Growth-factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development
Figures
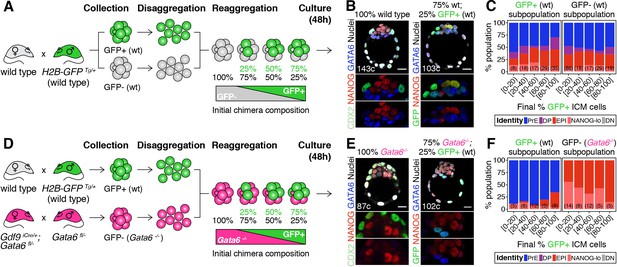
Cell fate decisions in the ICM of the blastocyst are made at the population level.
(A) Experimental design to test independence of cell fate decisions in the ICM. Wild type embryos (H2B-GFP+ or GFP-) were collected at the 8-cell stage, disaggregated into single cells or clumps, re-aggregated in the combinations indicated and allowed to develop in culture for 48 hr, until the late blastocyst stage (equivalent to ~4.5 days post-fertilization). For further experimental details and information on alleles, see Materials and methods. (B) Optical cross-sections through representative immunofluorescence images of a non-chimeric, wild-type control (100% wt) and a chimera made with 75% GFP-; 25% GFP+ cells (all wild type). Magnifications of the ICM are shown below for each marker. (C) Stacked bar plots showing the lineage distribution of GFP+ (left) and GFP- (right) wild-type ICM cells in embryos, stratified by the final % of H2B-GFP+ ICM cells, as indicated on the x-axis. Number of embryos in each group is indicated in parentheses. (D) Experimental design to test independence of cell fate decisions in the ICM. Wild-type embryos (H2B-GFP+) and Gata6−/− embryos were collected at the 8-cell stage, disaggregated into single cells or clumps, re-aggregated in the combinations indicated and allowed to develop in culture for 48 hr, until the late blastocyst stage (equivalent to ~4.5 days post-fertilization). For further experimental details and information on alleles, see Materials and methods. (E) Optical cross-sections through representative immunofluorescence images of a non-chimeric, Gata6−/− control (100% Gata6−/−) and a chimera made with 75% Gata6−/−; 25% GFP+ (wt) cells. Magnifications of the ICM are shown below for each marker. (F) Stacked bar plots showing the lineage distribution of GFP+ (wt, left) and GFP- Gata6−/− (right) ICM cells in embryos, stratified by the final % of H2B-GFP+ ICM cells, as indicated on the x-axis. Number of embryos in each group is indicated in parentheses. Color coding is indicated. All embryos labeled for NANOG (red), GATA6 (blue) and either CDX2 (controls) or GFP (chimeras) (green). All optical cross-sections are 5 µm maximum intensity projections. Total cell counts are indicated for each embryo within the merged images. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast, NANOG-lo: low NANOG epiblast, DN: Double Negative (for NANOG and GATA6). Scale bars = 20 µm.
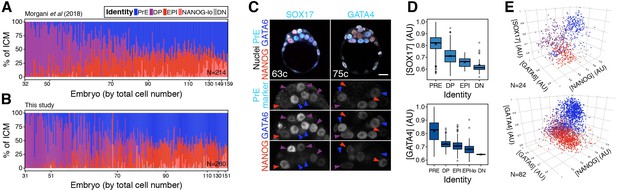
Cell type classification in the ICM of the mouse blastocyst.
(A) Stacked bar plots showing progression of lineage composition in the ICM of wild type blastocysts fixed upon collection analyzed in Morgani et al., 2018b. ICM cell types were re-assigned using hierarchical clustering, as described in the Materials and methods. (B) Stacked bar plots showing progression of lineage composition in the ICM of wild-type blastocysts fixed upon collection generated in this study. Color coding for cell types is indicated. Each bar represents the ICM of a single embryo. Embryos are arranged by ascending total cell number (from 32 up to >150 cells, comprising all the stages of blastocyst development). N indicates number of embryos in the plot. (C) Optical cross-sections through representative immunofluorescence images of blastocysts stained for NANOG (red), GATA6 (blue) and either SOX17 or GATA4 (cyan), as indicated. Arrowheads point at cells classified as PrE (blue), epiblast (red) or DP (purple). (D) Average SOX17 and GATA4 levels in each ICM cell type in a cohort of 24 embryos (SOX17) and 82 embryos (GATA4). Values are natural log re-scaled to 0–1 for each dataset. (E) 3D scatterplots for the three markers shown in (C, D). Each spot represents one cell. Spots are color-coded for cell identities as indicated in (A). Cells were classified using only relative NANOG and GATA6 levels, as described in the Materials and methods. In all box plots, whiskers span 1.5x the inter quartile range (IQR), open circles represent outliers (values beyond 1.5x IQR) and cross represents the arithmetic mean. All optical cross-sections are 5 µm maximum intensity projections. Total cell counts are indicated for each embryo within the merged images. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast, NANOG-lo: low NANOG epiblast, DN: Double Negative (for NANOG and GATA6). Scale bars = 20 µm.
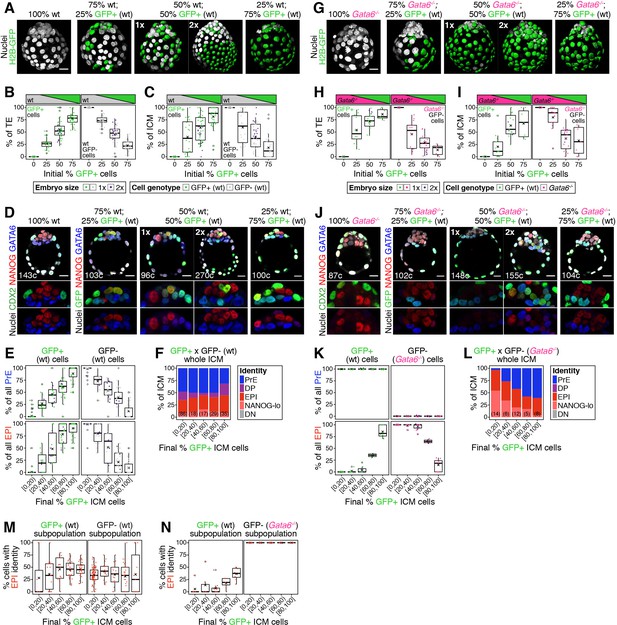
Cell fate decisions in the ICM of the blastocyst are made at the population level.
(A) Maximum intensity projections of all nuclei of a series of representative wild-type control and wt ↔ GFP+ chimeras made with increasing % of wild type GFP+ cells, as described in Figure 1A. 1x indicates normal size embryo, resulting from aggregating 8 cells, 2x indicates double-sized embryo, resulting from aggregating 2x intact 8-cell stage embryos. (B) Box plots showing the relative contribution of GFP+ (green) and GFP-, wild-type (gray) cells to the embryo for each of the initial % of GFP+ cells, as shown in (A) Dark green spots represent double embryos (2x). (C) Box plots showing the relative contribution of GFP+ (green) and GFP-, wild-type cells (gray) to the ICM for each of the initial % of GFP+ cells, as shown in (A). (D) Optical cross-sections through embryos shown in (A) and immunostained for the markers indicated. (E) Box plots showing the relative contribution of GFP+ (green) and GFP-, wild type cells (gray) to each the PrE and the epiblast. Embryos are grouped by the final fraction of GFP+ ICM cells, as indicated on the x-axis. (F) Stacked bar plots showing the ICM composition in wt ↔ GFP+ chimeras, grouped by the final fraction of GFP+ ICM cells, as in (E) and as indicated on the x-axis. Number of embryos in each group is indicated in parentheses. Color coding is indicated. (G) Maximum intensity projections of all nuclei of a series of representative Gata6−/− control and Gata6−/− ↔ GFP+ chimeras made with increasing % of GFP+ (wt) cells, as described in Figure 1D. 1x indicates normal size embryo, resulting from aggregating 8 cells, 2x indicates double-sized embryo, resulting from aggregating 2x intact 8 -cell stage embryos. (H) Box plots showing the relative contribution of GFP+ (green, wt) and GFP-, Gata6−/− cells (magenta) to the embryo for each of the initial % of GFP+ cells, as shown in (G) Dark green spots represent double embryos (2x). (I) Box plots showing the relative contribution of GFP+ (green, wt) and GFP-, Gata6−/− cells (magenta) to the ICM for each of the initial % of GFP+ cells, as shown in (G). (J) Optical cross sections through embryos shown in (G) and immunostained for the markers indicated. (K) Box plots showing the relative contribution of GFP+ (green, wt) and GFP-, Gata6−/− (magenta) cells to each the PrE and the epiblast for embryos grouped by the final fraction of GFP+ ICM cells, as in (E) and as indicated on the x-axis. (L) Stacked bar plots showing the ICM composition in Gata6−/− ↔ GFP+ chimeras, grouped by the final fraction of GFP+ ICM cells, as in (K) and as indicated on the x-axis. Number of embryos in each group is indicated in parentheses. Color coding is indicated. (M) Box plots showing the % of GFP+ or GFP- cells (both wild type) that adopt EPI/NANOG-lo identity in wt ↔ GFP+ chimeras, grouped by the final fraction of GFP+ ICM cells, as in (E, K) and as indicated on the x-axis. (N) Box plots showing the % of GFP+ (wt) or GFP-, Gata6−/− cells that adopt EPI/NANOG-lo identity in Gata6−/− ↔ GFP+ chimeras, grouped by the final fraction of GFP+ ICM cells, as in (M). In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. All embryos labeled for NANOG (red), GATA6 (blue) and either CDX2 (controls) or GFP (chimeras) (green). All optical cross-sections are 5 µm maximum intensity projections. Total cell counts are indicated for each embryo within the merged images. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast, NANOG-lo: low NANOG epiblast, DN: Double Negative (for NANOG and GATA6). Scale bars = 20 µm.
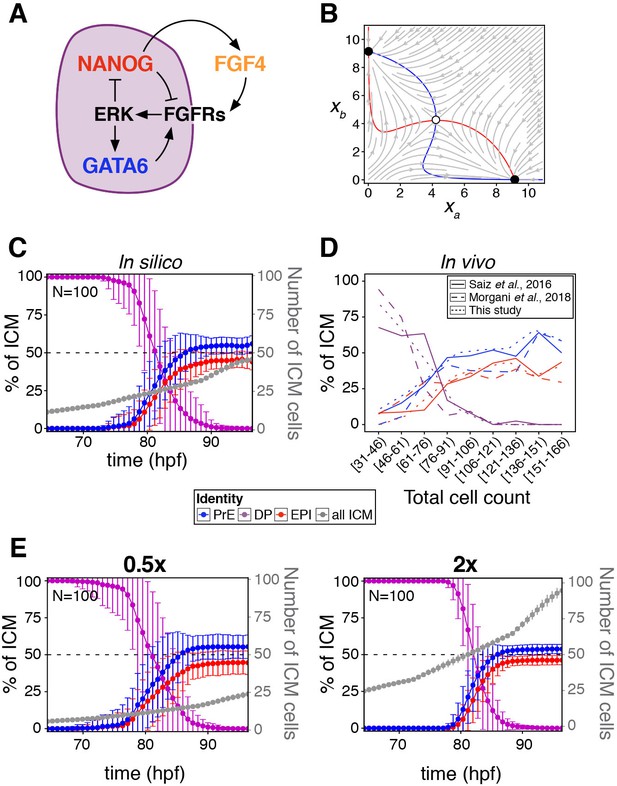
A minimal model of cell fate decisions solely mediated by growth factor signaling explains robust lineage specification in the ICM.
(A) Diagram of our proposed model of molecular control of cell fate in the ICM. (B) Phase plane of our model for the case of a two-cluster state. Each axis shows the NANOG levels in one of the two clusters. Red and blue lines show the nullclines of the system (Box 1); the gray arrows depict typical trajectories of the system; and black (and white) circles correspond to stable (and unstable) equilibria of the two-cluster system. (C) Lineage dynamics in in silico simulations of ICM development using our proposed model. (D) In vivo ICM lineage dynamics from three experimental datasets, as indicated. (E) Lineage dynamics in in silico simulations of scaling experiments (to be compared with the experimental results of Saiz et al., 2016b). Absolute ICM size was modified to 0.5 or 2x the normal size, as indicated.
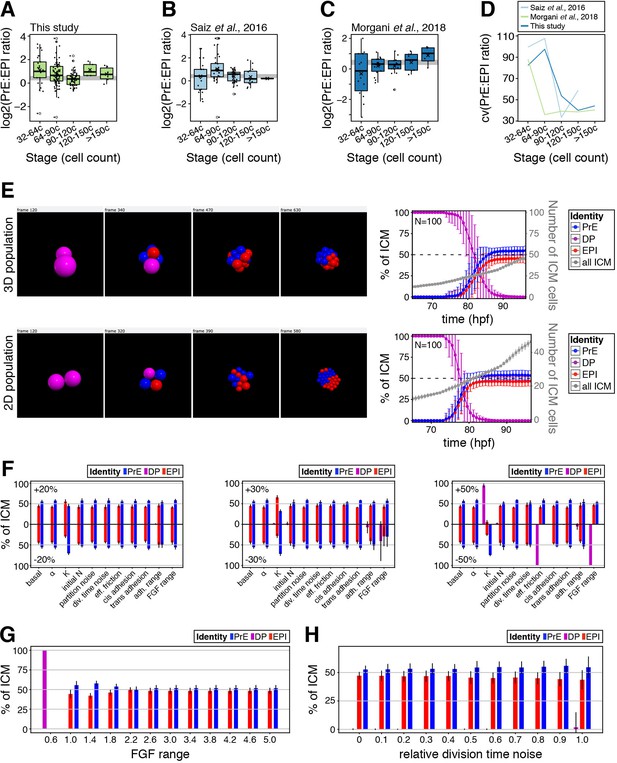
A minimal model of cell fate decisions solely mediated by growth factor signaling explains robust lineage specification in the ICM.
(A–C) Box plots showing the ratio of PrE to epiblast cells in embryos at sequential stages of blastocyst development for embryos collected in this study (A), those analyzed in Saiz et al., 2016b (B) and those analyzed in Morgani et al., 2018b (C) (shown also in Figure 1—figure supplement 1A–B). In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Crosses indicate the arithmetic mean and each dot represents one embryo. (D) Coefficient of variation of the PrE:epiblast ratio over developmental time in the datasets shown in (A), (B) and (C), as indicated. (E) Comparison between the behavior of our agent-based biochemical model in three (top) and two (bottom) dimensions. Filmstrips of the dynamics are shown at the left, and the time evolution of the cell-fate ratio is shown at the right. (F) Sensitivity analysis showing the cell-fate composition of the simulated embryos at the end point of the simulation, when perturbing several of the model parameters, as shown in the x-axis labels. Three different relative perturbation levels are shown. (G) Final cell-fate composition in the model as a function of the range of FGF signaling in the embryo. (H) Final cell-fate composition in the model for increasing noise in the time of cell division.
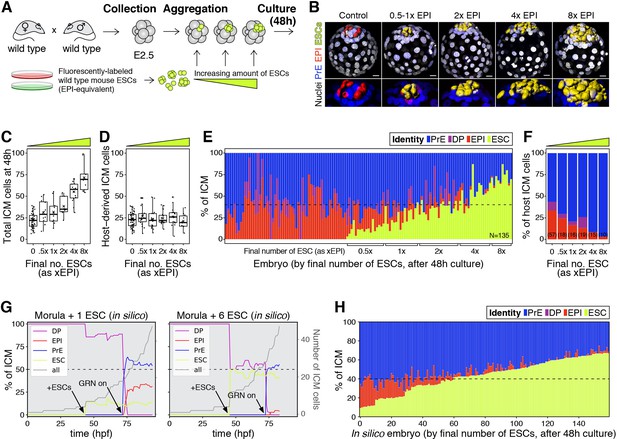
The lineage composition of the ICM is robust to expansion of the epiblast.
(A) Experimental design. 8- or 8–16 cell stage embryos (2.5 days post-fertilization) were recovered from CD1 (wild type) females crossed with CD1 males. Embryos were denuded, aggregated with clumps of fluorescently labeled ESCs and cultured for 48–56 hr, until the late blastocyst stage (equivalent to ~4.5 days post-fertilization). (B) 3D renders of a series of control (no ESCs) and chimeras, generated as indicated in (A), carrying increasing amounts of ESCs (indicated as EPI-equivalent size (x EPI)). Control epiblast and ESCs in chimeras are highlighted as computer-rendered volumes and color coded as indicated. GATA6+ PrE is shown in blue, NANOG+ host epiblast is shown in red where applicable. (C) Box plot showing the total size of the ICM (host-derived + ESCs) at the end of the experiment in each group of embryos, as defined by the size of the ESC compartment. (D) Box plot showing the size of the host-derived ICM component at the end of the experiment in each group of embryos, as in (C). (E) Stacked bar plot showing the relative ICM composition at the end of the experiment for all embryos analyzed. Each bar represents the ICM of one embryo, ordered by increasing absolute number of ESCs at the end of the experiment. Dashed line indicates the normal ratio of 60% PrE:40% epiblast found in intact wild-type embryos. Number of embryos analyzed is indicated (N). Brackets on x-axis indicate the number of ESCs in those embryos, relative to the size of the average wt control epiblast (xEPI). (F) Stacked bar plot showing the relative contribution of host cells to each of the ICM lineages in each group of embryos. Yellow wedge represents the increasing amount of ESCs in each group. Number of embryos in each group is indicated in parentheses. (G) Growth curves showing lineage dynamics in in silico simulations of the aggregation experiments shown in (A). Left Y-axis and curves for each lineage indicate relative size (as % of ICM). Right Y-axis and gray curves indicate total number of ICM cells (including ESCs). (H) Stacked bar plot showing the relative ICM composition at the end of the experiment in in silico simulations of the experiments shown in (A) and (E). Each bar represents the ICM of one simulated embryo (i.e., a single iteration), and bars are arranged by increasing absolute number of ESCs at the end of the simulation, as in (E). Dashed line indicates the normal ratio of 60% PrE:40% epiblast found in intact wild-type embryos. Color coding is indicated for (E, G, H). In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Crosses indicate the arithmetic mean and each dot represents one embryo. Yellow wedges represent the increasing amount of ESCs in each group. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast, ESC: embryonic stem cell. Scale bars = 20 µm.
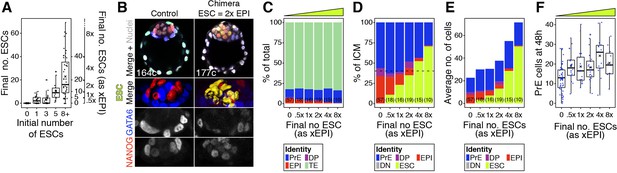
The lineage composition of the ICM is robust to expansion of the epiblast.
(A) Box plots indicating ESC contribution to chimeras. Number of ESCs aggregated with morulae is shown against the final number of ESC in the chimeric embryo after 48 hr in culture. Right y axis shows epiblast-equivalent size bins used to categorize chimeric embryos (xEPI). (B) Optical cross-sections through representative immunofluorescence images of a control and chimera with a 2xEPI-equivalent ESC compartment. Embryos are labeled for NANOG (red) and GATA6 (blue) to identify all ICM cell types. ESCs are shown in yellow. Lower panels show magnifications of the ICM, with all markers overlaid, for each individual marker as grayscale and for ESCs as 3D surface renders. Total cell count for each embryo is shown within the merged panel. (C) Stacked bar plot showing the overall lineage distribution of host cells in each group of embryos shown in Figure 3, grouped by the final size of the ESC compartment (also represented by the yellow wedge) (D) Stacked bar plot showing the average relative ICM composition of each group of chimeras shown in (C). (E) Stacked bar plot showing the average number of cells in each ICM population for each group of embryos shown in (D). (F) Box plot showing the size of the PrE at the end of the experiment in each group of embryos. Yellow wedge represents the increasing amount of ESCs in each group. Color coding is indicated. Number of embryos in each group is indicated in parenthesis. In all box plots, whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. All optical cross-sections are 5 µm maximum intensity projections. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast, ESC: embryonic stem cell. Scale bars = 20 µm.
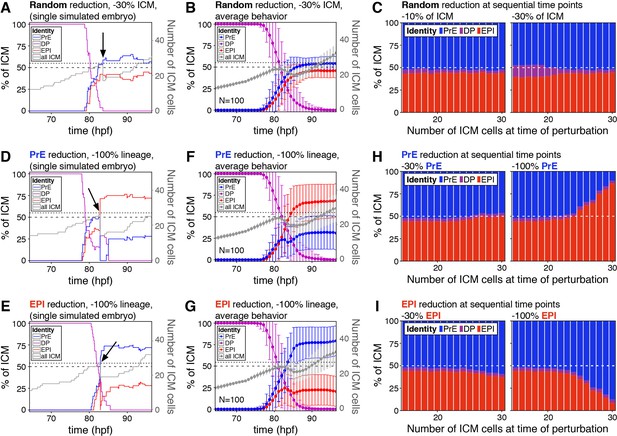
The lineage composition of the ICM is robust to in silico reduction of lineage size.
(A) Growth curves for each ICM lineage after simulation of a 30% reduction in ICM size (3 PrE, 3 DP and three epiblast cells removed from a 27-cell ICM) using our model described in Figure 2. Arrow indicates the time of cell elimination. Lines are color-coded for each lineage, as indicated and represent relative lineage size (scale on the left Y-axis). Grey line indicates the absolute size of the ICM, as shown on the right Y-axis. Dotted line indicates 27 ICM cells, the point at which cells were eliminated. Dashed line indicates 50% of the ICM, for reference. (B) Growth curves as those in (A) showing the average behavior for 100 simulations. Error bars indicate the standard deviation. (C) Stacked bar plots showing the final ICM composition after simulating the elimination of 10% (left) or 30% (right) of ICM cells at sequential points in embryo development. Developmental stage at the time of cell elimination is indicated on the x-axis as number of ICM cells (15–30 ICM cells, equivalent to ~50–100 total cells). Each bar represents the result of 100 simulations. (D, E) Growth curves for each ICM lineage after simulation of a 100% reduction in PrE (D) or epiblast (E), when the ICM reaches 27 cells, as shown in (A) and indicated by the arrow. Lines are color-coded for each lineage, as indicated and represent relative lineage size (scale on the left Y-axis). Grey line indicates the absolute size of the ICM, as shown on the right Y-axis. Dotted line indicates 27 ICM cells. Dashed line indicates 50% of the ICM, for reference. (F, G) Growth curves as those in (D, E) showing the average behavior for 100 simulations of PrE (F) and epiblast (H) reduction. Error bars indicate the standard deviation. (H, I) Stacked bar plots showing the final ICM composition after simulating the elimination of 30% (left) or 100% (right) of the PrE (H) or the epiblast (I) at sequential points in embryo development. Developmental stage at the time of cell elimination is indicated on the x-axis as number of ICM cells (15–30 ICM cells, equivalent to ~50–100 total cells). Each bar represents the result of 100 simulations. Color coding is indicated. hpf: hours post-fertilization, PrE: Primitive Endoderm, EPI: epiblast, DP: double positive.
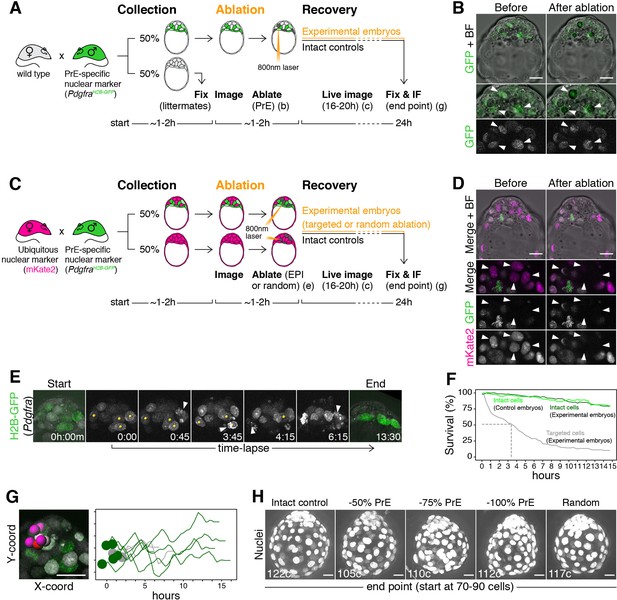
Laser ablation enables alteration of lineage size with high spatiotemporal control in mouse embryos.
(A) Experimental design for PrE ablation. Blastocysts recovered from CD1 (wild type) females crossed with PdgfraH2B-GFP/+ or PdgfraH2B-GFP/+; R26:CAG:3x-nls-mKate2Tg/Tg males were sorted for GFP. GFP- embryos were fixed as reference littermates to estimate the developmental stage of the entire litter. GFP+ embryos were used for the experiment and subject to ablation of different amounts of PrE cells (GFP+) followed by 16–20 hr of live imaging to visualize the response to cell ablation, followed by 8–4 hr of in vitro culture (for a total of 24 hr). (B) Representative images of live GFP+ embryos before and after ablation of PrE cells. Bottom panels show magnifications of the ICM with GFP alone on grayscale, as indicated. Arrowheads point at targeted PrE cells. (C) Experimental design for epiblast ablation. Blastocysts were recovered from R26:CAG:3x-nls-mKate2Tg/Tg females crossed with PdgfraH2B-GFP/+; R26:CAG:3x-nls-mKate2Tg/Tg males. GFP-, mKate2+ embryos were used as either intact or random ablation controls. GFP+, mKate2+ embryos were used for the experiment and subject to ablation of different amounts of epiblast cells (GFP-) or PrE cells (GFP+), followed by 16–20 hr of live imaging to visualize the response to cell ablation, followed by 8–4 hr of in vitro culture (for a total of 24 hr). (D) Representative images of live GFP+ embryos before and after ablation of epiblast cells. Bottom panels show magnifications of the ICM, for both markers together and each of them on grayscale, as indicated. Arrowheads point at targeted epiblast cells. (E) Still images of a representative embryo in the hours after ablation. See also Video 3. Yellow spots on grayscale images mark targeted cells. Arrowheads point at cell death of each targeted cell. All images are timestamped as h:mm. (F) Survival curves for targeted (gray) and intact cells in both experimental (dark green) and intact control embryos (light green). Dashed line marks half-life of targeted cells (~3 hr) (G) Survival of intact cells neighboring targeted cells. Image shows selected ICM cells (intact DP and epiblast cells, color coded, and targeted PrE cells, gray). Graph shows X-Y coordinates of cells shown in picture and survival (hours) for each cell. Time scale for each cell is shifted based on their initial X position, for visualization purposes. (H) Maximum intensity projections of representative embryos fixed after 24 hr in culture, showing all nuclei over bright field image. Treatment was done at the 70–90 cell stage and is indicated above. Total cell count for each embryo is shown within each image. PrE: Primitive Endoderm, EPI: Epiblast. Scale bars = 20 µm.
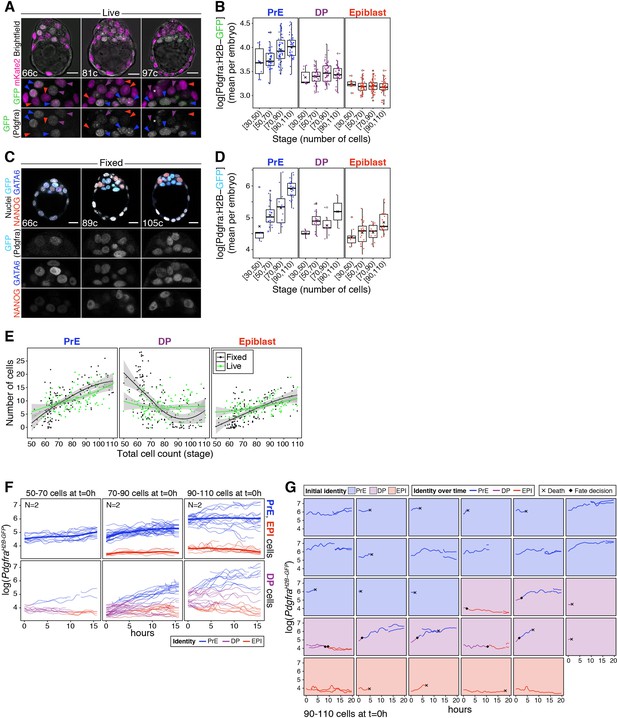
Lineage composition and PrE reporter dynamics in fixed and live embryos.
(A) Optical cross-sections through representative live blastocysts at sequential developmental stages (total cell count is indicated within each image). Lower panels show magnifications of the ICM for both markers, as indicated, and for GFP alone in grayscale. Colored arrowheads point at nuclei considered to be PrE (blue arrowheads), DP (purple arrowheads) or epiblast (red arrowheads) based on GFP level. (B) Box plots showing the average level of GFP (Pdgfra) per embryo, in each ICM cell type, for each developmental stage considered. (C) Optical cross-sections through representative immunofluorescence images of reference PdgfraH2B-GFP/+ littermates fixed upon collection and labeled for NANOG (red) and GATA6 (blue) to identify ICM cell types. Lower panels show ICM magnifications for each marker in grayscale, as indicated. (D) Box plots showing the average level of GFP (Pdgfra) per embryo, in each ICM cell type (as determined by NANOG and GATA6 expression), for each developmental stage considered – for embryos like those in (C). (E) Growth curves for each ICM population over time. Black lines and dots show numbers corresponding to fixed samples, where cell identities were assigned automatically based on relative NANOG and GATA6 levels, as described in the Materials and methods. Green lines and dots show numbers corresponding to live samples, where cell identities were assigned manually based on GFP (Pdgfra) levels alone, as described in the Materials and methods. Curves are local regression lines for each subset of data, fitted using the LOESS method. (F) Temporal dynamics of cell fate specification in live embryos. Expression levels of the PrE reporter allele PdgfraH2B-GFP (Hamilton et al., 2003) are shown over time for individual ICM cells in intact embryos at sequential stages of development. Reporter expression was assessed using time lapse imaging over a 15 hr time window. Top panels show dynamics of reporter expression in cells classified as PrE or epiblast at the beginning of the movie. Smoothing curves for each PrE and epiblast are shown as thicker lines and color coded. Bottom panels show dynamics of reporter expression in cells classified as DP (progenitors) at the beginning of the movie. DP cells become PrE or epiblast over the course of the movie, as determined by PdgfraH2B-GFP expression. (G) Pdgfra expression dynamics for individual cells in one embryo from (F) imaged from the 90–110 cell stage onwards. Panels are color coded for the initial identity of the cell. Traces are color coded for identity over time. Acquisition of PrE or epiblast identity is denoted by a black diamond. Black cross indicates cell death. In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast. Scale bars = 20 µm.
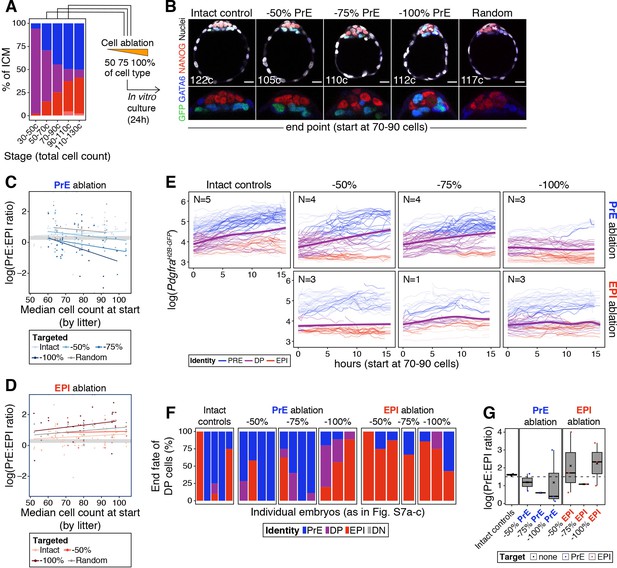
The cell fate choice of uncommitted ICM progenitors is dictated by lineage size.
(A) Stacked bar plot showing ICM composition at sequential stages of blastocyst development. Embryos at each of these stages were subject to laser ablation of different fractions of either the PrE or the epiblast and allowed to recover in vitro for 24 hr (see Figure 5A,C). (B) Optical cross sections through representative immunofluorescence images of embryos subject to ablation at the 70–90 cell stage and fixed at the end of the experiment (24 hr later) (same embryos as in Figure 5G). Embryos are labeled for NANOG (red) and GATA6 (blue) to identify all ICM cell types. H2B-GFP, indicating Pdgfra expression, is shown in green where applicable. Lower panels show magnifications of the ICM Treatment for each embryo is indicated over each image. (C) PrE:epiblast ratio (shown as natural logarithm) at the end of the experiment (24 hr) in embryos where fractions of the PrE were eliminated at sequential stages of blastocyst development, as indicated on the x-axis. Shades of blue indicate the magnitude of the reduction in the PrE. Intact controls are embryos in which no cells were killed, Random controls are embryos in which randomly chosen ICM cells were killed without knowing their identity, in equivalent numbers to the −100% group. (D) PrE:epiblast ratio (shown as natural logarithm) at the end of the experiment (24 hr) in embryos where fractions of the epiblast were eliminated at sequential stages of blastocyst development, as indicated on the x-axis. Shades of red indicate the magnitude of the reduction in the epiblast. (E) Dynamics of PdgfraH2B-GFP expression in progenitor cells (DP) of experimental embryos targeted at the 70–90 cell stage. Each line represents one cell. Color coding indicates cell identity, as inferred from reporter expression (see Methods). Number of embryos per plot is indicated. Cells classified as PrE or epiblast at the beginning of the experiment are shown as color-coded semi-transparent lines behind progenitor cells, for reference. Smoothing curves for Pdgfra expression in progenitor cells are shown as thick purple lines. Fraction of the PrE or epiblast eliminated is indicated above each panel, lineage targeted is indicated to the right of each panel. (F) Stacked bar plots showing the final identity adopted by progenitor (DP) cells in each of the embryos plotted in (E). (G) Box plots showing the PrE:epiblast ratio (shown as natural logarithm) at the end of the movie, in embryos where all or most of the ICM cells could be tracked after cell ablation at the 70–90 cell stage (subset of embryos shown in (E) and (F)). Compare to panels (C) and (D). Treatment is indicated on the x-axis. In box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast. Scale bars = 20 µm.
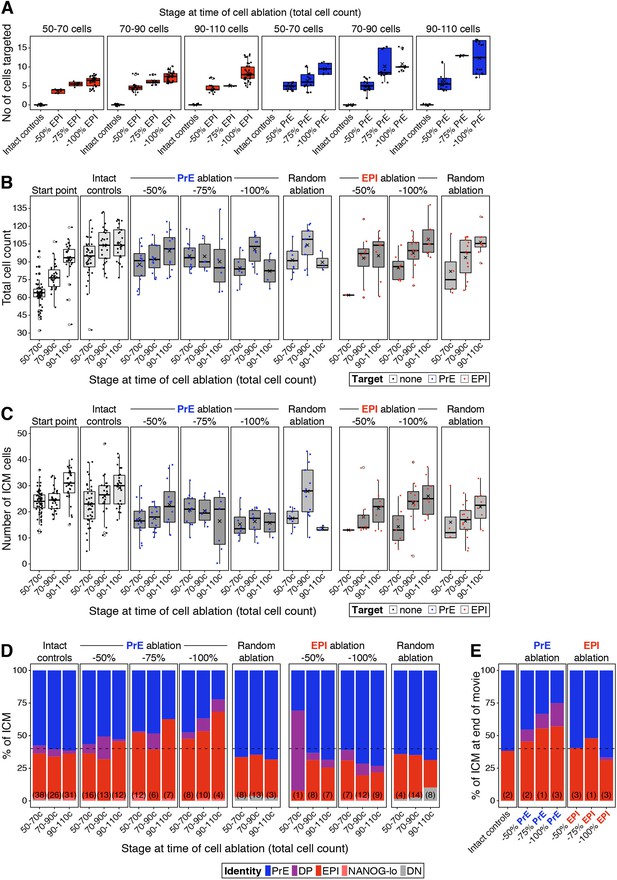
Population sizes and lineage composition in ablation experiments.
(A) Box plots showing the absolute number of cells targeted in ablation experiments, at each developmental stage. The experimental group is indicated on the x-axis (as % of the lineage targeted. Intact controls = 0%). Each box shows embryos at one developmental stage bin, for either epiblast (red) or PrE (blue). (B) Box plots showing the total cell count of embryos in each of the experimental groups shown in Figure 6, at the end of the experiment. Embryos are binned by developmental stage at the start of the experiment, as shown on the x-axis. Start point comprises reference littermates fixed at the beginning of the experiment. Intact controls are embryos in which no cell was targeted. Random controls are embryos in which randomly chosen ICM cells were targeted, irrespective of their identity, in equivalent numbers to the −100% group for each lineage targeted (see Methods). Embryos in which the PrE or epiblast were targeted are split by the fraction of the lineage eliminated (50–100%, as indicated). (C) Box plots showing the total number of ICM cells in each of the experimental groups shown in (B). (D) Stacked bar plots showing the relative ICM composition in each of the experimental groups shown in Figure 6C–D and in (B–C) above. The number of embryos in each group is indicated in parenthesis. Cell types are color-coded as indicated. (E) Stacked bar plots showing the relative ICM composition at the end of the movie in embryos shown in Figure 6E, for each of the treatments indicated on the x-axis. Data in (E) corresponds to embryos in which ablation was performed at the 70–90 cell stage and in which all or most of the ICM cells could be tracked throughout the movie. Cell types are color-coded as indicated. In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast.
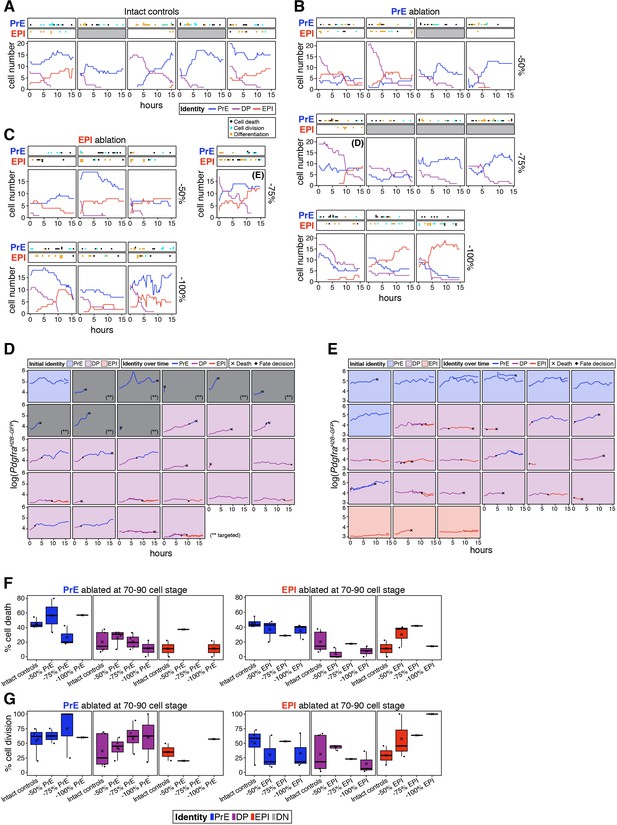
Analysis of cell behaviors in ablation experiments.
(A) Growth curves for each of the ICM populations in each of the intact embryos shown in Figure 6E. Cell types are color-coded. Top panels indicate cellular events in the PrE or epiblast populations: black dots represent cell death, cyan dots represent cell divisions, and orange dots represent a progenitor cell adopting either PrE or epiblast identity, respectively. Gray panels indicate no data is available. (B) Growth curves for each of the ICM populations in each of the embryos shown in Figure 6E in which the PrE was targeted, in the fractions indicated. (C) Growth curves for each of the ICM populations in each of the embryos shown in Figure 6E in which the epiblast was targeted. All embryos in (A–C) were manipulated at the 70–90 cell stage and live imaged for the first 15 hr of the 24 hr culture after ablation. (D) Temporal changes in Pdgfra expression and cell identity for all ICM cells tracked in an embryo in which 75% of the PrE was eliminated when it had 70–90 cells – labeled as (D) in panel B. (E) Temporal changes in Pdgfra expression and cell identity for all ICM cells tracked in an embryo in which 75% of the epiblast was eliminated when it had 70–90 cells – labeled as (E) in panel C. Each panel displays one cell. Panels are color coded for the initial identity of the cell, as indicated. Traces are color coded for identity over time. Acquisition of PrE or epiblast identity is denoted by a black diamond. Black cross indicates cell death. (F) Box plots showing frequency of cell death among intact cells of each ICM cell type, as color-coded. Each box represents one experimental group (Intact controls, or embryos in which increasing fractions of the corresponding lineage were eliminated, as indicated on the x-axis). Left plots correspond to PrE ablation, right plots to epiblast ablation, as indicated. (G) Box plots showing the frequency of cell division among intact cells of each ICM cell type, as color coded. Each box represents one experimental group (Intact controls, or embryos in which increasing fractions of the corresponding lineage were eliminated, as indicated on the x-axis). Left plots correspond to PrE ablation, right plots to epiblast ablation, as indicated. Color coding is indicated. In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast.
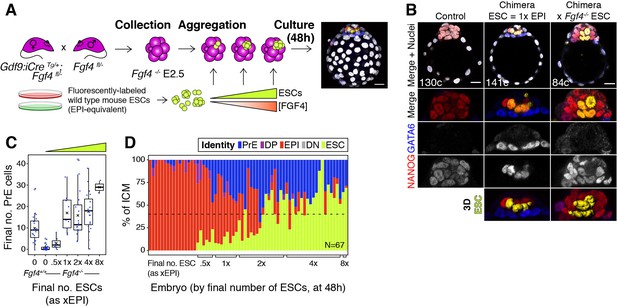
FGF4 provides the dynamic readout of lineage size that determines cell fate specification.
(A) Experimental design. Maternal zygotic (mz) Fgf4−/− Embryos were recovered from Gdf9iCre/+; Fgf4fl/− females crossed with Fgf4fl/− males at the 8cell stage (2.5 days post-fertilization). Embryos were aggregated with clumps of fluorescently labeled wild-type ESCs and cultured for 48–56 hr, until the late blastocyst stage (equivalent to ~4.5 days post-fertilization). Control embryos were allowed to develop without adding ESCs. Both chimeric and non-chimeric control embryos were fixed at the end of the culture period and labeled with lineage markers to assess ICM composition. (B) Optical cross-sections through representative chimeras carrying either wild type or Fgf4−/− fluorescently labeled ESCs (as indicated) and non-chimeric control embryos labeled for NANOG and GATA6 to identify all ICM cell types. The progeny of the introduced ESCs is labeled in yellow. Total cell count is indicated for each embryo. Lower panels show magnifications of the ICM, with all markers overlaid and for each individual marker in grayscale. Surface renders of ESC compartment within the ICM are shown below. (C) Box plots showing absolute number of PrE cells after 48 hr in wild type, control embryos (no ESCs), Fgf4−/− control embryos (no ESCs) and Fgf4−/− chimeric embryos, grouped by the size of the ESC compartment, as in Figure 3. (D) Stacked bar plot showing the relative ICM composition in individual embryos (controls or chimeras). Each bar represents the ICM of one embryo and bars are arranged by absolute number of ESCs present in the embryo. Brackets on x-axis indicate the number of ESCs in those embryos, relative to the size of the average wt control epiblast (xEPI), same groups as in (C). Color coding is indicated. All optical cross-sections are 5 µm maximum intensity projections. In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. Yellow wedges represent the increasing amount of ESCs in each group. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast, DN: Double Negative (for NANOG and GATA6), ESC: embryonic stem cell. Scale bars = 20 µm.
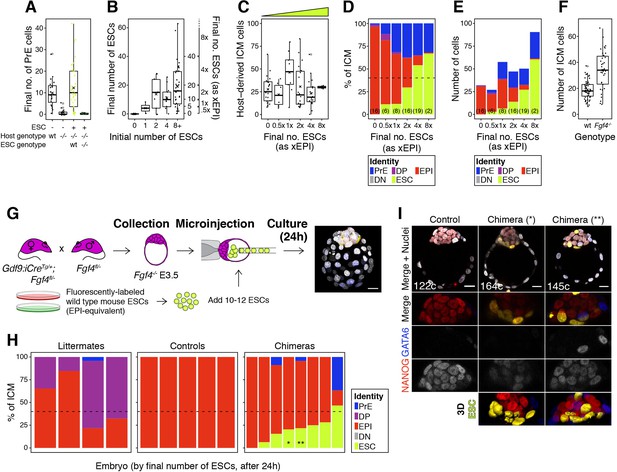
FGF4 provides the dynamic readout of lineage size that determines cell fate specification.
(A) Box plot indicating the number of PrE cells in wild type control embryos, Fgf4−/− controls, Fgf4−/− chimeras carrying wild type ESCs and Fgf4−/− chimeras carrying GFP-tagged, Fgf4−/− ESCs, as indicated. (B) Box plots indicating ESC contribution to chimeras. Number of ESCs aggregated with morulae is shown against the final number of ESC in the chimeric embryo after 48 hr in culture. Right y-axis shows epiblast-equivalent size bins used to categorize chimeric embryos. (C) Box pot showing the size of the host-derived ICM component at the end of the experiment in each group of embryos. (D) Stacked bar plots showing the average relative ICM composition of Fgf4−/− embryos carrying wild-type ESCs (shown in Figure 7D), binned by the final size of the ESC compartment. (E) Stacked bar plots showing the average number of each ICM cell type for each group of chimeras shown in (D) and Figure 7D. Number of embryos in each group is indicated in parentheses. (F) Box plot showing the ICM size of wild-type control embryos and Fgf4−/− control embryos. (G) Experimental design for blastocyst injection. Embryos were recovered from crosses equivalent to those in Figure 7A, at the mid-blastocyst stage (~60–80 cells) and 10–12 ESCs injected into the blastocyst cavity before allowing the embryos to develop for 24–30 hr in culture, until a stage equivalent to ~E4.5 days post-fertilization. (H) Stacked bar plots showing ICM composition for individual embryos like those shown in (I), as indicated. Each bar represents the ICM of one embryo and bars are arranged by absolute number of ESCs present. Stars (*, **) denote the bars corresponding to the chimeras shown in (I). Color coding is indicated. (I) Optical cross-sections through representative chimeras and control embryos cultured from the blastocyst stage and labeled for NANOG and GATA6 to identify all ICM cell types. The progeny of the introduced ESCs is labeled in yellow. These cells failed to mix with host ICM cells and tended to remain at the ICM surface, suggesting that differences in adhesion between cell types are necessary for cell mixing (see also Videos 9–10). Lower panels show magnifications of the ICM, with all markers overlaid, for each individual marker in grayscale and for ESCs as 3D surface renders. Total cell count is indicated for each embryo. All optical cross-sections are 5 µm maximum intensity projections. In all box plots whiskers span 1.5x the inter quartile range (IQR) and open circles represent outliers (values beyond 1.5x IQR). Cross indicates the arithmetic mean and each dot represents one embryo. Yellow wedges represent the increasing amount of ESCs in each group. PrE: Primitive Endoderm, DP: Double Positive (for NANOG and GATA6), EPI: Epiblast, DN: Double Negative (for NANOG and GATA6), ESC: embryonic stem cell. Scale bars = 20 µm.
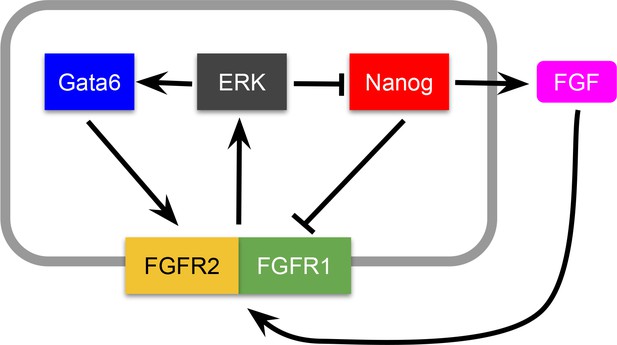
Potential molecular mechanism underlying the Epi-PrE decision.
We note, nevertheless, that the minimal model obtained below could also represent other molecular architectures of the Nanog-Gata6 circuit, and thus can be interpreted as a general representation of a growth-factor-mediated mutual inhibition.
Videos
Simulation of ICM formation and lineage specification using our minimal mathematical model.
Wild-type chimera with an ESC compartment equivalent to 2x control epiblast (added at morula stage).
Yellow surface: ESCs, blue nuclei: GATA6, white nuclei: Hoechst.
Wild-type chimera with an ESC compartment equivalent to 8x control epiblast (added at morula stage).
Yellow surface: ESCs, blue nuclei: GATA6, white nuclei: Hoechst.
Movie of experimental embryo after ablation highlighting targeted PrE cells (gray) and intact PrE cells (blue).
Movie of experimental embryo at the 67 cell stage after ablation of 50% PrE cells, highlighting all tracked cells – PrE (yellow: targeted, blue: intact), DP (magenta), epiblast (red).
Green nuclei are H2B-GFP expressed from the Pdgfra locus, white nuclei are nuclear mKate expressed from the ROSA26 locus (see text).
Movie of experimental embryo at the 97-cell stage after ablation of 50% epiblast cells, highlighting all tracked cells – PrE (blue), DP (magenta), epiblast (red).
Green nuclei are H2B- GFP expressed from the Pdgfra locus, white nuclei are nuclear mKate expressed from the ROSA26 locus (see text). Targeted epiblast cells cannot be followed because of bleaching of nuclear mKate reporter.
Fgf4- / - control blastocyst.
Red surface: epiblast (NANOG+), blue nuclei: GATA6, white nuclei: Hoechst.
Fgf4- / - chimera with an ESC compartment equivalent to 1x control epiblast (added at morula stage).
Yellow surface: ESCs, red nuclei: NANOG, blue nuclei: GATA6, white nuclei: Hoechst.
Fgf4- / - chimera with Fgf4- / - ESCs added at morula stage.
Yellow surface: ESCs, red nuclei: NANOG, blue nuclei: GATA6, white nuclei: Hoechst.
Fgf4- / - chimeras injected with ESCs at the blastocyst stage.
Yellow surface: ESCs, red nuclei: NANOG, blue nuclei: GATA6, white nuclei: Hoechst.
Fgf4-/-chimerasinjectedwith ESCs at the blastocyst stage.
Yellow surface: ESCs, red nuclei: NANOG, blue nuclei: GATA6, white nuclei: Hoechst.
Tables
Parameter values.
The parameters of the biochemical circuit (top four parameters, above the line) are given in dimensionless units. The parameters of the agent-based model (below the line) are given in arbitrary units, with time being rescaled a posteriori to match the experimental observations approximately.
| Parameter | Description | Value |
|---|---|---|
| α | maximum expression strength | 10 |
| K | inhibition threshold | 0.9 |
| N | Nanog inhibition cooperativity | 2 |
| m | Gata6 inhibition cooperativity | 2 |
| FGF coupling range factor | 1.2 | |
| initial mass | 10−6 | |
| initial radius | 5 | |
| b | effective friction coefficient | 10−6 |
| K0 | adhesion strength | 10−4 |
| adhesion strength reduction for different fates | 1.5 | |
| µ | adhesion range | 2 |
| average division time | 10 | |
| relative division time noise | 0.5 | |
| initial x | 3.0 | |
| relative partition noise | 0.01 | |
| circuit turn-on average ICM size | 20 | |
| circuit turn-on relative noise | 0.1 | |
| circuit turn-off lower factor | 0.05 | |
| circuit turn-off upper factor | 0.95 | |
| PrE fate factor | 0.2 | |
| EPI fate factor | 0.8 | |
| circuit turn-on average time (chimera sims.) | 45.0 | |
| circuit turn-on relative noise (chimera sims.) | 0.02 | |
| ESC radius (chimera sims.) | 2.0 |
Additional files
-
Supplementary file 1
List of alleles used in the study.
- https://cdn.elifesciences.org/articles/56079/elife-56079-supp1-v3.xlsx
-
Supplementary file 2
List of primers used for genotyping.
- https://cdn.elifesciences.org/articles/56079/elife-56079-supp2-v3.xlsx
-
Supplementary file 3
List of antibodies used for immunofluorescence.
- https://cdn.elifesciences.org/articles/56079/elife-56079-supp3-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56079/elife-56079-transrepform-v3.pdf





