A conserved and regulated mechanism drives endosomal Rab transition
Figures
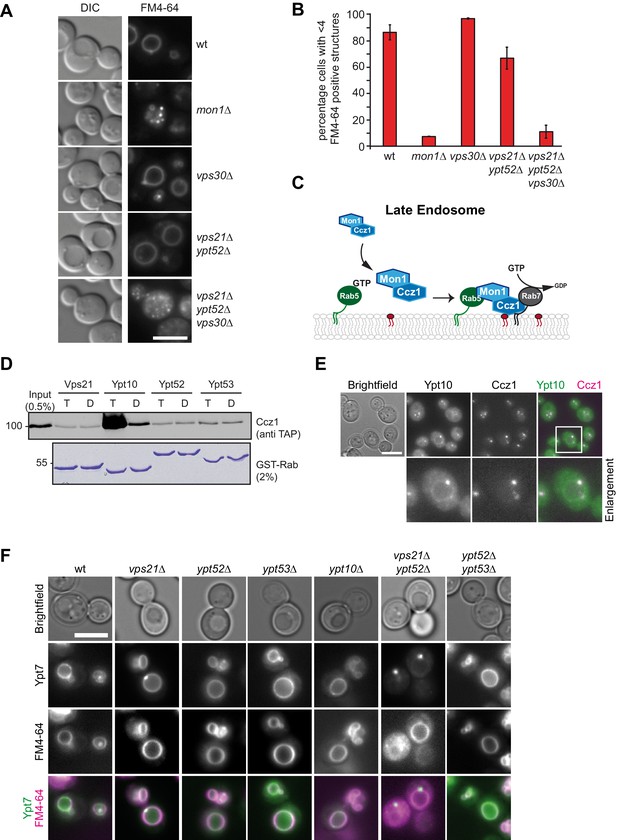
Rab5 effect on Mon1-Ccz1 function.
(A) Vacuole morphology in Rab5 mutants. Cells with the indicated mutations were grown in the presence of 10 µM FM4-64 and analyzed by fluorescence microscopy. Size bar, 5 µm. (B) Quantification of vacuole morphology. Percentage of cells with less than four vacuoles is shown. Error bars represent standard deviation. (C) Model of cooperation of Rab5-GTP and PI-3-P for Mon1-Ccz1 recruitment to late endosomes. PI-3-P is indicated as red lipid. (D) Interaction of yeast Rab5-like proteins with Mon1-Ccz1. Purified GST-tagged Rab5 proteins (Vps21, Ypt10, Ypt52, and Ypt53) were loaded with GTP (T) or GDP (D) and incubated with purified Mon1-Ccz1 complex. Eluates were analyzed on SDS-PAGE by Western blotting with an antibody against the TAP-tag on Ccz1 (top) and Coomassie staining (bottom). For details see methods. (E) Localization of Ypt10 in yeast. Cells expressing endogenously GFP-tagged Ypt10 and mKate-tagged Ccz1 were analyzed by fluorescence microscopy. Size bar, 5 µm. (F) Analysis of Ypt7 localization and vacuole morphology in Rab5 deletion strains. mNeon-tagged Ypt7 was expressed under the control of the Ypt7 promoter in cells the indicated Rab5 proteins. Cells were stained with FM4-64, and analyzed by fluorescence microscopy. Size bar, 5 µm.
-
Figure 1—source data 1
Quantification of vacuole number in FM4-64 stained wild-type and mutant strains in Figure 1A.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig1-data1-v2.xlsx
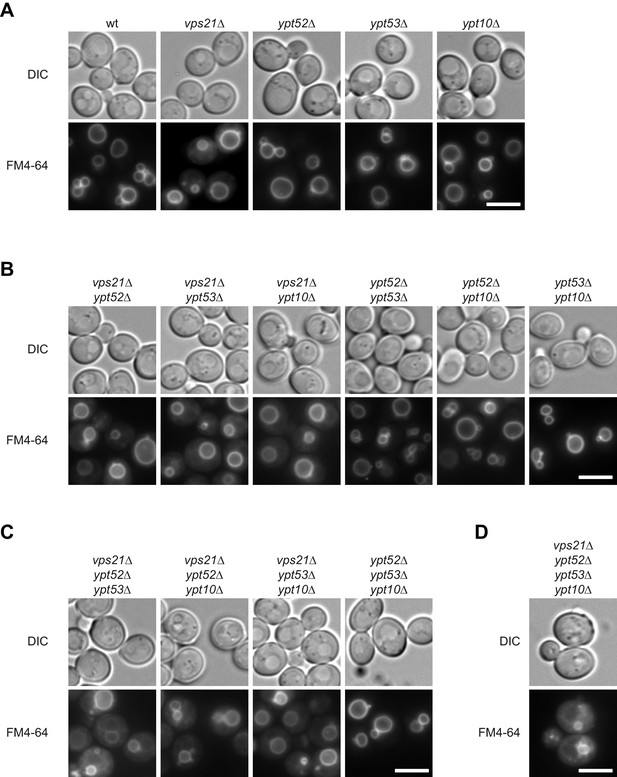
Vacuole morphology in Rab5 mutants.
Cells with the indicated (A) single (B) double (C) triple and (D) quadruple mutations were grown in the presence of 10 µM FM4-64 and analyzed by fluorescence microscopy. Size bar, 5 µm.
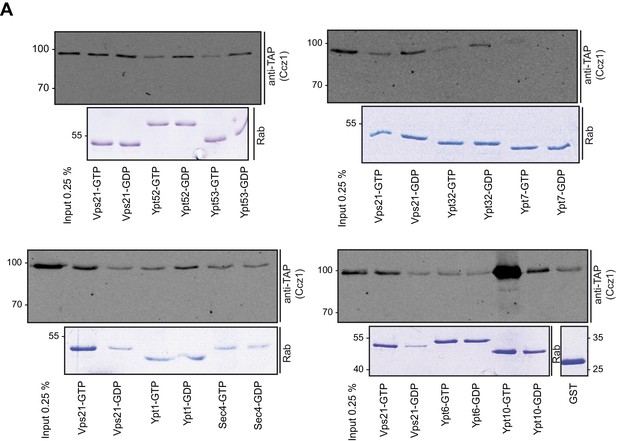
Interaction of yeast Rab-GTPases with Mon1-Ccz1.
(A) Purified GST-tagged Rab-GTPases were loaded with GTP (T) or GDP (D) and incubated with purified Mon1-Ccz1 complex. Eluates were analyzed on SDS-PAGE by Western blotting with an antibody against the TAP-tag on Ccz1 (top) and Coomassie staining (bottom). For details see methods.
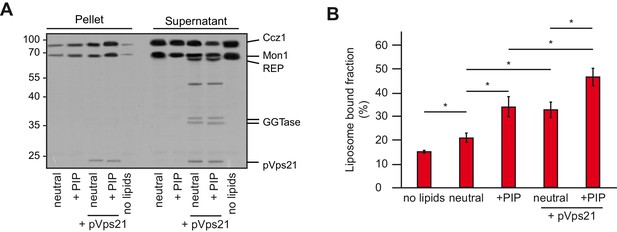
Membrane recruitment of Mon1-Ccz1.
(A) Liposome binding assay. Liposomes were either composed of 82% PC, 18% PE (neutral), or 79% PC, 18% PE, 2% PI-3-P and 1% PI-3,5-P2 (PIP). Where indicated, prenylated Vps21, which was initially in complex with REP, was activated in the presence of liposomes. Mon1-Ccz1 was added to liposomes, and incubated at room temperature for 20 min. Liposomes were separated by centrifugation into pellet and supernatant (20 min, 20,000 g, 4°C), the supernatant was acetone-precipitated. Proteins in pellet and supernatant fraction were analyzed by SDS-PAGE and Coomassie staining. (B) Quantification of pellet fractions from three independent assays. Error bars represent standard deviation. Significance analysis were performed by two-tailed heteroscedastic t-test statistics (*, p≤0.05).
-
Figure 2—source data 1
Quantification of Figure 2A.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig2-data1-v2.xlsx
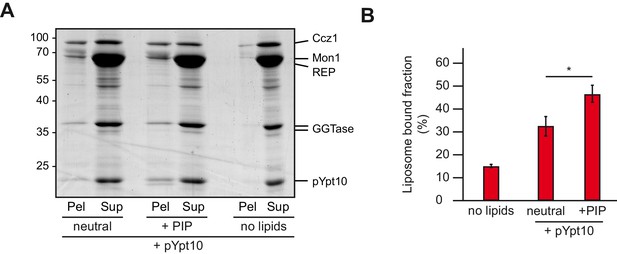
Membrane recruitment of Mon1-Ccz1.
(A) Liposome binding assay. Liposomes were either composed of 82% PC, 18% PE (neutral), or 79% PC, 18% PE, 2% PI-3-P and 1% PI-3,5-P2 (PIP). Where indicated, prenylated Ypt10, which was initially in complex with REP, was activated in the presence of liposomes. Mon1-Ccz1 was added to liposomes, and incubated at room temperature for 20 min. Liposomes were separated by centrifugation into pellet and supernatant (20 min, 20,000 g, 4°C), the supernatant was acetone-precipitated. Proteins in pellet and supernatant fraction were analyzed by SDS-PAGE and Coomassie staining. (B) Quantification of pellet fractions from three independent assays. Error bars represent standard deviation. Significance analysis were performed by two-tailed heteroscedastic t-test statistics (*, p≤0.05).
-
Figure 2—figure supplement 1—source data 1
Quantification of Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig2-figsupp1-data1-v2.xlsx
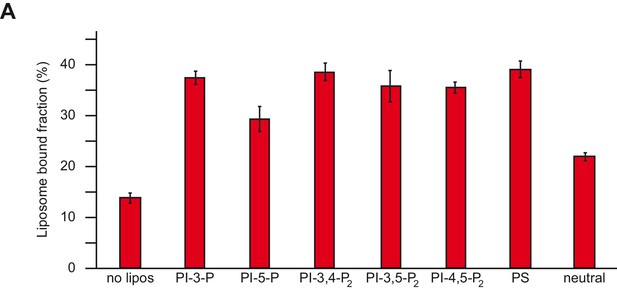
Membrane recruitment of Mon1-Ccz1.
(A) Liposome binding assay. Liposomes were composed of PC, 18% PE (neutral), and 2 mol% PI(X)P, 1 mol% PI(X,X)P2, 4 mol% PS, respectively, for a total of 4 mol% negative charge each. Concentration of PC was adjusted to get to a total of 100%. Mon1-Ccz1 was added to liposomes, and incubated at room temperature for 20 min. Liposomes were separated by centrifugation into pellet and supernatant (20 min, 20,000 g, 4°C), the supernatant was acetone-precipitated. Proteins in pellet and supernatant fraction were analyzed by SDS-PAGE and Coomassie staining. Quantification of pellet fractions from three technical repeats. Error bars represent standard deviation. d.
-
Figure 2—figure supplement 2—source data 1
Quantification of Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig2-figsupp2-data1-v2.xlsx
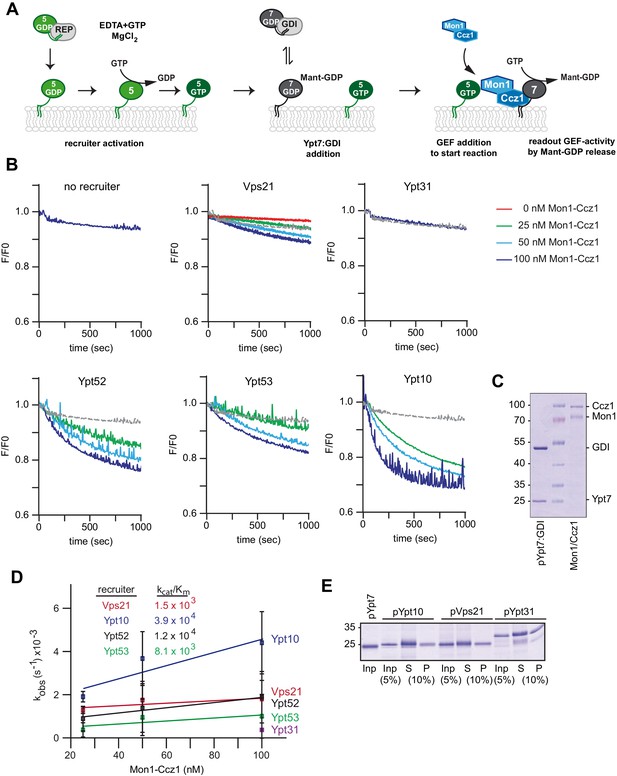
Rab5 proteins can activate the Mon1-Ccz1 GEF on membranes.
(A) Scheme of Rab5-dependent Rab7 activation. Rab5-proteins in complex with the REP were chemically activated to bring protein to membranes. Addition of MANT-GDP-loaded Ypt7:GDI was followed by Mon1-Ccz1 addition. For details see text. (B) Recruiter GEF-assay of Mon1-Ccz1. 250 nM MANT-GDP-loaded Ypt7 activation was followed by change in fluorescence over time. Liposomes composed of a vacuolar mimicking lipid mix were incubated with 1.5 µM recruiter GTPase as indicated or buffer as a control (no recruiter). For activation of the recruiter, 200 µM GTP was added in the presence of 1.5 mM EDTA for 15 min at room temperature. Afterwards the EDTA was quenched by addition of 3 mM MgCl2. After pre-activation of the recruiter, 250 nM prenylated Ypt7:GDI was added. Increasing amounts of Mon1-Ccz1 (as indicated) were added to start the reaction. Trace of no recruiter sample was plotted into all graphs for reference (grey line) (C) Protein complexes used for recruiter assay. Prenylated Ypt7 in complex with GDI and heterodimeric Mon1-Ccz1 complex were purified as described in the method section, and analyzed by SDS-PAGE and Coomassie staining. (D) Enzymatic parameters as derived from the recruiter assays in (B). Values were calculated from at least two independent measurements. Error bars represent standard deviation. For details see methods. (E) Membrane association of Rab-GTPases in the recruiter-assay. Samples from the recruiter-assay were recovered after 1000 s, and soluble and membrane fraction were separated by centrifugation for 20 min at 20,000 g. Fractions were analyzed by SDS-PAGE and Coomassie staining.
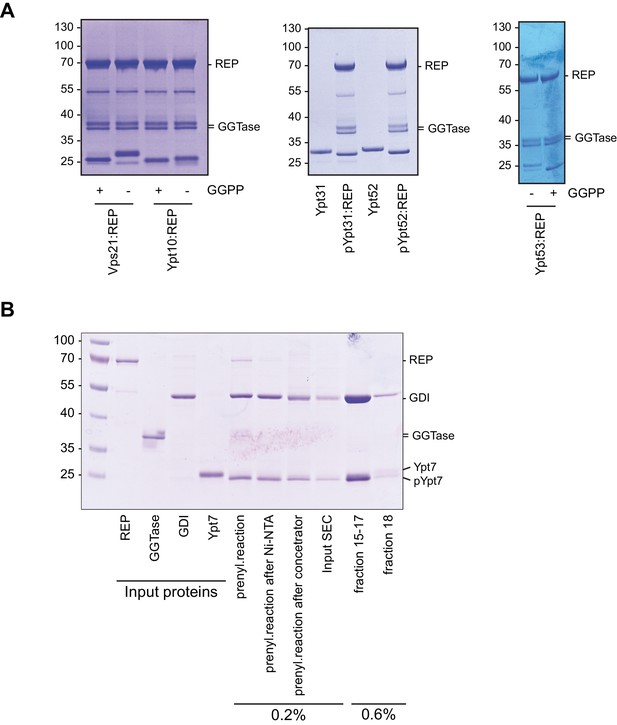
Prenylation of yeast Rab-GTPases for the Recruiter GEF-assay.
(A) Rab-GTPases were preloaded with GDP (Sigma Aldrich, Germany) and incubated with equimolar amounts of Rab Escort Protein (REP) in the presence of Geranylgeranyltransferase and Geranylgeranylpyrophosphat for 1 hr at 30°C. Successful prenylation was judged by downshift of the Rab-GTPase by SDS-PAGE analysis and Coomassie staining. (B) Prenylation of Ypt7 and complex formation with GDI. Ypt7 was prenylated in the presence of GDI and substoichiometric amounts REP as described above. Prenylated Ypt7:GDI was further purified by size exclusion chromatography (SEC). Fractions of Ypt7:GDI were pooled.
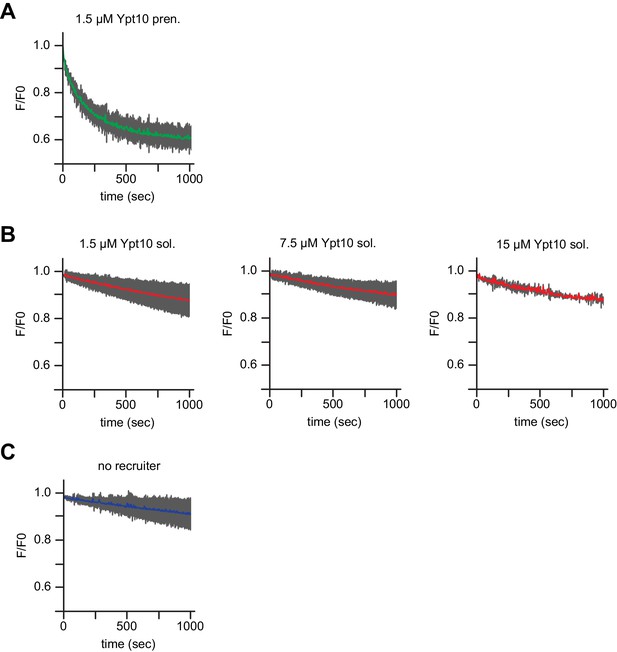
Membrane bound but not soluble Ypt10 activates Mon1-Ccz1.
Recruiter GEF-assay of Mon1-Ccz. 250 nM MANT-GDP-loaded Ypt7 activation was followed by change in fluorescence over time. Liposomes composed of a vacuolar mimicking lipid mix were incubated with (A) 1.5 µM prenylated Ypt10, (B) increasing amounts of soluble Ypt10 as indicated or (C) buffer as a control (no recruiter). For activation of Ypt10, 200 µM GTP was added in the presence of 1.5 mM EDTA for 15 min at room temperature. Afterwards the EDTA was quenched by addition of 3 mM MgCl2. After pre-activation of the recruiter or mock treatment (no recruiter), 250 nM prenylated Ypt7:GDI was added. 100 nM Mon1-Ccz1 was added to start the reaction.
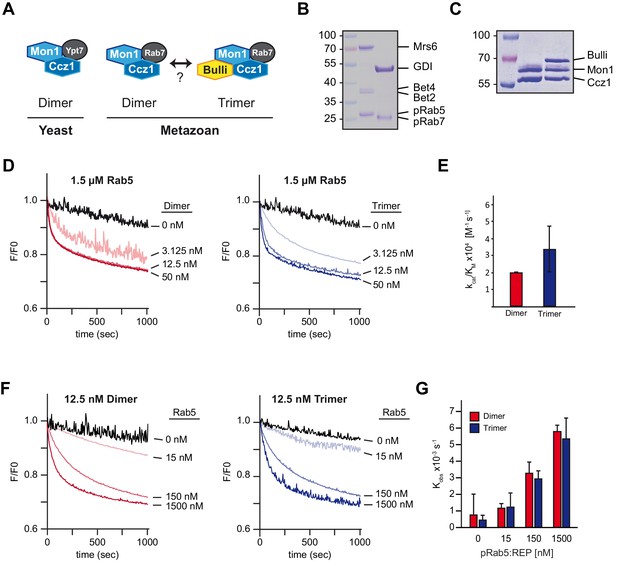
Mon1-Ccz1 activation by Rab5 is conserved in metazoan cells.
(A) Schematic representing the architecture of yeast and metazoan Rab7-GEF-complexes. Yeast Mon1-Ccz1 consists of a heterodimer whereas metazoan GEF-complex is a heterotrimer with Bulli (in Drosophila) or RMC1 in mammalian cells as a third subunit. A dimer of metazoan Mon1-Ccz1 may exist as well. (B) Purification and prenylation of Drosophila Rab-GTPases. Rab5 and Rab7 were prenylated and complexed with REP and GDI, respectively. (C) Purification of Drosophila dimeric and trimeric Rab7-GEF-complexes from insect cells as used for GEF-assays. (D) Recruiter GEF-assay with Drosophila dimeric and trimeric (Bulli-)Mon1-Ccz1. The recruiter GEF-assay was performed as described in Figure 3 using Drosophila protein complexes and increasing amounts of either Mon1-Ccz1 (Dimer) or Bulli-Mon1-Ccz1 (Trimer). (E) Enzymatic parameters as measured in (D). Values were calculated of two independent experiments. Error bars represent standard deviation. (F) Recruiter GEF-assay with decreasing amounts of Rab5. The recruiter-assay was performed as described above with 12.5 nM Mon1-Ccz1 and Bulli-Mon1-Ccz1, respectively, and the indicated amounts of recruiter Rab5. (G) Rate constants as derived from (F) in dependence of concentration of Rab5. Error bars represent standard deviation. For details see Methods.
-
Figure 4—source data 1
Quantification Figure 4E.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Quantification Figure 4G.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig4-data2-v2.xlsx
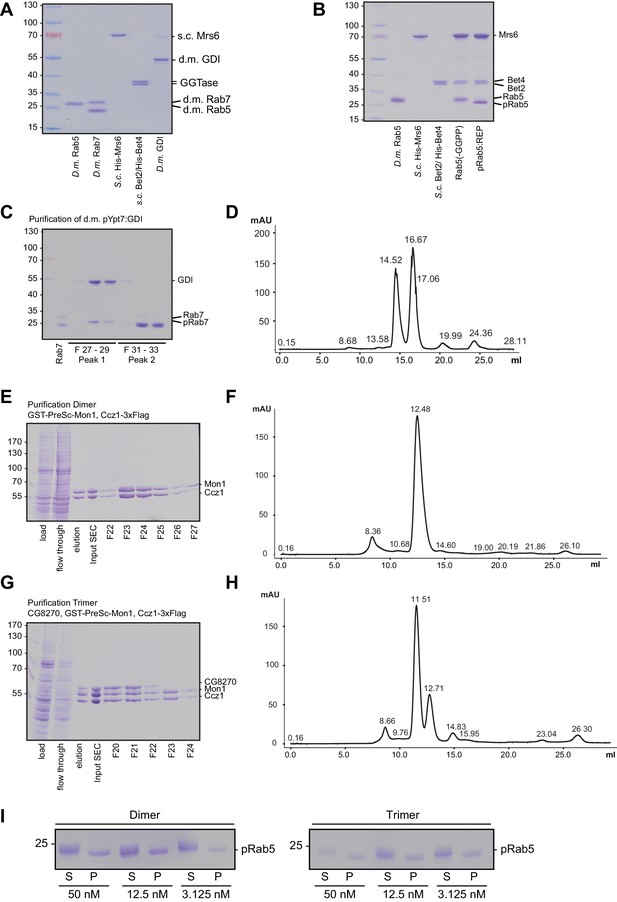
Purification and prenylation of Drosophila proteins.
(A) Proteins as used for prenylation of Drosophila Rab5 and Rab7. For purification details see methods. (B) Prenylation of Drosophila Rab5. Drosophila Rab5 was incubated with equimolar amounts of yeast REP in the presence of yeast GGTase and GGPP for 1 hr at 30°C. As a control GGPP was omitted. Downshift of the Rab5 band indicates successful prenylation. (C) Prenylation of Drosophila Rab7 in complex with Drosophila GDI. Rab7 was prenylated as described in the method section and was further purified by (D) size exclusion chromatography. (E) Purification of Drosophila dimeric Mon1-Ccz1 complex. For details see text. (F) Size exclusion chromatography of purified d.m. Mon1-Ccz1. Mon1-Ccz1 eluted as one peak at a retention volume of 12.5 ml. (G) Purification of Drosophila trimeric Bulli-Mon1-Ccz1 complex. For details see text. (H) Size exclusion chromatography of purified d.m. Mon1-Ccz1. Bulli-Mon1-Ccz1 eluted as one peak at a retention volume of 11.5 ml. (I) Membrane association of recruiter GTPase Rab5 in assays shown in Figure 4D.
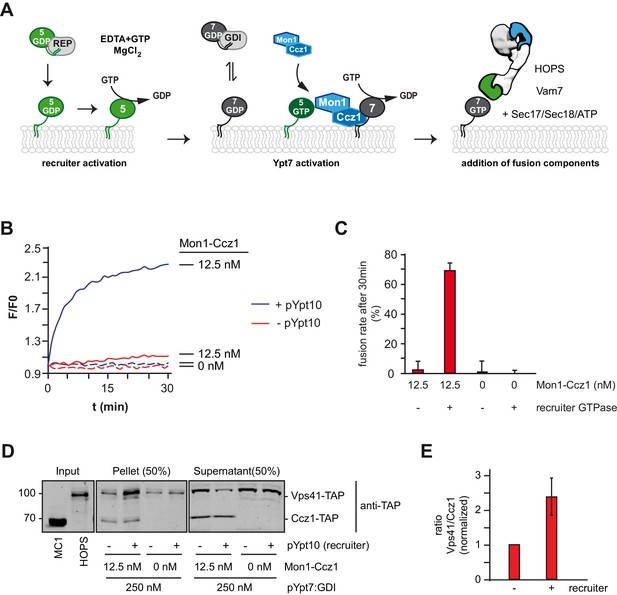
Rab5 can trigger Rab7-dependent fusion.
(A) Scheme of the Rab5-dependent fusion assay. Rab5-proteins in complex with the REP protein were chemically activated. Addition of MANT-GDP-loaded Ypt7:GDI was followed by Mon1-Ccz1 addition. After allowing for nucleotide exchange of Ypt7, fusion machinery was added and fusion reaction was finally triggered by addition of Vam7. (B) Fusion depends on the recruiter GTPase. SNARE-bearing proteoliposomes were primed either with chemically activated pYpt10 or mock-treated for 10 min at 27°C. 250 nM Ypt7:GDI and 12.5 nM Mon1-Ccz1 was added, and the reaction was incubated for another 15 min, allowing for nucleotide exchange of Ypt7. Fusion reaction was triggered by addition of 50 nM HOPS, 0.6 µM Sec17, 50 nM Sec18 and 20 nM Vam7. Fusion rate was followed by a content mixing assay, where FRET of enclosed fluorophores is followed (see Methods). (C) Fusion rates of (B) as measured after 30 min. Error bars represent standard deviation. (D) Membrane association of Mon1-Ccz1 and HOPS in fusion assay. Samples of fusion experiments as in (B) were recovered after 30 min of measurement. Membrane-bound and soluble fraction were separated by centrifugation for 20 min at 20,000 g, and analyzed by SDS-PAGE and Western-Blot. Proteins were detected by using an antibody against the TAP-tag on Vps41 and Ccz1 in the HOPS- and GEF-complex, respectively. (E) Densiometric analysis of Western-Blot signals of Vps41 and Ccz1 as shown in (D). The ratio of Vps41 over Ccz1-signal is shown for fusion reactions in the presence or absence of a recruiter-GTPase. Signals have been normalized to the Ccz1 signal in the pellet fraction. Error bars represent standard deviation.
-
Figure 5—source data 1
Quantification Figure 5C and Figure 5—figure supplement 1B.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Quantification Figure 5E.
- https://cdn.elifesciences.org/articles/56090/elife-56090-fig5-data2-v2.xlsx
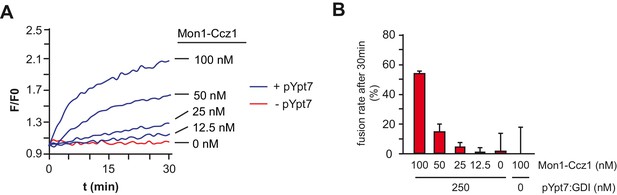
Fusion of reconstituted proteoliposomes in dependence of Mon1-Ccz1.
(A) SNARE-bearing proteoliposomes were incubated with 250 nM pYpt7:GDI in presence of indicated amounts of Mon1-Ccz1 and GTP. After allowing for nucleotide exchange for 15 min, fusion was triggered by addition of 50 nM HOPS, 0.6 µM Sec17, 50 nM Sec18 and 20 nM Vam7. Fusion was followed by a content mixing assay, where FRET of enclosed fluorophores is followed (see Materials and methods). (B) Fusion rates of (A) as determined after 30 min.
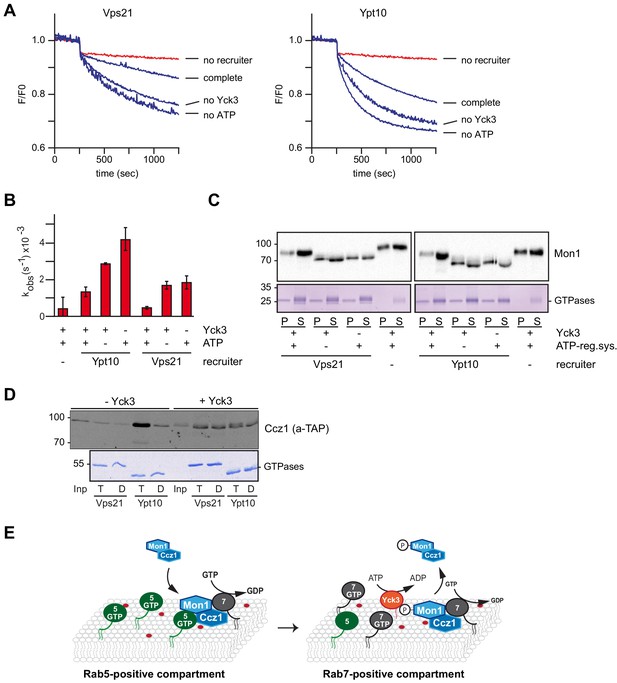
The casein kinase Yck3 regulates interaction of Rab5 with Mon1-Ccz1 in yeast.
(A) Recruiter GEF-assay of phosphorylated Mon1-Ccz1. 50 nM Mon1-Ccz1 were pretreated either with Yck3 and ATP (complete) or with either one of them (no Yck3/no ATP) before it was used in the recruiter GEF assay as described in Figure 3A. 1.5 µM GTP-loaded pVps21 or pYpt10 were used as a recruiter GTPase, as control proteoliposomes were mock treated (no recruiter). (B) Rate constants as calculated from (A). Error bars represent standard deviation. (C) Soluble and membrane fraction of recruiter assays in (A) were recovered and separated by centrifugation for 20 min at 20,000 g, 4°C. Samples were analyzed by SDS-PAGE and Western-Blot for presence of Mon1, and by Coomassie staining for used Rab-GTPases. (D) Interaction of yeast Rab5-like proteins with Mon1-Ccz1. Purified GST-tagged Rab5 proteins (Vps21, Ypt10) were loaded with GTP (T) or GDP (D) and incubated with purified Yck3 or mock-treated Mon1-Ccz1 complex. Eluates were analyzed on SDS-PAGE by Western blotting with an antibody against the TAP-tag on Ccz1 (top) and by Coomassie staining (bottom). For details see methods. (E) Working model. Mon1-Ccz1 is recruited to charged membranes. On a Rab5-positive compartment, where Mon1-Ccz1 interacts with Rab5, its GEF-activity towards Rab7 is stimulated. This would lead to a fast transition from a Rab5 to a Rab7-positive compartment. The Rab7-positive compartment converts from an endosomal to a lysosomal/vacuolar membrane. After fusion with the lysosome-like vacuole in yeast, Mon1-Ccz1 is phosphorylated by Yck3. This abolishes the interaction with Rab5 and may in addition lead to a release of Mon1-Ccz1 from the vacuole (Lawrence et al., 2014).
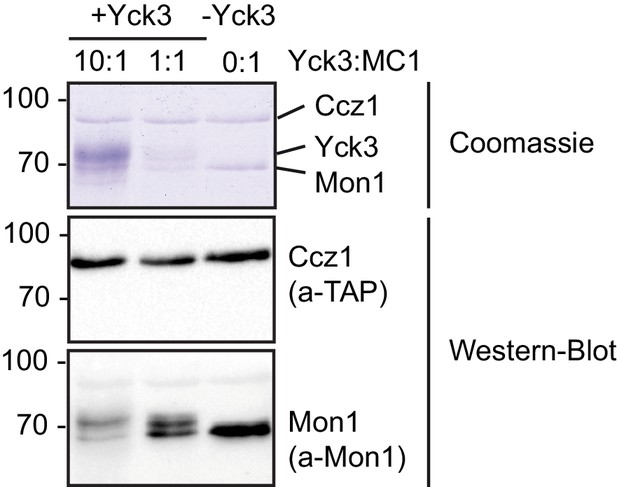
Phosphorylation of yeast Mon1-Cz1 by Yck3 in vitro.
(A) Mon1-Ccz1 was incubated with different amounts of recombinantly produced Yck3 in the presence of ATP regenerating system for 30 min at 27°C. As a control Yck3 was omitted. Samples were analyzed by SDS-PAGE and Western-Blot using antibodies against Mon1 and the TAP-tag on Ccz1. Phosphorylation resulted in an upshift of the band of Mon1.
Additional files
-
Source data 1
- https://cdn.elifesciences.org/articles/56090/elife-56090-data1-v2.xlsx
-
Supplementary file 1
Strains used in this study.
- https://cdn.elifesciences.org/articles/56090/elife-56090-supp1-v2.docx
-
Supplementary file 2
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/56090/elife-56090-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56090/elife-56090-transrepform-v2.docx





