Emergence and diversification of a highly invasive chestnut pathogen lineage across southeastern Europe
Figures
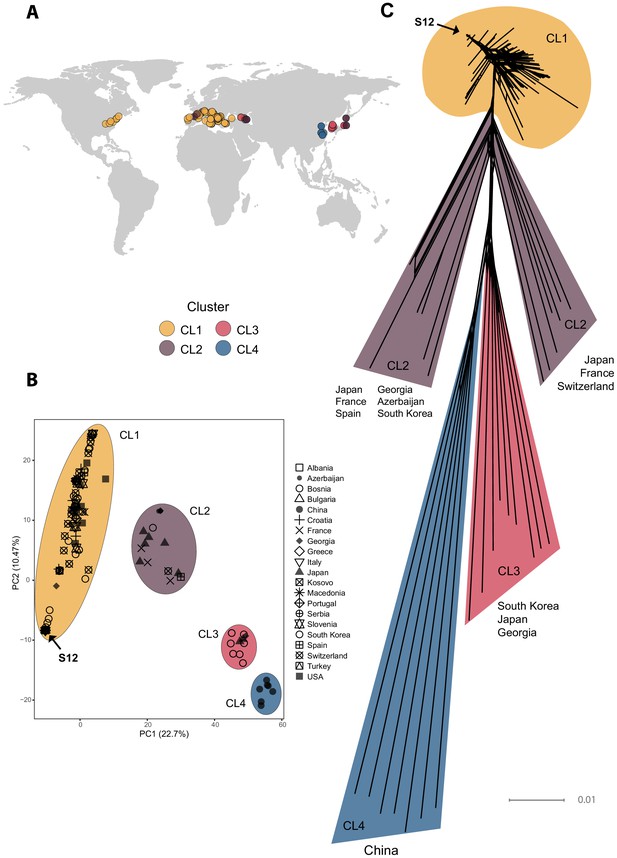
Genome-wide analyses of global Cryphonectria parasitica isolates.
(A) Map of global sampling locations and (B) principal component analysis (PCA) of the 230 sequenced C. parasitica isolates. Colors indicate PCA clusters (CL1–CL4) and are as in (A). (C) SplitsTree phylogenetic network of the global C. parasitica sample set. Colors are as in (A) and (B). Isolates belonging to the S12 lineage are marked with an arrow in (B) and (C).
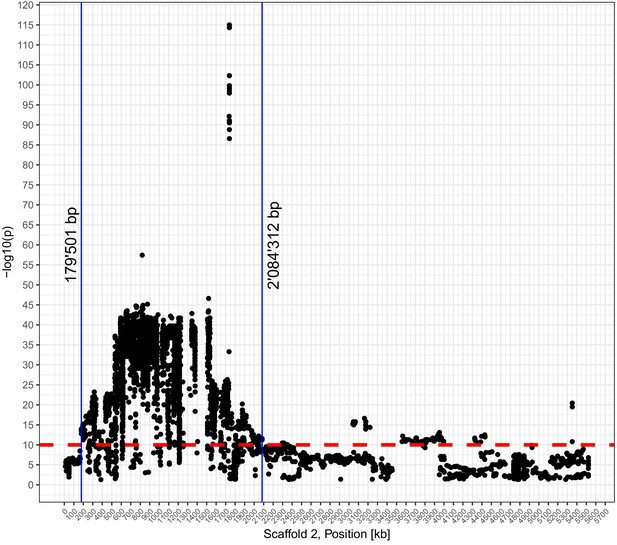
Genome-wide association mapping (GWAS) outcome for the mating type (MAT) locus.
The red line shows the p-value-based threshold to exclude MAT-locus-associated single nucleotide polymorphisms (SNPs) for population structure analyses.
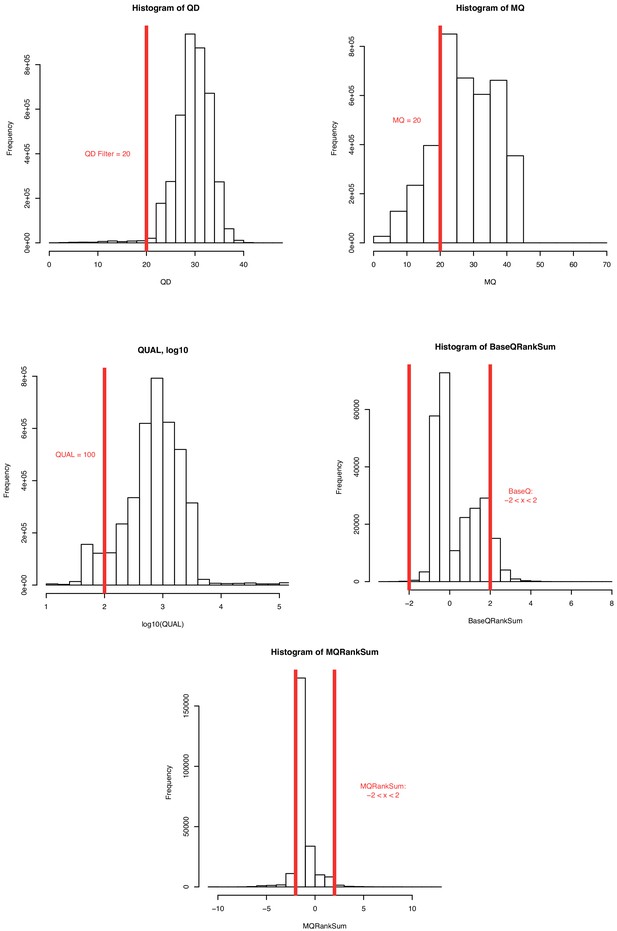
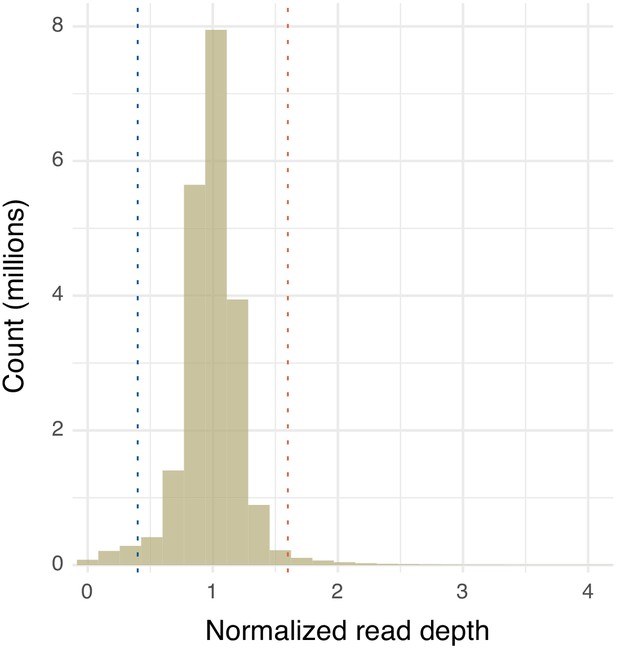
Distribution of observed single nucleotide polymorphism (SNP) filtration quality values.
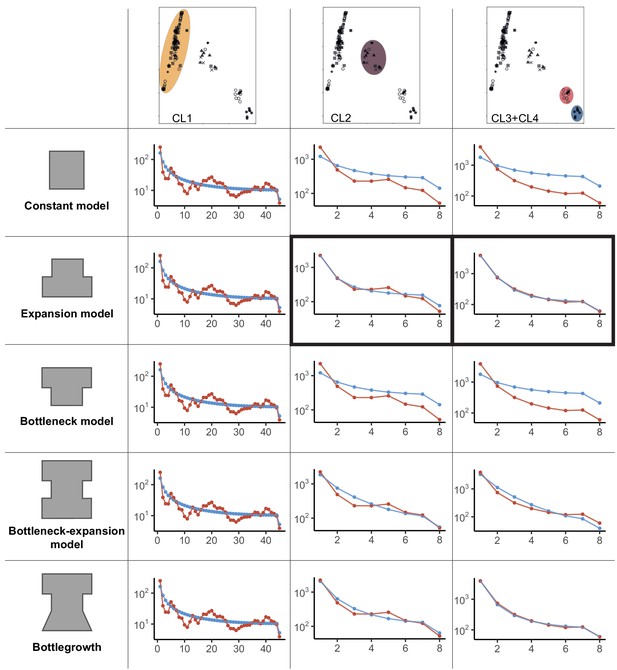
Demographic models for global Cryphonectria parasitica populations.
Constant, expansion, bottleneck, bottleneck-expansion, and bottlegrowth population models were tested for three clusters (i.e., European/North American CL1, mixed European/Asian CL2, and Asian CL3 and CL4 clusters shown by color highlights). Clusters were inferred by principal component analysis (PCA) (see Figure 1B). The two smaller Asian clusters (top-right PCA panel) highlighted in red and blue were combined for demographic analyses to obtain a sufficient sample size. Panels in columns 2–4 show model—data comparison plots for the three tested scenarios (i.e., constant, expansion and bottleneck, bottleneck-expansion and bottlegrowth) as inferred by dadi. Blue data points and line represent the model, and red lines show the empirical data. Best fitting models according to the Akaike information criterion (AIC) model comparisons are marked in black boxes. A detailed list of initial and best-fit parameters, as well as likelihood and AIC values, is shown in Supplementary file 2.
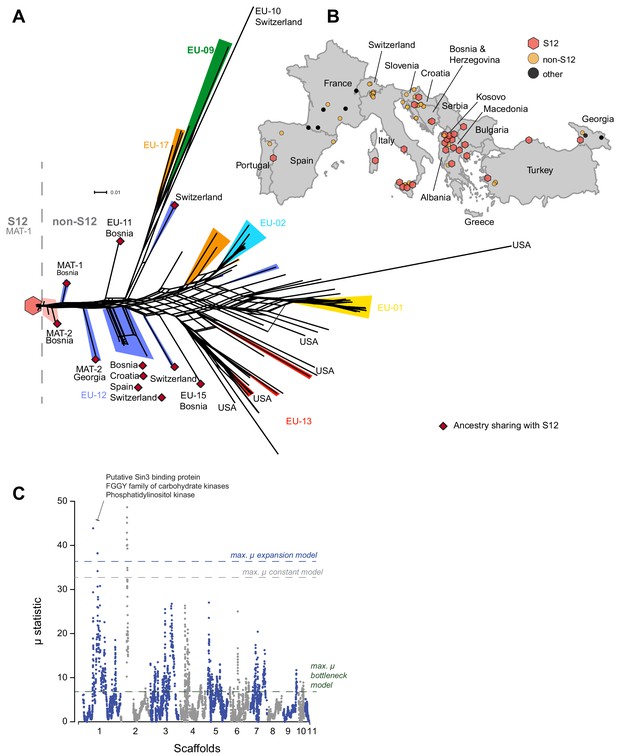
Phylogenetic network structure of the main European/North American subgroup and spatial distribution of S12 across Europe.
(A) The highlighted branches represent the most abundant vegetative compatibility types. Isolates belonging to the S12 outbreak lineage (EU-12; mating type MAT-1, n = 105) are marked with a red hexagon and match symbols in (B). S12 isolates of mating type MAT-2 are highlighted in light red. Additional EU-12 isolates not belonging to the S12 lineage are highlighted in blue with information on the country of origin. Isolates sharing ancestry with the S12 lineage as inferred by fineSTRUCTURE (Figure 4, Figure 4—figure supplement 2) are marked with dark red diamonds. (B) Geographic map of European sampling locations. Light red hexagons mark locations where S12 was found, and yellow circles indicate locations containing other genotypes from the main European/North American CL1 cluster (Figure 1B). Black circles show the location of highly distinct genotypes outside of the CL1 cluster (Figure 1A, B). (C) Genome-wide scan for selective sweeps (RAiSD). Effects due to the unknown demographic history of the European/North American CL1 cluster (Figure 1B) were mitigated by implementing population simulations under constant, expansion, and bottleneck scenarios. The three dashed lines show the maximum values observed in the simulated datasets following constant (gray) expansion (blue) and bottleneck (green) demographic models. Encoded protein functions overlapping with the top regions are shown as summaries.
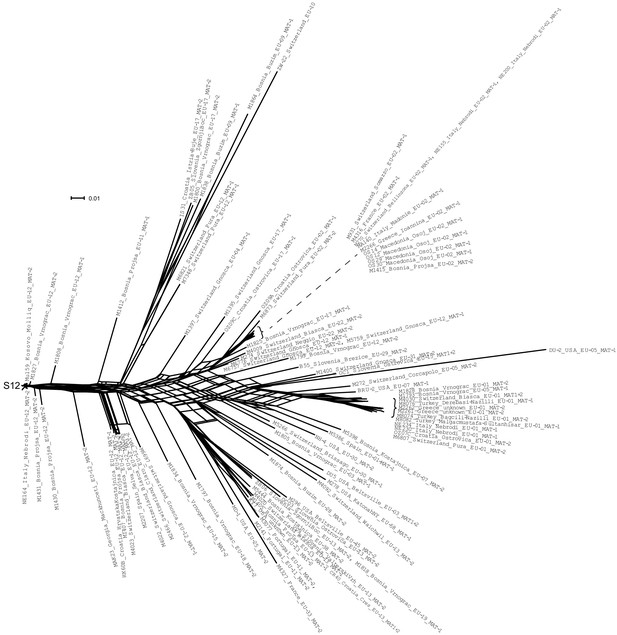
SplitsTree of all non-S12 isolates showing full isolate identifiers.
Isolates identified as S12 are listed in Supplementary file 1.
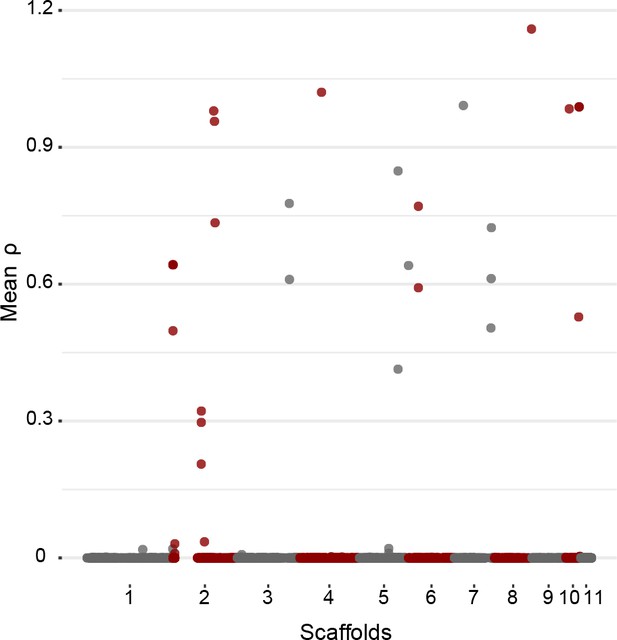
Per site population recombination rate ρ of European/North American Cryphonectria parasitica populations as inferred with LDhat.
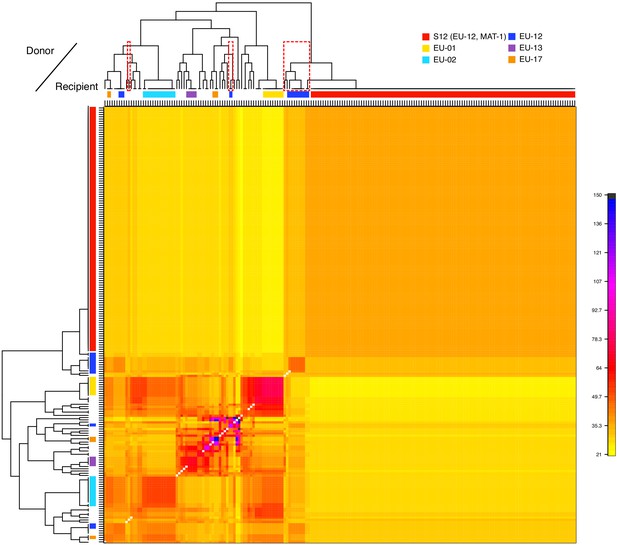
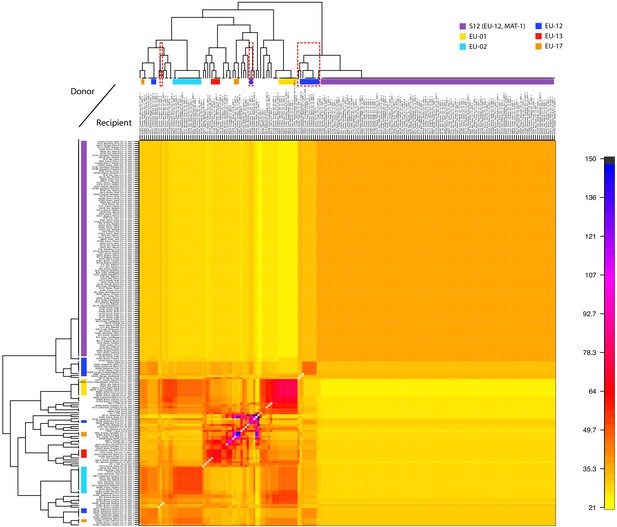
Analysis of donors to the S12 lineage genotypes.
Averaged coancestry matrix and population tree of the North American/European subgroup estimated by fineSTRUCTURE. The heatmap indicates averaged coancestry between populations. Populations sharing ancestry with S12 are marked in red-dashed boxes. S12 ancestry-sharing populations are additionally marked in Figure 3A. Detailed coancestry matrix with extensive population information is shown in Figure 4—figure supplement 2.
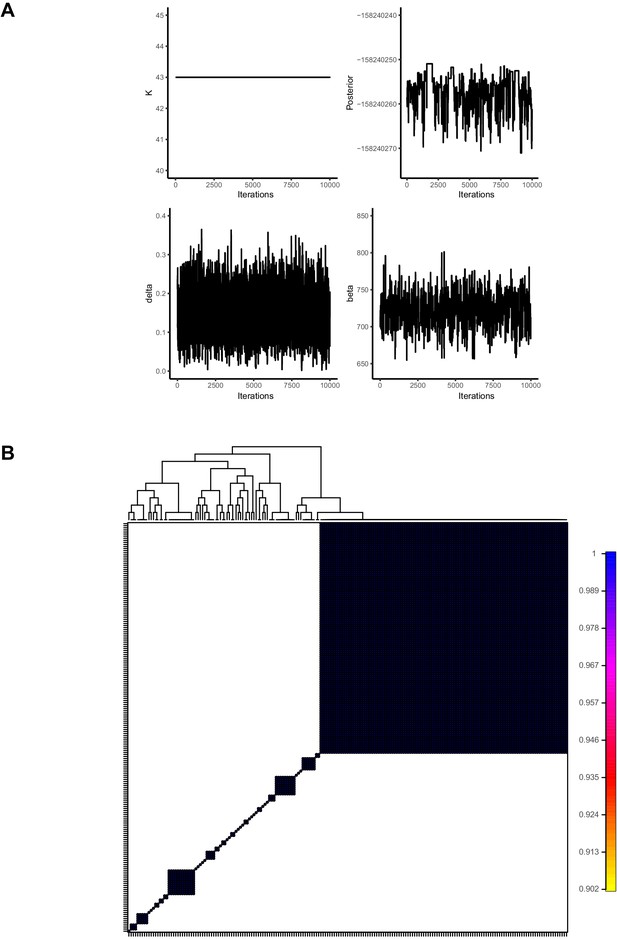
fineSTRUCTURE convergence plots.
(A) FineSTRUCTURE Markov Chain–Monte Carlo (MCMC) traces. (B) Pairwise coincidence matrix of population assignments.
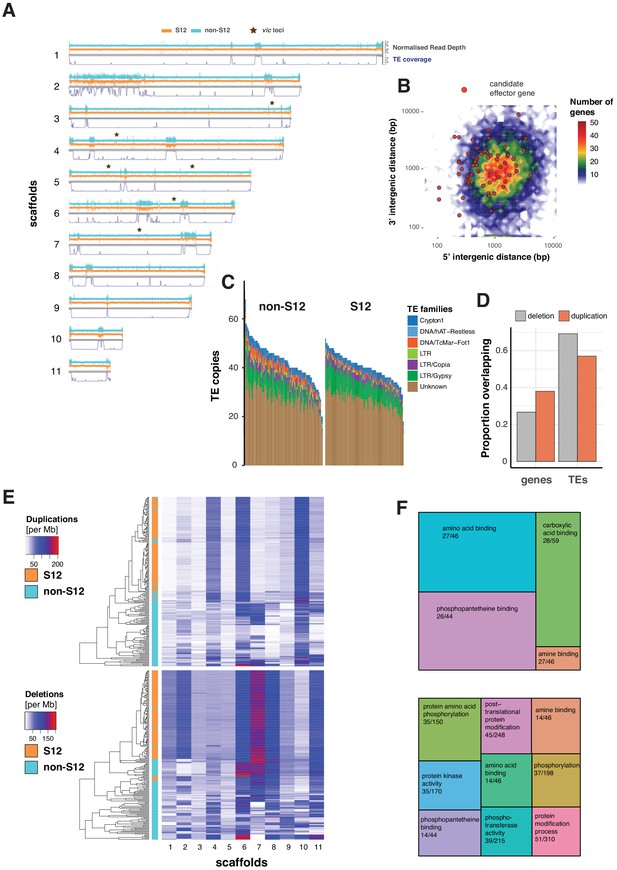
Transposable element (TE) landscape and copy number variation.
(A) Genome-wide coverage of transposable elements (in 10 kb windows) matched by with normalized read depth for the S12 and non-S12 lineages (North America and Europe only). vic loci: vegetative incompatibility loci. (B) Genome-wide distribution of intergenic distances according to the length of 5′ and 3′ flanking regions. Red dots represent genes encoding predicted effector proteins. (C) Counts of detected TE sequences across S12 and non-S12 isolates using split reads and target site duplication information. (D) Proportion of normalized read depth windows (800 bp) with evidence for duplications (normalized read depth >1.6) or deletions (<0.4) overlapping with genes and TEs. (E) Heatmap showing the number of windows (800 bp) with duplications and deletions. The dendrogram shows the similarity in duplication or deletion profiles for S12 and non-S12 isolates (North America and Europe only). (F) Molecular functions (based on gene ontology) enriched in duplicated and deleted regions (upper and lower panel, respectively). Enrichment was tested by hypergeometric tests, and significance was established for a Bonferroni threshold at alpha = 0.05. The numbers represent the number of genes with the matching gene ontology term in a duplicated or deleted region, and across the genome, respectively.
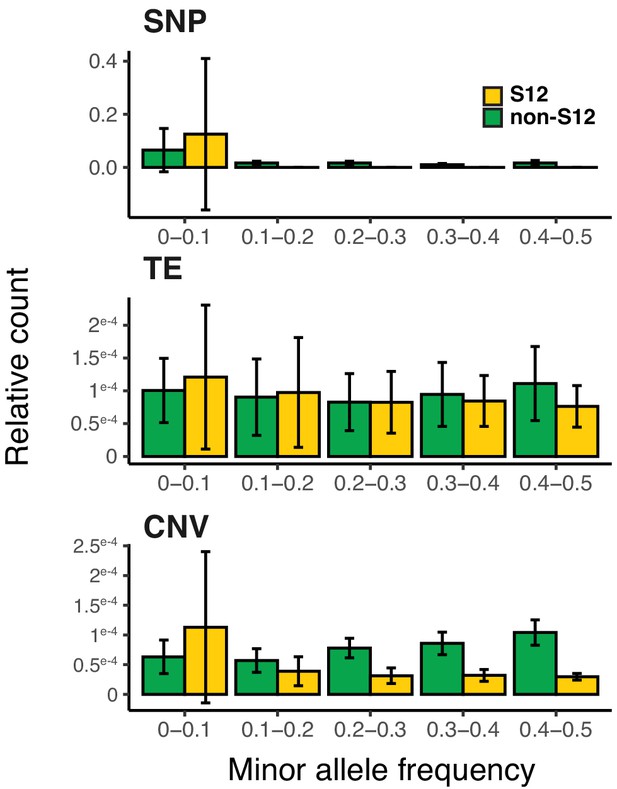
Genetic diversity within the S12 lineage.
Minor allele frequency distribution spectrum for three classes of variants. The number of polymorphic positions was normalized to the total number of polymorphic positions in S12 and non-S12 isolates, respectively. Error bars represent the standard deviation estimated from a resampling (n = 100) of 80 isolates in each group. We evaluated the difference between the S12 and non-S12 populations using two-way analysis of variance (number of sites ~ minor allele frequency*population). Both single nucleotide polymorphism (SNP) and copy number variant (CNV) frequencies show strong contrast (p<1e-16) while we find only minor differences for transposable elements (TEs) between S12 and non-S12 isolates (p<0.05).
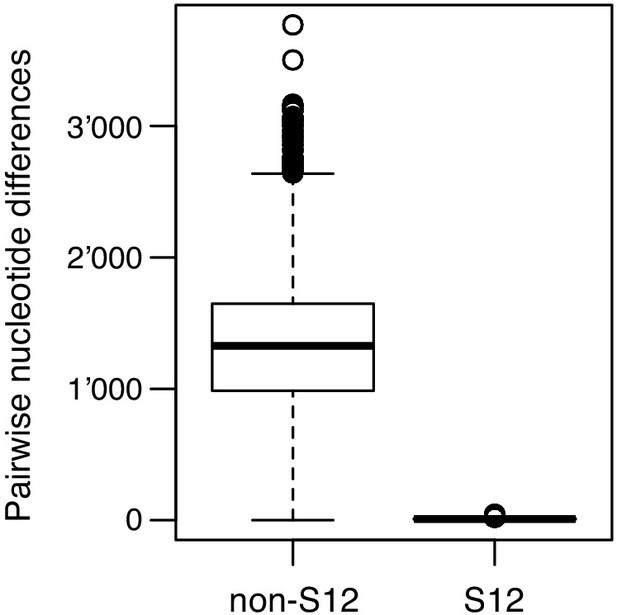
Pairwise genome-wide nucleotide differences between S12 and non-S12 isolates.
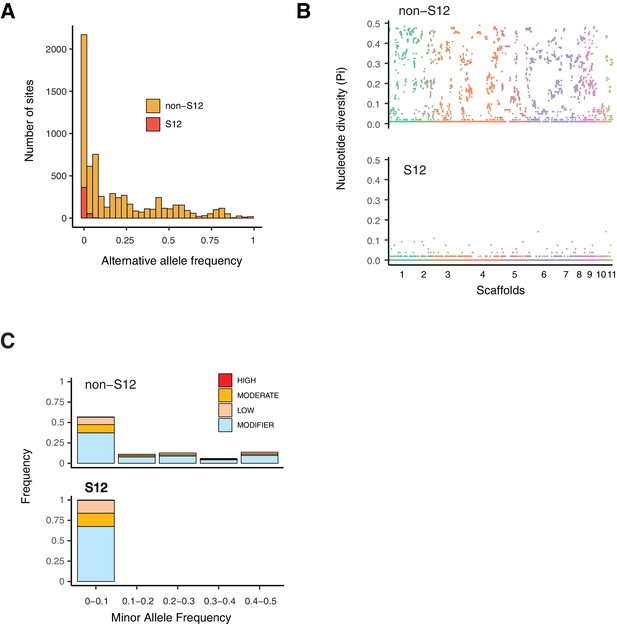
Polymorphism segregating in the S12 lineage.
(A) Alternative allele frequencies spectra across the genome for the S12 lineage (n = 105) compared to all other analyzed European (non-S12; n = 92). (B) Genome-wide nucleotide diversity (Pi) for the S12 lineage and non-S12 lineages in 10 kb windows. (C) Minor allele frequency spectra of high, moderate, and modifier (i.e., near neutral) impact mutations as identified by SnpEff.
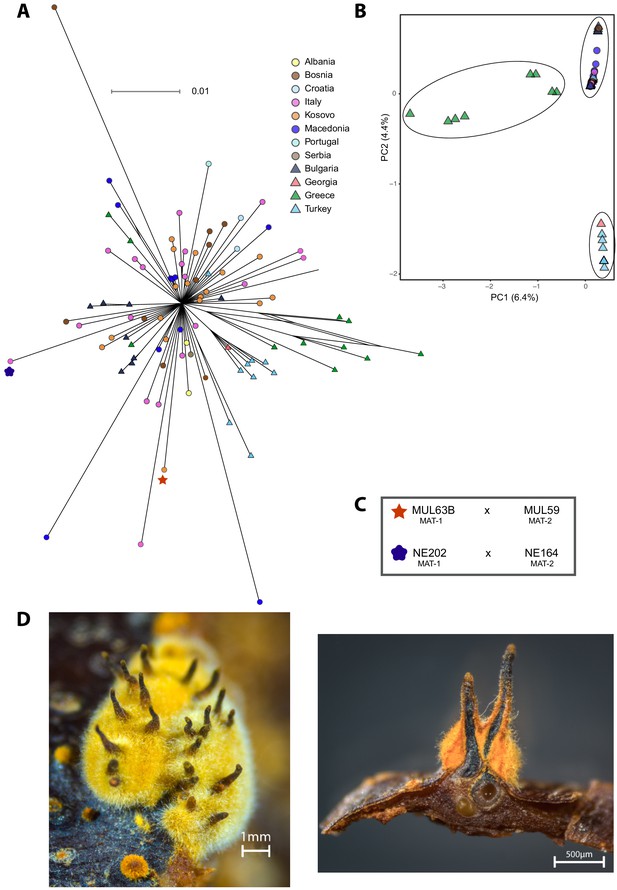
Fine-scale genetic diversity analyses of the S12 lineage.
(A) SplitsTree and (B) principal component analysis (PCA) of the S12 mating type MAT-1 outbreak isolates (n = 105). Symbols and colors are as in (A). (C) Scheme of successful mating pairs of S12 mating type MAT-1 isolates crossed with isolates from the opposite mating type of the same geographic origin. Symbols are as in (A). (D) Photographic images of sexual C. parasitica fruiting bodies (i.e., perithecia) emerging from crosses of S12 mating type MAT-1 isolates with isolates of the opposite mating type after 5 months of incubation under controlled conditions. Left: perithecia embedded in a yellow-orange stromatic tissue. Right: cross-section of perithecia and chestnut bark. Flask-shaped structures with a long cylindrical neck develop in yellow-orange stromatic tissue and are embedded in the bark (except for the upper part). The ascospores are formed in sac-like structures (asci) in the basal part of the perithecium. When mature, the ascospores are actively ejected into the air through a small opening (ostiole) at the end of the perithecial neck.
Additional files
-
Supplementary file 1
Information on all analyzed isolates including sampling location, vegetative compatibility type (EU-type), (SSR) simple sequence repeats genotyping outcome, collector information, collection years, and NCBI accessions.
- https://cdn.elifesciences.org/articles/56279/elife-56279-supp1-v1.xlsx
-
Supplementary file 2
Initial and best-fit parameters as well as likelihoods and Akaike information criterion (AIC) values as inferred by dadi for demographic analyses.
Cluster names correspond to clusters as shown in Figure 1A.
- https://cdn.elifesciences.org/articles/56279/elife-56279-supp2-v1.xlsx
-
Supplementary file 3
List of the genes in regions with signatures of recent positive selection (based on RAiSD).
Putative functions are shown by (PFAM) Protein families (database) and Gene Ontology (GO) annotations. Single nucleotide polymorphisms (SNPs) and the corresponding composite μ statistic are shown.
- https://cdn.elifesciences.org/articles/56279/elife-56279-supp3-v1.xlsx
-
Supplementary file 4
Summary of MAT-1 and MAT-2 isolate pairings used for testing sexual recombination in S12 populations.
*Isolates from the S12 MAT-1 lineage.
- https://cdn.elifesciences.org/articles/56279/elife-56279-supp4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56279/elife-56279-transrepform-v1.pdf