peptide binding was monitored by SPA. Signals were background-corrected and multiplied by the dilution factor of radiolabeled peptides. (c) Peptide binding after the IF-to-OF switch. The fluorescence anisotropy of C4F peptide (50 nM) was determined at λex/em = 485/520 nm in the absence and presence of TmrAEQB in Nds (1 µM). To distinguish between the IF and OF conformation, TmrAEQB was incubated for 5 min at 45 °C with and without Mg2+-ATP (1 mM). (d) Nucleotide-dependent peptide binding. TmrAEQB in Nds (1 µM) was incubated for 5 min at 45 °C with increasing concentrations of nucleotide. (e) Equilibrium binding affinity of ATP or ADP analyzed by scintillation proximity assay. TmrAEQB (0.2 µM) was incubated with increasing concentration of ATP or ADP supplemented with 3H-ATP or 3H-ADP, respectively. The mean of three independent experiments was analyzed by a one-site binding model, KD, ATP = 0.10 ± 0.03 mM, KD, ADP = 0.25 ± 0.10 mM. In (b–e) the mean ± SD (n = 3) is displayed.
-
Figure 1—source data 1
Source Data to Figure 1b-e and Figure 1—figure supplement 1 and 2.
- https://cdn.elifesciences.org/articles/55943/elife-55943-fig1-data1-v2.docx
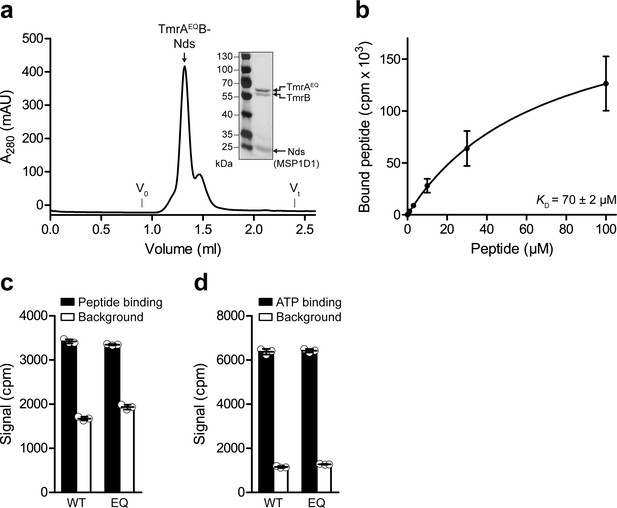
Peptide binding to TmrAB reconstituted in lipid nanodiscs.
(a) TmrAEQB reconstituted in lipid nanodiscs (Nds, MSP1D1) was isolated by size-exclusion chromatography (SEC) and analyzed by SDS-PAGE (silver stain). (b) Peptide equilibrium binding affinity determined by scintillation proximity assay (SPA). TmrAB (0.2 µM) was incubated with increasing concentration of R9LQK supplemented with 3H-R9L. Data were analyzed by the one-site binding model (KD = 70 ± 2 µM). (c), Equilibrium peptide binding monitored by SPA. Wildtype TmrAB (WT) or TmrAEQB (EQ, 0.2 µM each) were immobilized on SPA beads and incubated with R9LQK peptide (10 µM, traced with 3H-R9L). (d), Equilibrium ATP binding. TmrAB (WT) or TmrAEQB (EQ, 0.2 µM each) were immobilized on SPA beads and incubated with ATP (3 µM, traced with 3H-ATP). In (b–d) the mean values ± SD (n = 3) are shown. The background was determined after releasing His-tagged TmrAB from the beads by adding 200 mM imidazole. In (c and d) original cpm values are displayed which were not corrected by the dilution factor of radiolabeled compound.
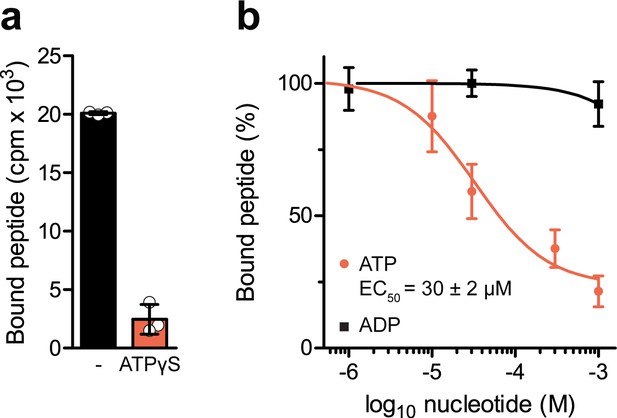
Nucleotide-dependent peptide binding of TmrAEQB in lipid nanodiscs.
(a) Nucleotide trapping using ATPγS. TmrAEQB (0.2 µM) was incubated with R9LQK peptide (10 µM, supplemented with 3H-R9L), 1 mM ATPγS and 3 mM MgCl2 for 5 min at 45 °C. The equilibrium peptide binding was determined by SPA. (b), TmrAEQB (0.2 µM) was incubated with R9LQK peptide (10 µM, supplemented with 3H-R9L), 3 mM MgCl2, and increasing concentration of ATP or ADP for 5 min at 45 °C. The equilibrium peptide binding was determined by SPA. In a–b), data are shown as mean values ± SD (n = 3).
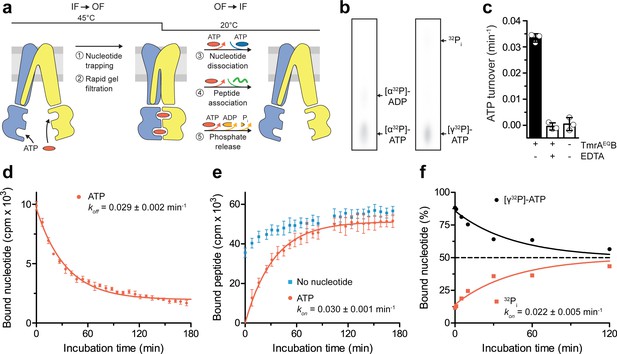
A single ATP turnover and phosphate release launches the OF-to-IF return.
(a) Experimental scheme monitoring the OF-to-IF transition. (b) Nucleotide occlusion after the IF-to-OF transition. TmrAEQB in Nds (2 µM) was incubated for 5 min at 45 °C with Mg2+-ATP (1 mM, traced with [α32P]-ATP or [γ32P]-ATP). ATP (2 mM) was added and unbound nucleotides were removed by rapid gel filtration. The radiolabeled nucleotides occluded by TmrAEQB were identified by thin layer chromatography and autoradiography. Representative autoradiograms for three independent triplicates are shown. (c) ATPase activity. TmrAEQB (1 µM) was incubated with Mg2+-ATP (1 mM, traced with [γ32P]-ATP) for 5 min at 45 °C and 15 min at 20 °C. Control reactions were performed in the presence of EDTA (10 mM). ATP autohydrolysis was recorded in the absence of TmrAEQB. The release of [γ32P] was quantified by thin layer chromatography and autoradiography. Data were normalized to autohydrolysis and the mean values ± SD (n = 3) are displayed. (d) Nucleotide dissociation upon the OF-to-IF return. TmrAEQB in Nds (0.2 µM) was incubated with Mg2+-ATP (3 µM, traced with 3H-ATP) for 5 min at 45 °C. Unbound nucleotides were removed by rapid gel filtration. An excess of Mg2+-ATP (1 mM) was added to prevent any reassociation of released nucleotides. Nucleotide dissociation was followed by SPA at 20 °C. The mean ± SD (n = 3) is displayed and monoexponentially fitted, leading to a dissociation rate constant koff of 0.029 ± 0.002 min−1 (τ1/2 = 21–26 min 95% confidence interval). (e) Peptide rebinding along the OF-to-IF return. TmrAEQB (0.2 µM) complexes were converted to the OF state as described in a). The excess of ATP was removed by rapid gel filtration. R9LQK peptide binding (10 µM, traced with 3H-R9L) was monitored by SPA at 20 °C. The mean ± SD (n = 3) is displayed and monoexponentially fitted, leading to an apparent association rate kon of 0.030 ± 0.001 min−1 (τ1/2 = 21–25 min 95% confidence interval). (f) Release of inorganic phosphate along the OF-to-IF return. TmrAEQB in Nds (2 µM) was incubated with Mg2+-ATP (1 mM, traced with [γ32P]-ATP) and converted to the OF state as described in b). ATP (2 mM) was added and unbound nucleotides were removed by rapid gel filtration. Subsequently, a large excess of unlabeled ATP (2 mM) was added to prevent reassociation of [γ32P]-ATP and thus to guarantee a single-turnover round of ATP hydrolysis. Nucleotides and released phosphate were analyzed as in panel c). It is important to note that some ATP is hydrolyzed before the TLC analysis. The mean ± SD (n = 3) is displayed and monoexponentially fitted, leading to inversely correlated rates of ATP turnover and phosphate release (kon (32Pi)=koff ([γ32P]-ATP)=0.022 ± 0.005 min−1).
-
Figure 2—source data 1
Source Data to Figure 2c-f and Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/55943/elife-55943-fig2-data1-v2.docx
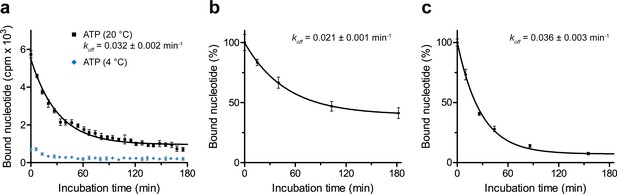
Dissociation of occluded ATP.
(a) Nucleotide occlusion at different temperatures and subsequent dissociation. TmrAEQB in Nds (0.2 µM) was incubated with Mg2+-ATP (3 µM, traced with 3H-ATP) for 5 min at 20 °C or 4°C. Unbound nucleotides were removed by rapid gel filtration. Subsequently, ATP (1 mM) was added to prevent any reassociation of released ATP. Nucleotide dissociation was followed by SPA at 20 °C. Data shown as mean ± SD (n = 3) are monoexponentially fitted, leading to a dissociation rate constant koff of 0.032 ± 0.002 min−1 (τ1/2 = 19–24 min 95% confidence interval). (b), Nucleotide dissociation at 4 °C. TmrAEQB (0.2 µM) was incubated with Mg2+-ATP for 5 min at 45°C as described in a). After rapid gel filtration and ATP addition, nucleotide dissociation was followed by SPA at 4 °C. The mean ± SD (n = 3) is displayed and monoexponentially fitted, leading to a dissociation rate constant koff of 0.021 ± 0.001 min−1 (τ1/2 = 30–38 min 95% confidence interval). (c), Nucleotide dissociation at 45 °C. TmrAEQB in Nds (0.2 µM) was incubated with Mg2+-ATP for 5 min at 45 °C as described in a). After rapid gel filtration and ATP addition, nucleotide dissociation was followed by SPA at 45 °C. The mean ± SD (n = 3) is displayed and monoexponentially fitted, leading to a dissociation rate constant koff of 0.036 ± 0.003 min−1 (τ1/2 = 16–24 min 95% confidence interval).
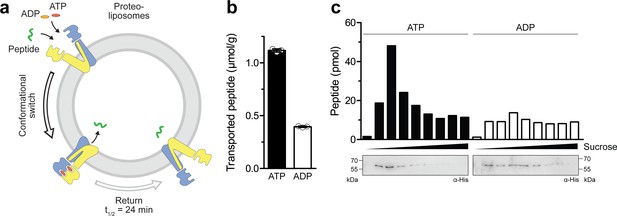
Single-conformational switch by ATP binding is coupled to an efficient substrate translocation.
(a) Experimental scheme. TmrAEQB was reconstituted in liposomes and incubated with C4F peptide and Mg2+-ATP or Mg2+-ADP. For single-turnover, transport was performed for 5 min at 45 °C to allow a single IF-to-OF transition. After solubilization of proteoliposomes, transported peptides were quantified by fluorescence detection (λex/em = 485/520 nm). (b) Single-turnover transport studies. Liposomes containing reconstituted TmrAEQB were mixed with C4F peptide (50 µM) and Mg2+-ATP or Mg2+-ADP (3 mM each) for 5 min at 45 °C. Liposomes were extensively washed. After solubilization, peptides were fluorescently quantified. Data are shown as mean ± SD (n = 3). (c) Single-turnover transport and liposome flotation assays. C4F peptide translocation was performed as described in b). Proteoliposomes were floated by sucrose density gradient centrifugation to remove aggregates and unspecifically adsorbed C4F peptides. Transported peptides were fluorescently quantified. TmrAEQB was detected using immunoblotting (α-His).
-
Figure 3—source data 1
Source Data to Figure 3b and c.
- https://cdn.elifesciences.org/articles/55943/elife-55943-fig3-data1-v2.docx
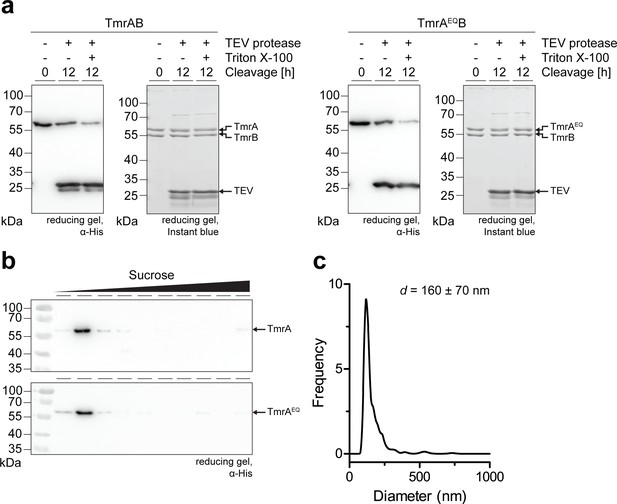
Orientation and incorporation of TmrAEQB in proteoliposomes.
(a) Orientation of TmrAB and TmrAEQB in proteoliposomes. Proteoliposomes were incubated with TEV protease in the presence and absence of Triton X-100 for 16 hr at 4 °C. Fraction of cleaved TmrAB was quantified by immunoblotting (α-His). An equal load of proteoliposomes was confirmed using SDS-PAGE. (b), Incorporation of TmrAB and TmrAEQB in proteoliposomes. Proteoliposomes were resuspended in 100 mM Na2CO3 pH 11.5, 30% sucrose for 30 min at 4 °C. Proteoliposomes and aggregates were separated by sucrose density gradient centrifugation for 16 hr at 270,000 g. TmrAB was quantified by immunoblotting (α-His). (c), Size distribution of proteoliposomes. Proteoliposomes were analyzed by nanoparticle tracking at 25 °C. The average diameter of three independent measurements is shown.
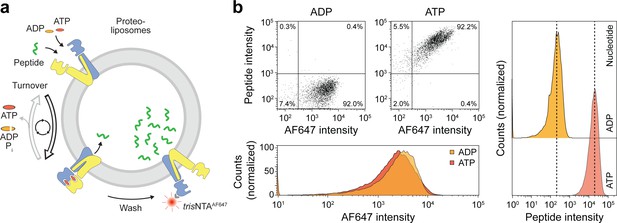
Sequential rounds of peptide translocation monitored by flow cytometry.
(a) Experimental scheme. TmrAB-containing liposomes were incubated with C4F peptide (30 µM) and Mg2+-ATP or Mg2+-ADP (3 mM each) for 5 min at 45 °C. Proteoliposomes were extensively washed by centrifugation, and His-tagged TmrAB complexes were labeled with trisNTAAF647 (150 nM). The mean fluorescence intensities of correctly oriented TmrAB complexes and translocated peptides were examined by flow cytometry. (b), Multiple rounds of C4F peptide translocation. Proteoliposomes were incubated with C4F peptide and nucleotides as described in a). Proteoliposomes were washed and correctly-oriented TmrAB complexes were labeled with trisNTAAF647 as described in a). The mean fluorescence intensities of 100,000 events were analyzed.
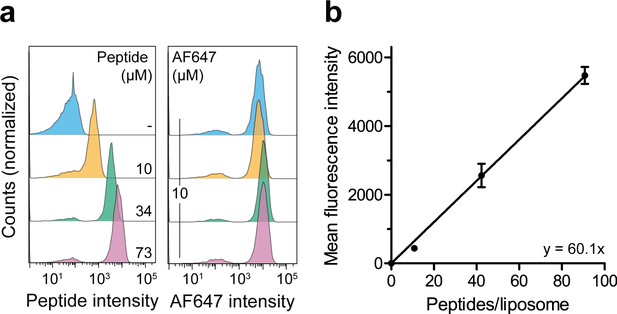
Correlation of the lumenal peptide amount and fluorescence intensity by quantitative flow cytometry.
(a) Liposomes were loaded with defined amounts of C4F peptide and dextranAF647 serving as loading control. After extensive washing, the mean fluorescence intensities of 100,000 liposomes were quantified by flow cytometry. (b), Linear regression. Encapsulated C4F peptide intensities were correlated with the average number of peptides per liposome. Data are shown as mean ± SD (n = 3) and were fitted by linear regression.
-
Figure 4—figure supplement 1—source data 1
Source Data to Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/55943/elife-55943-fig4-figsupp1-data1-v2.docx
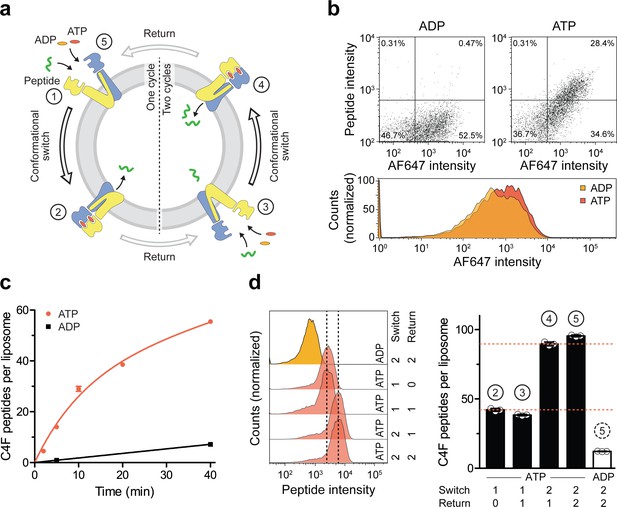
A single IF-to-OF transition driving substrate translocation.
(a) Experimental setups. Liposomes containing reconstituted TmrAEQB were incubated with C4F peptide (30 µM) and Mg2+-ATP or Mg2+-ADP (3 mM each). To guarantee a single-turnover, transport was performed for 5 min at 45 °C (switch 1). For two consecutive cycles, proteoliposomes were incubated for 1 hr at 4 °C (return) followed by 5 min at 45 °C (switch 2). Proteoliposomes were extensively washed, and His-tagged TmrAEQB complexes were labeled with trisNTAAF647 (150 nM). Proteoliposomes were washed by centrifugation. The mean fluorescence intensities of correctly oriented TmrAEQB complexes and translocated peptides were examined by flow cytometry. (b) Single-translocation events monitored by flow cytometry. TmrAEQB proteoliposomes were incubated with C4F peptide and nucleotide as described in a). The mean fluorescence intensities of 100,000 events were analyzed by flow cytometry. (c) Time-dependent translocation of peptides induced by ATP binding and IF-to-OF switching. TmrAEQB proteoliposomes were incubated with C4F peptide and nucleotides as described in a) for increasing time at 45 °C. For ATP, signals were subtracted by mean fluorescence intensities of untreated proteoliposomes and fitted by a burst equation (Equation 6), yielding ~35 peptides per liposome at the Y-intercept (Spanfast) and kslow ≈ 0.5 peptides liposome−1 min−1. Data are shown as mean ± SD (n = 3). (d), Two consecutive rounds of C4F peptide translocation driven by ATP binding and IF-to-OF switching. TmrAEQB proteoliposomes were incubated with C4F peptide and nucleotides as described in a). The mean fluorescence intensities of 105 events were analyzed by flow cytometry. Data are shown as mean ± SD (n = 3).
-
Figure 5—source data 1
Source Data to Figure 5c and d as well as Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/55943/elife-55943-fig5-data1-v2.docx
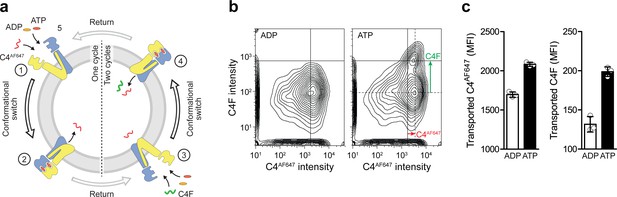
Single-turnover translocation of diverse substrates induced by ATP binding and IF-to-OF switching.
(a) Experimental scheme. (b) Dual-substrate translocation of TmrAEQB induced by sequential rounds of IF-to-OF switching. TmrAEQB containing proteoliposomes were incubated with C4AF647 peptide (30 µM), ATP or ADP (3 mM each), and MgCl2 (5 mM) for 5 min at 45 °C (cycle 1). ADP (20 mM) was added for 1 hr at 4 °C (return). Proteoliposomes were extensively washed and incubated with C4F peptide (30 µM), ATP or ADP (3 mM each), and MgCl2 (5 mM) for 5 min at 45 °C (cycle 2). Proteoliposomes were washed again, and mean fluorescence intensities of both peptides were determined by flow cytometry. (c) Data are shown as mean ± SD (n = 3).
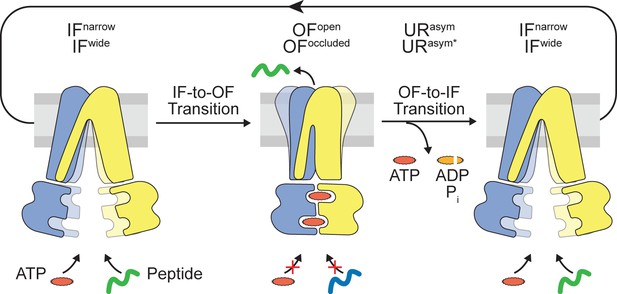
Translocation cycle.
IFwide conformation of TmrAB allows binding of nucleotides and peptide. ATP binding triggers the IF-to-OF conformational transition, which drives substrate translocation. ATP-occluded OF conformations represent pre-hydrolysis states, in which peptide binding and nucleotide exchange is abolished. After ATP hydrolysis at the consensus site, phosphate (Pi) release triggers the OF-to-IF transition via asymmetric unlocked-return (URasym and URasym*) conformers, revealed by cryo-EM (Hofmann et al., 2019), to reactivate peptide and nucleotide binding.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Monoclonal α-His | Sigma-Aldrich | SAB1305538 | Mouse origin; 1:2000 dilution |
| Antibody | Polyclonal α-mouse-HRP conjugate | Sigma-Aldrich | AP130P | Goat origin; 1:20,000 dilution |
| Chemical compound, drug | β-n-dodecyl β-D-maltoside (DDM) | Carl Roth | CN26.5 | |
| Chemical compound, drug | Bovine brain lipid extract | Sigma-Aldrich | B1502 | |
| Chemical compound, drug | E. coli polar lipids | Avanti | 100600 | |
| Chemical compound, drug | 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine (DOPC) | Anatrace | D518 | |
| Chemical compound, drug | [2,5’,8-3H(N)]-ATP (3H-ATP) | PerkinElmer | NET118900 | |
| Chemical compound, drug | [2,8-3H]-ADP (3H-ADP) | Biotrend | ART-0386 | |
| Chemical compound, drug | [α32P]-ATP | Hartmann Analytic | FP-207 | |
| Chemical compound, drug | [γ32P]-ATP | Hartmann Analytic | FP-201 | |
| Chemical compound, drug | Copper-chelated PVT SPA beads | PerkinElmer | RPNQ0095 | |
| Gene (Thermus thermophilus) | TmrA | Q72J05 | TTC0976 | |
| Gene (Thermus thermophilus) | TmrB | Q72J04 | TTC0977 | |
| Peptide, recombinant protein | RRY-[3H]L-KSTEL | Hartmann Analytic | Custom synthesis | |
| Peptide, recombinant protein | RRY-C*-KSTEL | This study (Peptide labeling and purification) | C* denotes fluorescein labeled Cys | |
| Peptide, recombinant protein | RRY-C*-KSTEL | This study (Peptide labeling and purification) | C* denotes Alexa Fluor 647 labeled Cys | |
| Recombinant DNA reagent | pET-22b | Merck Millipore | 69744 | Vector for protein expression in E. coli |
| Software, Algorithm | Prism 5 | GraphPad | ||
| Software, Algorithm | PyMOL | Schrödinger | ||
| Strain, strain background (Escherichia coli) | BL21(DE3) | Thermo Fisher | C600003 | Chemically competent cells |





