Emergence and diversification of a host-parasite RNA ecosystem through Darwinian evolution
Figures
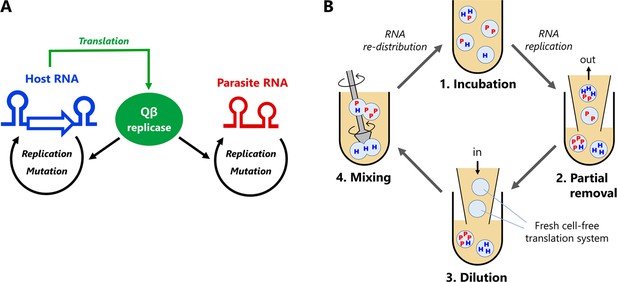
Host and parasitic RNA replication system.
(A) Replication scheme of the host and parasitic RNAs. The host RNA encodes the Qβ replicase subunit, whereas the parasitic RNA does not. Both RNAs are replicated by the translated Qβ replicase in the reconstituted translation system of E. coli. (B) Replication-dilution cycle for a long-term replication experiment. The host RNA is encapsulated in water-in-oil droplets with ~ 2 μm diameter. The parasitic RNA spontaneously appears. (1) The droplets are incubated at 37°C for 5 hr for translation and replication. (2) Eighty percent of the droplets are removed and (3) diluted with new droplets containing the translation system (i.e. five-fold dilution). (4) Diluted droplets are vigorously mixed to induce fusion and division among the droplets. We repeated this cycle for 120 rounds. The reaction volume was 1 mL, with 1% aqueous phase, corresponding to ~ 108 droplets.
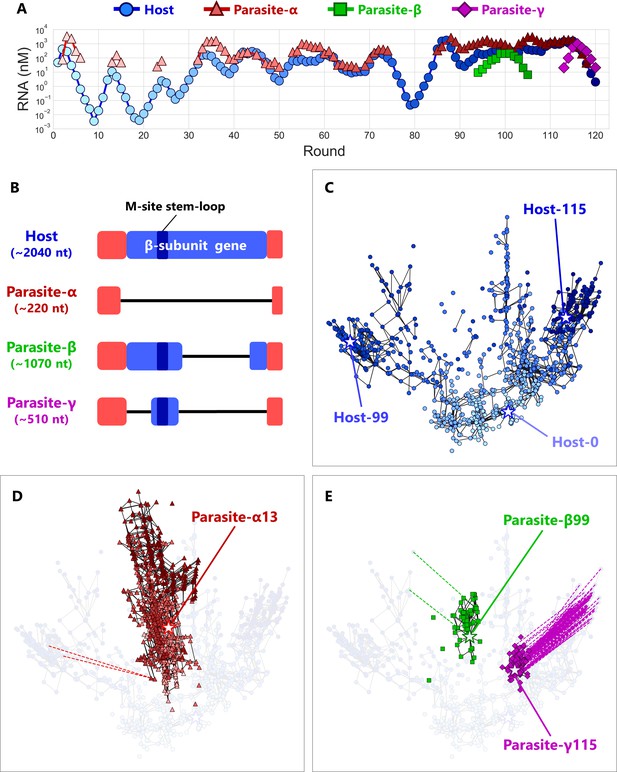
Coevolutionary dynamics of host and parasitic RNAs.
(A) Population dynamics of the host and parasitic RNAs during a long-term replication experiment. In the regions without points, parasitic RNA concentrations were under the detection limits (<30 nM) of the gel analysis. Three different parasitic species (α, β, and γ) are classified based on their sizes. (B) Schematic representation of the sequence alignments of the host and parasitic RNA species. The terminal regions (red) of all the RNA species are derived by the replicase MDV-1 (Mills et al., 1973), from a small replicable RNA. The β-subunit encoding regions are shown in blue, and the branched stem-loop of the M-site, one of the binding sites for Qβ replicase, is also indicated. Deleted regions are shown using black lines. (C, D, E) 2D maps of the dominant RNA genotypes for the host RNA (C), parasite-α (D), and parasite-β and parasite-γ (E). The top 90 dominant genotypes were plotted for each round. A point represents each genotype. The color depths are consistent with those in (A). Black lines connect pairs of points one Hamming distance apart in the same RNA species. A broken line connects a pair of points zero Hamming distance apart (perfect match) in the different RNA species, ignoring the large deletion between host and parasitic RNAs. Stars represent the genotypes of the evolved RNAs used for the competitive replication assay shown in Figure 4A. The original host RNA is Host-0. Round-by-round data are shown in Figure 3.
-
Figure 2—source data 1
Read numbers of deep sequencing.
Note that the coverage is 100% for all the reads.
- https://cdn.elifesciences.org/articles/56038/elife-56038-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Sequence data file after the alignment with the original host sequence, used to identify the 74 dominant mutations.
- https://cdn.elifesciences.org/articles/56038/elife-56038-fig2-data2-v1.zip
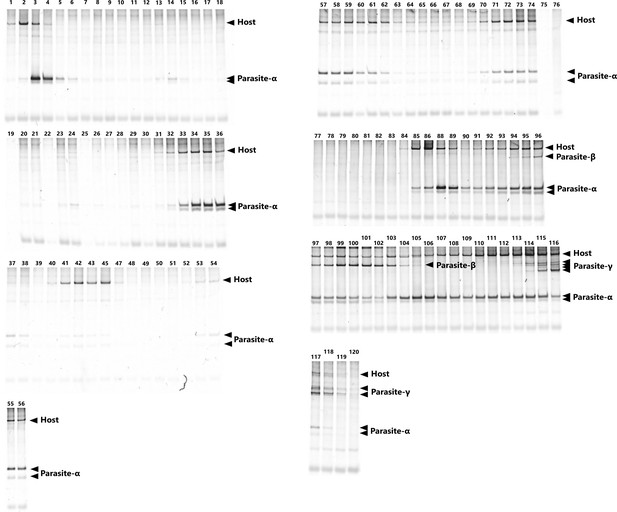
The native polyacrylamide gel electrophoresis of the RNA mixture during the long-term replication experiment.
The numbers above the gels indicate the sampled rounds. The parasitic RNAs exhibit multiple bands due to structural heterogeneity.
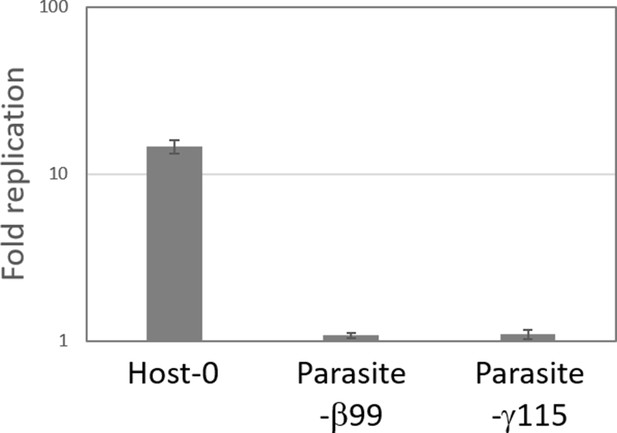
Replication of Parasite-β99 and Parasite-γ115 without host species.
The RNA replication reactions were performed with 10 nM of each RNA for 5 hr, and RNA concentration was measured by sequence-specific RT-qPCR. Error bars represent standard errors of three independent competition assays. Host-0 was also replicated for comparison.
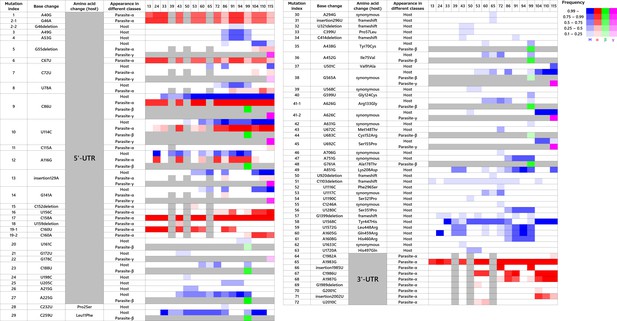
Dominant mutations and fixation dynamics among host and parasitic RNAs.
The base numbers are determined based on the single reference sequence (shown in Supplementary file 1), which contains all dominant insertions (129A, 296U, 1985T, and 2002T) in the original host sequence (Host-0). α, β, and γ in the ‘Appearance in different species’ are shorthand expressions for the parasite-α, -β, and -γ. The numbers above color boxes represent rounds of the long-term replication experiment. The host, parasite-α, -β, and -γ RNAs each corresponds to blue, red, green, and purple color. The intensity of colors represents fixation frequency. Gray boxes represent the rounds where parasitic RNAs were not recovered and sequenced.
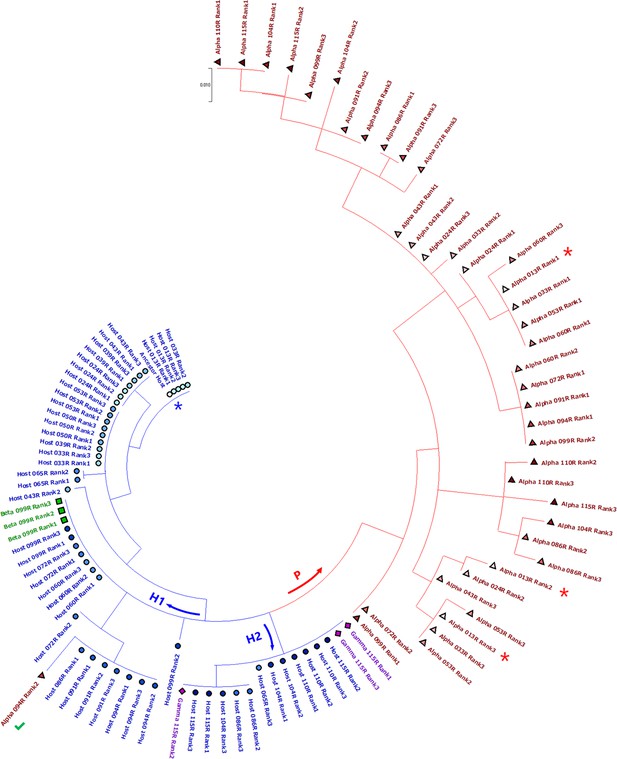
Phylogenic analysis of the host and parasite RNAs.
Top three most frequent of host and parasite sequences in all the sequenced rounds are shown. Αlpha, Βeta, and Gamma each represents parasite-α, parasite-β, and parasite-γ. The shapes and colors of markers correspond to those in Figure 2. The ancestral host and parasite-α of the earliest sequenced round (round 13) are marked with blue and red asterisks, respectively. A green tick indicates parasite-α that probably appeared by the deletion event of a host RNA in a later round. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
-
Figure 2—figure supplement 4—source data 1
Alignment data used for Figure 2—figure supplement 4.
- https://cdn.elifesciences.org/articles/56038/elife-56038-fig2-figsupp4-data1-v1.txt
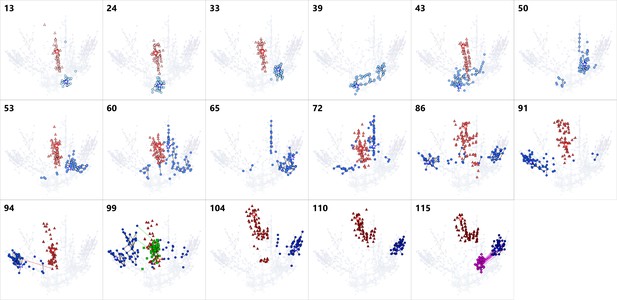
Series of snapshots of dominant RNA genotypes on 2D maps for the host RNA, parasite-α, -β and -γ.
The upper-left numbers indicate the round. The top 90 dominant genotypes of each RNA species were plotted for each round. A point represents each genotype. The colors of points are consistent with Figure 2, the host (blue), parasite-α (red), -β (green), and -γ (purple). A star in each figure represents the most frequent genotype of the host (blue), parasite-α (red), -β (green), and -γ (purple). Black lines connect pairs of points one Hamming distance apart in the same RNA species. A broken line connects a pair of points zero Hamming distance apart in the different RNA species, ignoring the large deletion, which represent a plausible generation route of each parasite. Colors of the broken lines correspond to the host and parasite-α (red), the host and parasite-β (green), and the host and parasite-γ (purple). Parasite-α is not shown in the round-39, 50, and 65 because they could not have been recovered and sequenced.
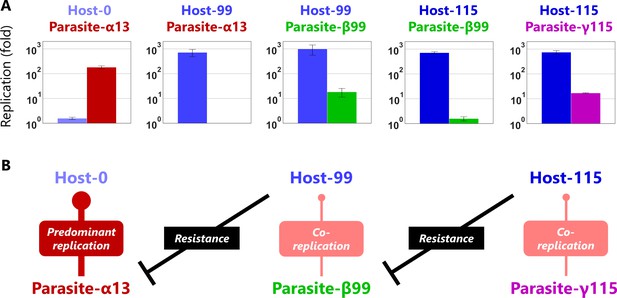
Evolutionary arms races between host and parasitic RNAs.
(A) Competitive replication assays of each pair of the evolved host and parasitic RNA clones. The RNA replication reactions were performed with 10 nM of the host and parasitic RNAs for 3 hr, and each concentration was measured by sequence-specific RT-qPCR. Error bars represent standard errors of three independent competition assays. (B) Schematic representation of the host-parasite relationships among the RNA clones.
-
Figure 4—source data 1
Dominant mutations in Host-99 and Host-115.
Host-99 and Host-115, which exhibited distinct parasite resistance (Figure 4), have very different mutation sets with only three redundant dominant mutations each other. Mutation indexes correspond to those in Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/56038/elife-56038-fig4-data1-v1.xlsx
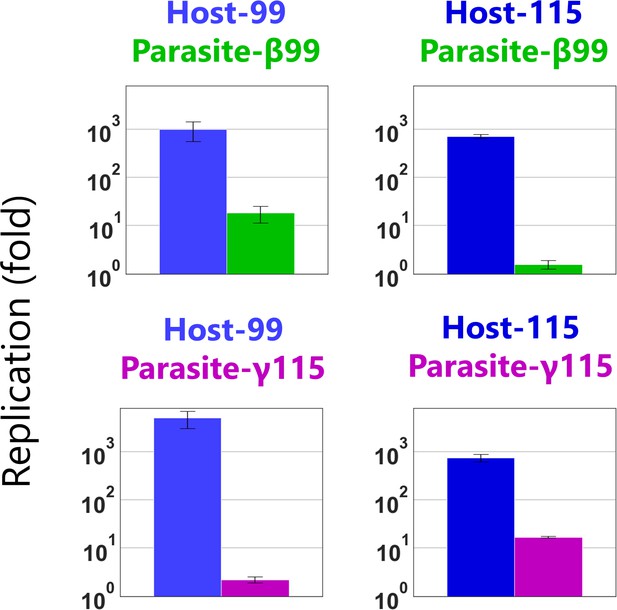
Combinatorial competitive replication assay of Hosts-99 and −115 with Parasites-β99 and -γ115.
The RNA replication reactions were performed with 10 nM of the host and parasitic RNAs for 3 hr and each concentration was measured by sequence-specific RT-qPCR. The three results (Host-99 vs parasite-β99, Host-115 vs parasite-β99, and Host-115 vs parasite-γ115) are the same as those in Figure 4. Error bars represent standard errors of three independent competition assays.
Additional files
-
Supplementary file 1
Clone sequence, primer list, the dominant genotypes and their frequencies.
- https://cdn.elifesciences.org/articles/56038/elife-56038-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56038/elife-56038-transrepform-v1.docx





