Alternative splicing of coq-2 controls the levels of rhodoquinone in animals
Figures
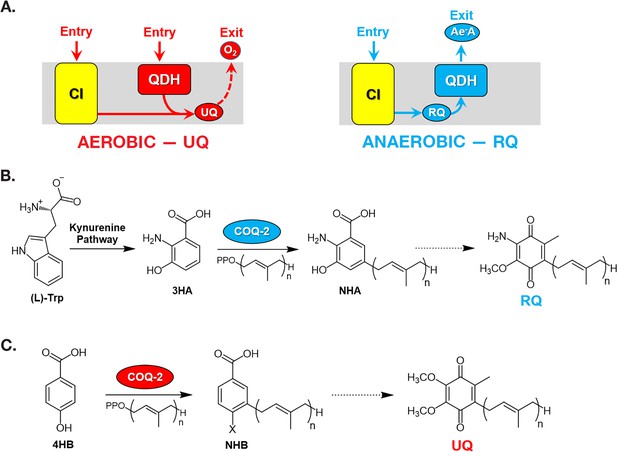
Rhodoquinone and ubiquinone biosynthesis and function in electron transport chains.
(A) In aerobic metabolism, ubiquinone (UQ) shuttles electrons in the ETC from Complex I (CI; yellow box) and quinone-coupled dehydrogenases (QDHs), such as Complex II. These electrons are ultimately transferred to oxygen. In anaerobic metabolism, rhodoquinone (RQ) reverses electron flow in QDHs and facilitates an early exit of electrons from the ETC onto anaerobic electron acceptors (Ae-A), such as fumarate. (B) The RQ biosynthetic pathway in C. elegans requires L-tryptophan, a precursor in the kynurenine pathway. L-Trptophan is transformed into 3-hydroxyanthranilic acid (3HA) in four steps. It is proposed that 3HA is a substrate for COQ-2, producing 3-hydroxy-5-nonaprenylanthranilic acid (NHA), where n=9. The transformation of NHA to RQ requires several shared proteins from the UQ biosynthetic pathway. (C) Eukaryotes can use either p-aminobenzoic acid (pABA) or 4-hydroxybenzoic acid (4HB) as precursors to UQ. Prenylation is facilitated by Coq2 to form 3-hexaprenyl-4-hydroxybenzoic acid (HHB) or 3-hexaprenyl-4-aminobenzoic acid (HAB), where n = varies between species. Further functionalization of these intermediates occurs through a Coq synthome (Coq3–Coq9 and Coq11) to yield UQ.
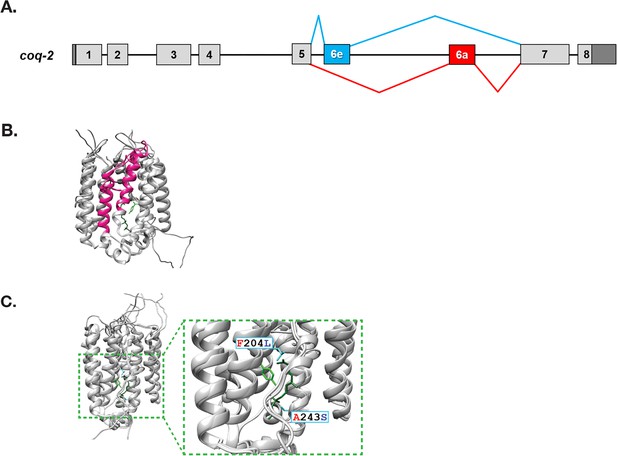
C. elegans coq-2 gene model.
(A) The coq-2 gene contains two mutually exclusive exons, 6e (blue box) and 6a (red box), that are alternatively spliced (blue and red lines, respectively) generating two COQ-2 isoforms. Light gray boxes represent coding sequences of exons 1–5 and 7–8, black lines represent introns, and dark gray boxes denote 5´and 3´ untranslated regions of exons 1 and 8. (B) Alternative splicing of COQ-2 changes the enzyme core. The sequences of C. elegans coq-2a and coq-2e were threaded onto the crystal structure of the apo-form of the Aeropyrum pernix COQ-2 homolog (PDB: 4OD5) in Chimera using Modeller. The region switched by mutually exclusive alternative splicing is shown magenta color. (C) The alternative exons found in all RQ-synthesizing species have two residues that are invariant (L204 and S243 show in cyan; C. elegans numbering) that are near the binding site of the two substrates. Substrates are the polyprenyl tail (dark green; geranyl S-thiolodiphosphate in the crystal structure), and the aromatic ring (light green; p-hydroxybenzoic acid [4HB] in the crystal structure). Note that COQ-2 is rotated from panel B to panel C for clarity.
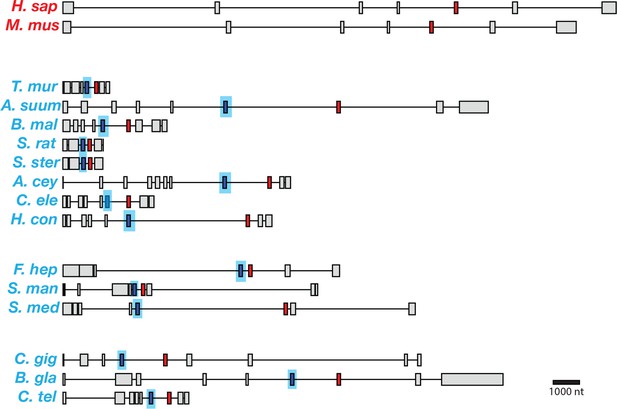
Gene models for coq-2 orthologs in various species.
Parasitic helminths, as well as annelids and mollusks, have two internal exons that are spliced in a mutually exclusive manner. By contrast, humans and other hosts only have one exon that is homologous to exon 6a of C. elegans coq-2. The a-form exon (red) shares greater similarity to the exon present in species that do not synthesize RQ, while the e-form (blue) is present only in RQ-producing species. The genes used for each species are listed in Supplementary file 3. The gene structures shown are based on genome annotations but in many cases include manual reannotations — in all such cases, the manual annotations are confirmed with RNA-seq data.

Conserved changes between a- and e-form exons across RQ-producing species.
Amino acid sequences of COQ-2 orthologs were aligned using Clustal Omega (Madeira et al., 2019). The sequences of exons homologous to exon 6a/e in C. elegans, as well as the flanking five amino acid sequences, were used to generate the alignment. Sequences of the mutually exclusive exons are shaded in red (a-form) or blue (e-form). Two residue changes between the a- and e- forms are highlighted (Phe to Leu, Ala to Ser) and are invariant across diverse species that synthesize RQ. The COQ-2 orthologs and exons used for each species are listed in Supplementary file 3.
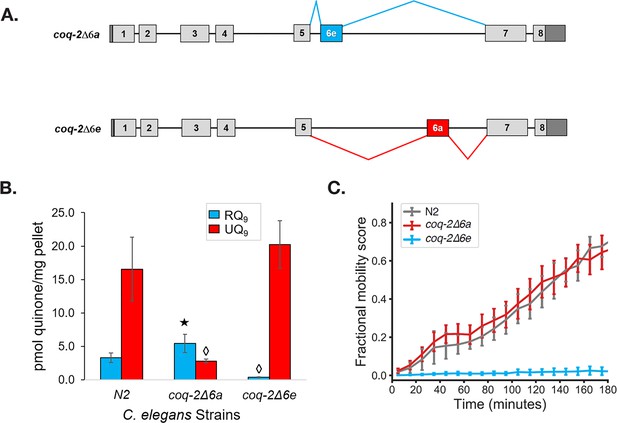
The C. elegans coq-2 edited strains and effects of exon 6a and 6e deletions on quinone biosynthesis.
(A) Mutant strains were generated in C. elegans by deletion of exon 6a (coq-2∆6a) or exon 6e (coq-2∆6e). (B) Deletion of exon 6a from the coq-2 gene significantly increased the level of RQ9 (p=0.013) and significantly decreased UQ9 (p<0.001) compared to the N2 control. By contrast, the deletion of exon 6e decreased RQ9 to a negligible level (p<0.001) and slightly increased the level of UQ9 (p=0.130) compared to N2. Statistically significant increases and decreases with respect to N2 levels are denoted with ★ and ◊, respectively; error bars reflect standard deviation where N = 4. (C) Deletion of coq-2 exon 6e affects the ability of worms to survive extended KCN treatment. Wild-type (N2) and coq-2 mutant L1 worms were exposed to 200 µM KCN for 15 hr. KCN was then diluted 6-fold and worm movement was measured over 3 hr to track recovery from KCN exposure (see Materials and methods). Worms without exon 6e could not survive extended treatment with KCN while deletion of exon 6a had little effect on KCN survival. Cyanide titration is shown in Figure 5—figure supplement 1. Curves show the mean of four biological replicates and error bars are standard errors of the mean.
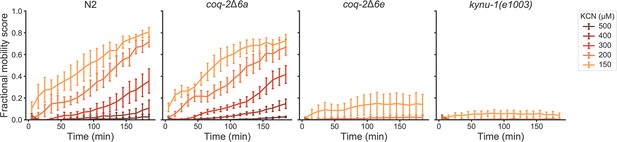
Deletion of coq-2 exon 6e affects the ability of worms to survive extended KCN treatment at various KCN concentrations.
Wild-type (N2), coq-2 and kynu-1(e1003) mutant L1 worms were exposed to 150–500 µM KCN for 15 hr. KCN was then diluted 6-fold and worm movement was measured over 3 hr to track recovery from KCN exposure (see Materials and methods). Worms without exon 6e could not survive extended treatment at KCN concentrations greater than 150 µM, and resembled kynu-1(e1003), a mutant devoid of RQ. Deletion of exon 6a had little effect on KCN survival and showed recovery patterns similar to wild-type worms. Curves show the mean of four biological replicates and error bars are standard errors of the mean.
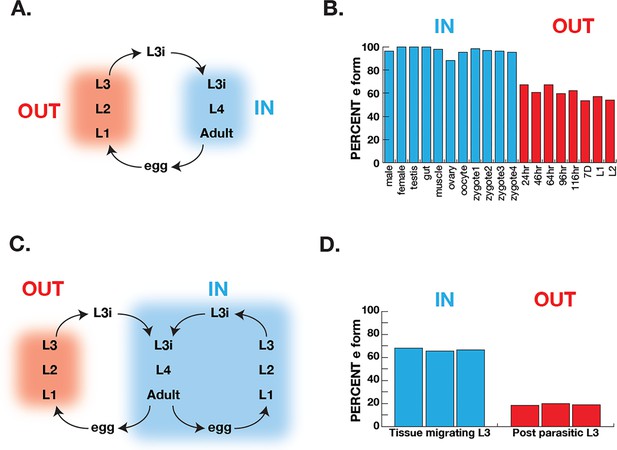
Correlation of COQ-2 splicing with change from aerobic to anaerobic life stages.
(A) Schematic of the life cycle of A. suum. ‘OUT’ denotes aerobically respiring free-living stages; ‘IN’ indicates stages living inside the host intestine. (B) Graph indicates the percentage of all COQ-2 transcripts that include the RQ-specific exon (Percent e-form) in a number of life cycle stages, sexes, and tissues. Timing of embryogenesis shows the post-fertilization time in hours. (C) Schematic of the life cycle of Strongyloides stercoralis. ‘OUT’ denotes aerobically respiring free-living stages; ‘IN’ indicates stages living inside the host. Note that egg, L1, L2, and L3 can either develop inside or outside the host. (D) Graph indicates the percentage of all COQ-2 transcripts that include the RQ-specific exon (Percent e-form) in L3 larvae that either developed outside the host (OUT) or inside the host (IN). Data were derived from three individual replicates taken from published RNA-seq data (Stoltzfus et al., 2012).
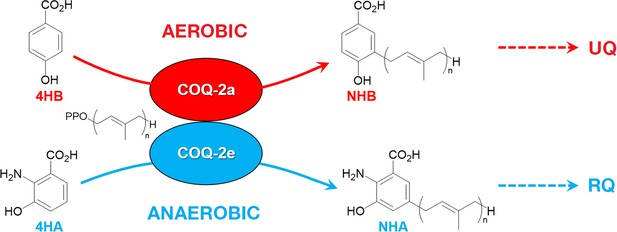
Discrimination between RQ and UQ biosynthesis in C.
elegans. There are two variants of exon 6 in the C. elegans coq-2 gene (6a and 6e) which undergo mutually exclusive alternative splicing leading to COQ-2a and COQ-2e isoforms, respectively. Synthesis of UQ originates from 4-hydroxybenzoic acid (4HB) and prenylation is catalyzed by COQ-2a (and marginally by COQ-2e) to form 4-hydroxy-3-nonaprenylbenzoic acid (NHB). By contrast, RQ is most likely synthesized from 3HA, and prenylation is facilitated by COQ-2e to form NHA. Several additional steps are required to convert NHB to UQ and NHA to RQ, respectively. Supplementary Documentation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | coq-2 | WormBase | WBGene00000762 | |
| Strain, strain background (Caenorhabditis elegans) | N2 | Caenorhabditis Genetics Center (CGC) | N2 | Wild-type |
| Strain, strain background (Caenorhabditis elegans) | coq-2 (syb1715) | This paper | PHX1715 | coq-2∆6a Supplementary file 1 |
| Strain, strain background (Caenorhabditis elegans) | coq-2 (syb1721) | This paper | PHX1721 | coq-2∆6e Supplementary file 1 |
| Strain, strain background (Escherichia coli) | OP50 | Caenorhabditis Genetics Center (CGC) | OP50 | |
| Sequence-based reagent | sgRNAs used in this study | This paper | Supplementary file 1 | |
| Sequence-based reagent | PCR primers used in this study | This paper | Supplementary file 1 | |
| Chemical compound, drug | Potassium cyanide (KCN) | Sigma-Aldrich | 60178–25G | Stock solution: 50 mM in PBS buffer |
| Other | Hexane, HPLC grade | Sigma-Aldrich | 650552–1L | |
| Other | Acetonitrile, LC-MS grade | Fisher Scientific | A955-4 | |
| Other | Ubiquinone-3 standard | Campbell et al., 2019 | ||
| Other | Ubiquinone-9 standard | Sigma-Aldrich | 27597–1 MG | |
| Other | Rhodoquinone-9 standard | Roberts Buceta et al., 2019 | Isolated from Ascaris suum | |
| Software, algorithm | Whippet | Sterne-Weiler et al., 2018 | RRID:SCR_018349 | https://github.com/timbitz/Whippet.jl |
| Software, algorithm | HISAT2 | Kim et al., 2019 | RRID:SCR_015530 | https://daehwankimlab.github.io/hisat2/ |
| Software, algorithm | UCSF Chimera | Resource for Biocomputing Visualization and Informatics Pettersen et al., 2004 | RRID:SCR_004097 | http://plato.cgl.ucsf.edu/chimera/ |
| Software, algorithm | MODELLER | University of California at San Francisco Webb and Sali, 2016 | RRID:SCR_008395 | https://salilab.org/modeller/ |
| Software, algorithm | Image analysis pipeline | Spensley et al., 2018 | https://github.com/fraser-lab-UofT/acute_assay | |
| Software, algorithm | Python | Python | RRID:SCR_008394 | https://www.python.org/ |
| Software, algorithm | NIS-Elements | Nikon | RRID:SCR_014329 | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| Software, algorithm | ImageMagick | ImageMagick | RRID:SCR_014491 | https://imagemagick.org/ |
| Software, algorithm | BioFormats | OME - Open Microscopy Environment | RRID:SCR_000450 | https://docs.openmicroscopy.org/bio-formats/5.7.1/users/comlinetools/index.html |
Additional files
-
Supplementary file 1
C. elegans strains.
- https://cdn.elifesciences.org/articles/56376/elife-56376-supp1-v2.elegansstrains.docx
-
Supplementary file 2
Statistical analysis of RQ9 and UQ9 levels in coq-2 mutant strains.
- https://cdn.elifesciences.org/articles/56376/elife-56376-supp2-v2.docx
-
Supplementary file 3
Genomic coordinates of known and predicted mutually exclusive coq-2 a/e exons.
- https://cdn.elifesciences.org/articles/56376/elife-56376-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56376/elife-56376-transrepform-v2.docx





