Chemical and structural investigation of the paroxetine-human serotonin transporter complex
Figures
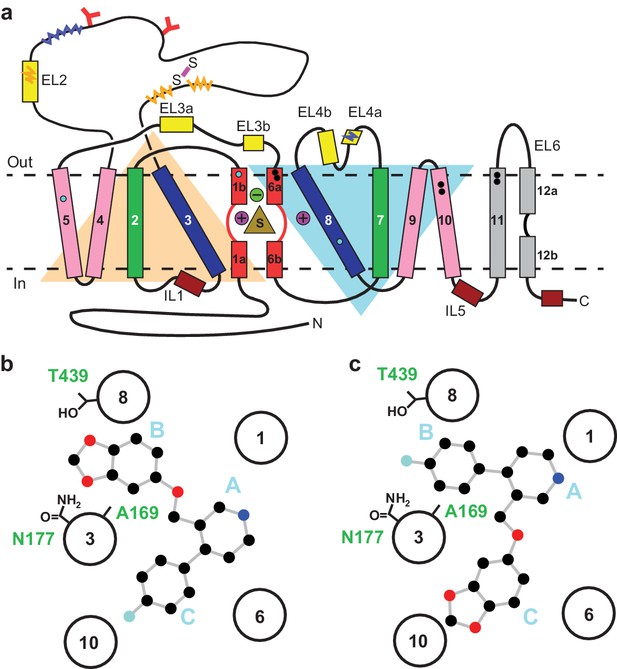
Topology of SERT.
(a) The substrate is bound at the central site (sand, triangle), near two sodium ions (purple, spheres +) and a chloride ion (green, sphere -). The light orange and light blue triangles depict pseudo two-fold symmetric helical repeats comprised of TM1-5 and 6–10, respectively. The disulfide bond (purple line) and N-linked glycosylation (red ‘Y’ shapes) in extracellular loop 2, along with sites of thermostable mutations (Tyr110Ala, TM1a; Ile291Ala, TM5; Thr439Ser, TM8) are also shown (cyan-filled circles). Structural elements involved in binding allosteric ligands are depicted as black-filled circles. Epitopes for the 8B6 and 15B8 Fab binding sites are in squiggly dark-blue and orange lines, respectively. (b) Schematic of the ABC pose of paroxetine bound to the central binding site, derived from the previously determined x-ray structures (Coleman and Gouaux, 2018; Coleman et al., 2016a). The transmembrane helices are shown with circles and mutated residues in subsite B are in sticks. c, The ACB pose of paroxetine bound to the central binding site of SERT predicted by molecular dynamics simulations and mutagenesis (Abramyan et al., 2019; Slack et al., 2019).
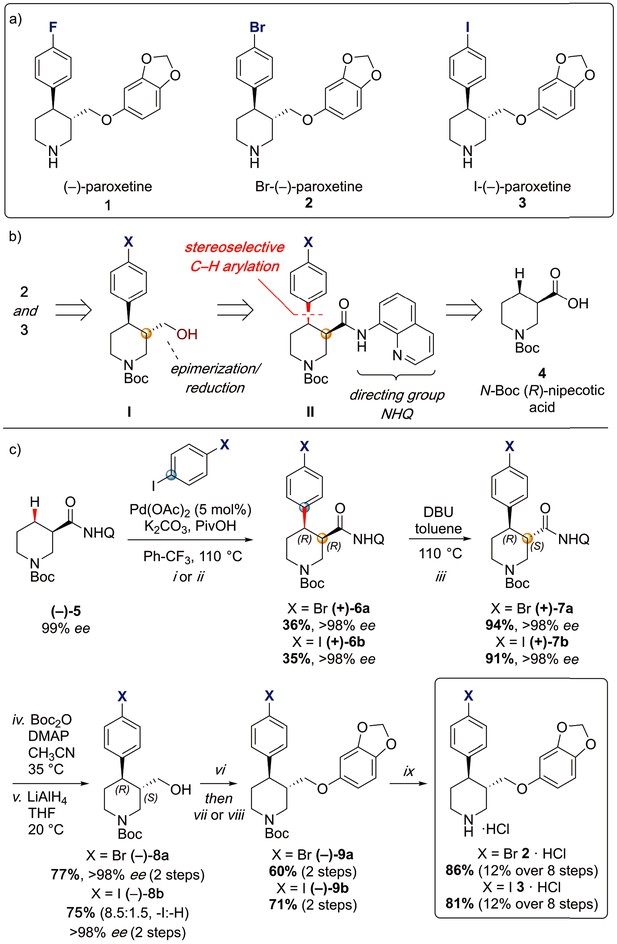
Synthesis of paroxetine analogues.
(a) Structures of (–)-paroxetine (1) and the targeted Br- (2) and I-analogues (3). (b) Retrosynthetic analysis of Br- and I-(–)-paroxetine. (c) Synthesis of Br- and I-(–)-paroxetine 2 and 3. Q = 8 quinolinyl-. Reaction conditions: i) X = Br: (–)−5 (4.0 mmol), 4-bromo iodobenzene (three equiv), Pd(OAc)2 (5 mol %), K2CO3 (one equiv), PivOH (one equiv), Ph-CF3 (2 mL, 2 M), 110°C, 18 hr; ii) X = I: (–)−5 (4.0 mmol), 1,4-diiodobenzene (four equiv), Pd(OAc)2 (5 mol %), K2CO3 (one equiv), PivOH (one equiv), Ph-CF3 (2 mL, 2 M), 110°C, 18 hr; iii) DBU (three equiv), toluene (1 M), 110°C, 24 hr; iv) Boc2O (four equiv), DMAP (20 mol %), CH3CN (0.5 M), 35°C, 22 hr; v) LiAlH4 (two equiv), THF, 20°C, 0.5 hr; vi) MsCl (1.3 equiv), Et3N (1.4 equiv), CH2Cl2, 0 to 25°C, 2 hr; vii) X = Br: sesamol (1.6 equiv), NaH (1.7 equiv), THF, 0°C to 70°C, 18 hr; viii) X = I: sesamol (2.0 equiv), NaH (2.2 equiv), DMF, 0°C to 90°C, 20 hr; ix) 4 N HCl in dioxane (10 equiv), 0°C to 25°C, 18 hr.
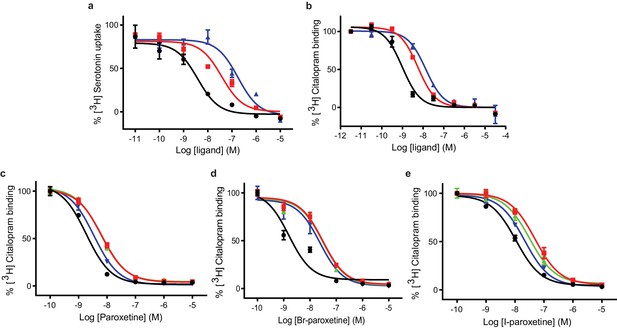
Inhibition of [3H]5-HT transport and [3H]citalopram binding by paroxetine and the Br- and I-derivatives.
(a) 5-HT-transport of wild-type SERT and its inhibition by paroxetine, Br-, and I-paroxetine. Data are mean ± s.e.m. (n = 6). (b) Competition binding of paroxetine and its derivatives to ts2-inactive SERT. In panels a and b, paroxetine, Br-paroxetine, and I-paroxetine curves are shown as black, red, and blue lines, respectively. Data are mean ± s.e.m. (n = 6). (c) Competition binding of paroxetine to ts2-active (black), Asn177Val (red), Asn177Thr (green), and Asn177Gln (blue). Data are mean ± s.e.m. (n = 3). (d) Competition binding of Br-paroxetine. Data are mean ± s.e.m. (n = 3). (e) Competition binding of I-paroxetine. Data are mean ± s.e.m. (n = 3). The values associated with these experiments are reported in Tables 2 and 3.
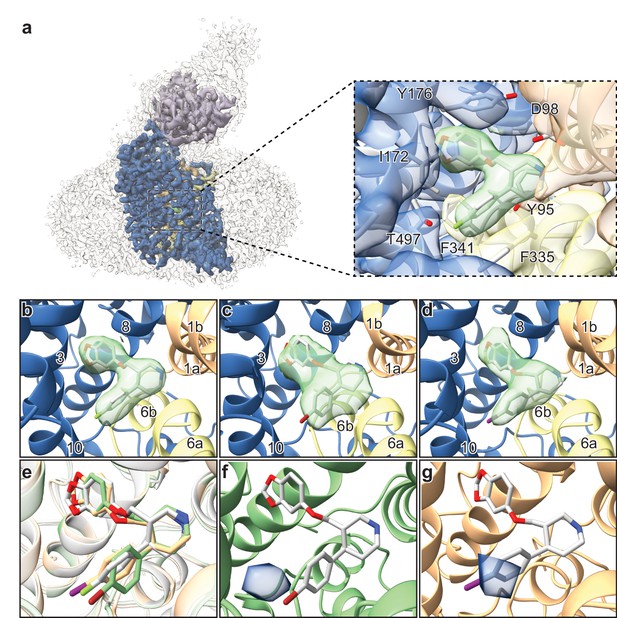
Structures of SERT-paroxetine complexes.
(a) Cryo-EM reconstruction of SERT bound to paroxetine where the shape of the SERT-8B6 Fab complex and detergent micelle is shown in transparent light grey. The density of SERT is shown in dark blue with TM1 and TM6 colored in orange and yellow, respectively, and the density for paroxetine in green. The variable domain of the 8B6 Fab is colored in purple. Inset shows the density features at the central site of paroxetine. (b) Density feature at the central site of paroxetine. (c) Density feature at the central site of Br-paroxetine. (d) Density feature at the central site of I-paroxetine. (e) Comparison of the binding poses of paroxetine (grey), Br-paroxetine (green), and I-paroxetine (orange). (f) Anomalous difference electron density (blue) derived from Br-paroxetine, contoured at 5.2σ. g, Anomalous difference electron density (blue) derived from I-paroxetine, contoured at 4.3σ.
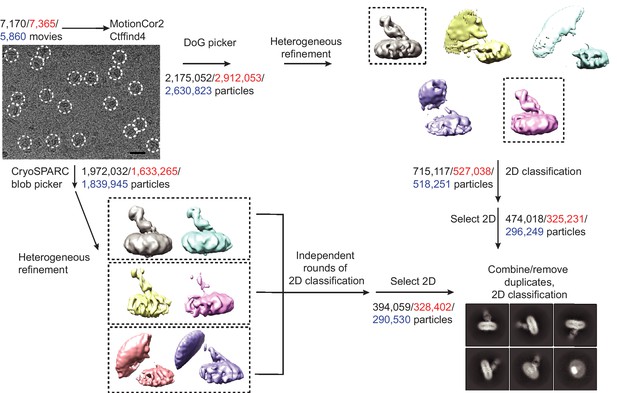
Work-flow of cryo-EM data processing of ΔN72/ΔC13 SERT/8B6 Fab/paroxetine complexes.
A representative zoomed, motion-corrected micrograph with individual single particles circled in white. Bar equals 20 nm. Motion-correction and CTF estimation was performed using MotionCor2 and Ctffind4. The number of movies/particles collected for each data set are shown in black (paroxetine), red (Br-paroxetine), and blue (I-paroxetine). After particle picking using either DoG picker or the blob picker in cryoSPARC, particles were sorted using heterogeneous refinement in cryoSPARC followed by 2D classification. For the DoG-picked particles, 3D classes containing SERT-Fab features (boxed) were combined and subjected to 2D classification. For cryoSPARC-picked particles, heterogeneous refinement was also used to initially sort particles in cryoSPARC. Classes with similar features (boxed) were combined, subjected to three independent 2D classifications, and 2D classes containing SERT-Fab features were combined. Particles picked by both methods were combined and duplicate particle-picks were removed in RELION (particle picks that are less than 100 Å of one another were considered duplicates).
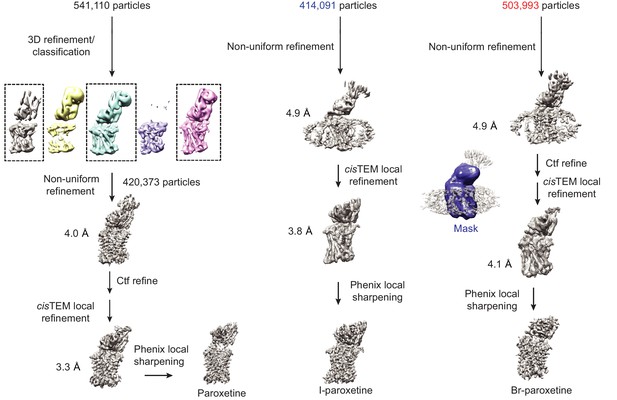
3D refinement of ΔN72/ΔC13 SERT/8B6 Fab/paroxetine complexes.
For the paroxetine complex, 3D refinement was performed in RELION followed by 3D classification without alignment and a mask which isolated SERT and Fab. 3D classification was not performed on the Br-paroxetine and I-paroxetine particles. Particles were further refined using non-uniform refinement in cryoSPARC, followed by local refinement in cisTEM with a mask which isolated SERT and the Fab variable domain and removed the Fab constant domain and micelle (mask is shown overlaid in blue on top of the Br-paroxetine reconstruction). The final reconstructed volume was sharpened using Phenix local sharpening.
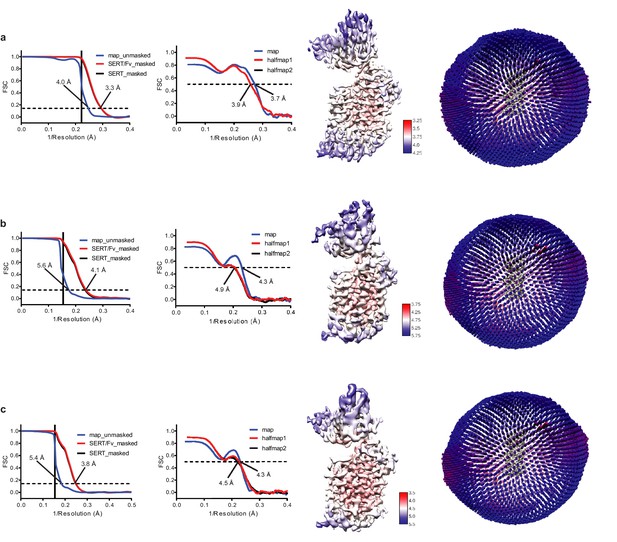
Cryo-EM reconstruction of ΔN72/ΔC13 SERT/8B6 Fab/paroxetine complexes.
(a) Reconstruction of SERT-8B6 paroxetine complex. Left panel, FSC curves for cross-validation, the final map (blue), masked SERT-Fv (red), and a mask which isolated SERT (black). The high-resolution limit cutoff for refinement was 4.5 Å. Middle left panel: model vs. half map 1 (working, red), half map 2 (free, black), model vs. final map (blue). Middle right panel: cryo-EM density map colored by local resolution estimation. Right panel: the angular distribution of particles used in the final reconstruction. (b) Reconstruction of the SERT-8B6 Br-paroxetine complex. The high-resolution limit cutoff for refinement was 6.5 Å. (c) Reconstruction of the SERT-8B6 I-paroxetine complex. The high-resolution limit cutoff for refinement was 6.5 Å.
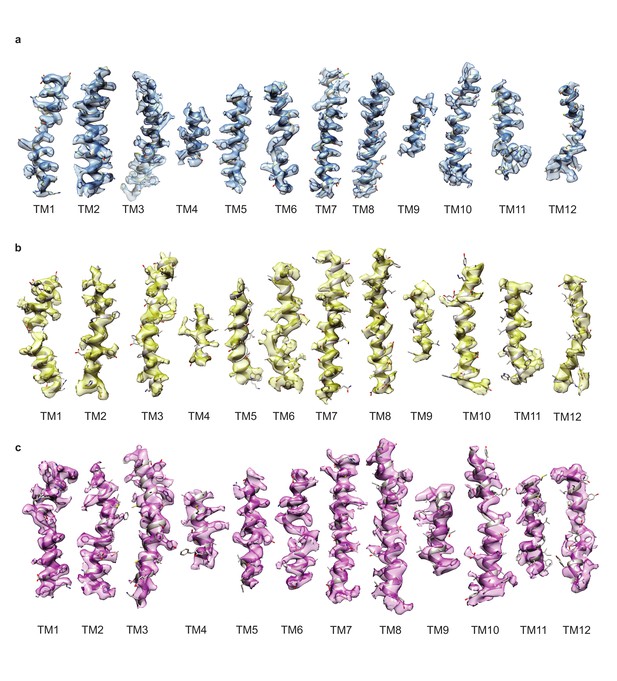
Cryo-EM density segments of the transmembrane helices.
(a) Density of TM1-12 of the paroxetine reconstruction, shown in blue. (b) Density of TM1-12 of the Br-paroxetine reconstruction, shown in yellow. (c) Density of TM1-12 of the I-paroxetine reconstruction, shown in purple.
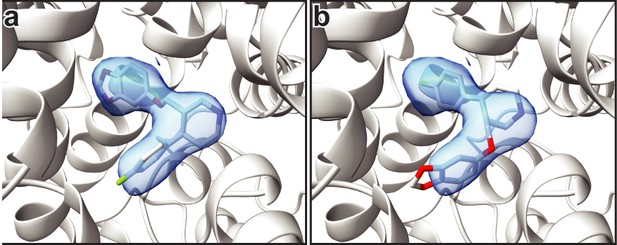
Comparison of the fit of paroxetine in the ABC and ACB poses.
(a) Shows the fit of paroxetine to the cryo-EM density in the ABC pose. (b) Shows the fit in the ACB pose.
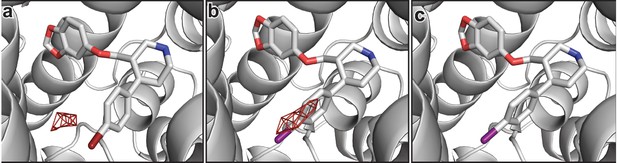
Isomorphous difference densities at the central site.
(a) A negative difference density feature (red mesh, 4σ) was observed in subsite C for the Fo(paroxetine)-Fo(Br-paroxetine) map. (b) A negative difference density feature (red mesh, 3.5σ) was observed in subsite C for the Fo(paroxetine)-Fo(I-paroxetine) map. (c) No significant difference densities for the Fo(Br-paroxetine)-Fo(I-paroxetine) map was observed at 3.5σ (shown).
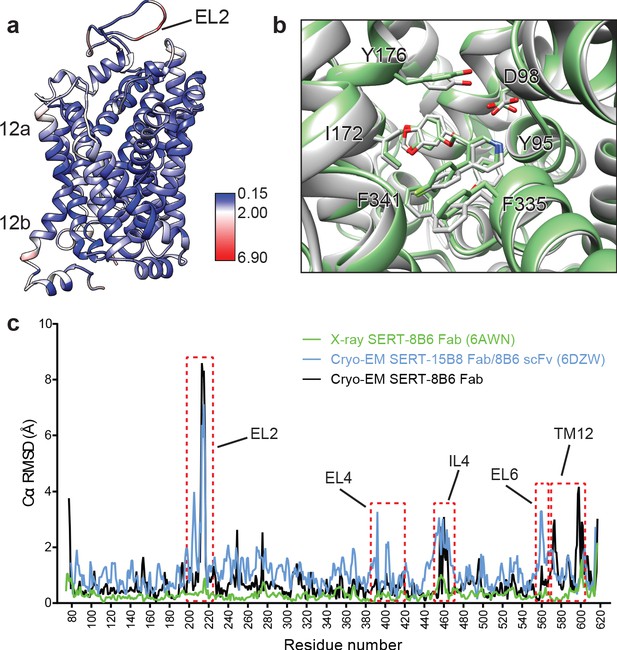
Comparison of the X-ray and cryo-EM structures of the SERT-paroxetine complex.
(a) Superposition of the x-ray ts3-SERT-8B6 paroxetine structure (PDB: 5I6X) with the SERT-8B6 paroxetine complex determined by cryo-EM. The root-mean-square-deviations (RMSD) for Cα positions were plotted onto the cryo-EM SERT-8B6 paroxetine structure. (b) Comparison of the central binding site of the x-ray (grey) and cryo-EM (green) paroxetine structures. (c) The structure of the ts2-inactive SERT-8B6 scFv/15B8 Fab paroxetine (cryo-EM, 6DZW), ts2-inactive SERT-8B6 Fab paroxetine (x-ray, 6AWN), and the SERT-8B6 paroxetine (cryo-EM, this work) complexes were superposed onto the ts3 SERT-8B6 paroxetine complex (x-ray, 5I6X) as a reference. The RMSD for Cα positions were calculated for each structure in comparison with the reference. Regions with RMSD > 3.0 Å are shown boxed in red.
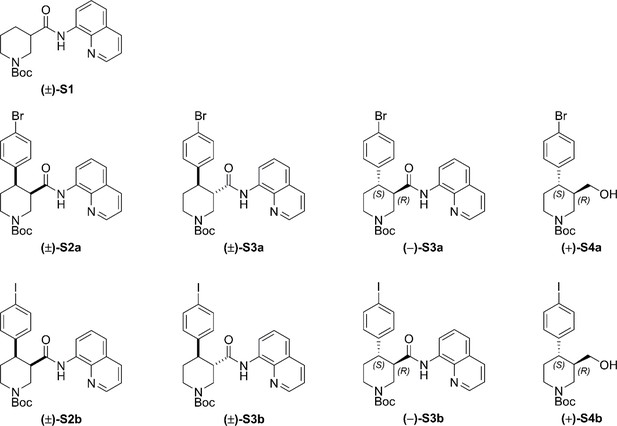
Full Synthetic Route to Racemic and Enantioenriched Br-Piperidine Derivatives (±)-S2a, (±)-S3a, (+)−6a, (+)−7a, (–)-S3a, (–)−8a, (+)-S4a, (–)−9a and Br-(–)-paroxetine 2.
In order to evaluate the enantiomeric excess of key intermediates (+)−6a and (+)−7a by chiral HPLC, the C–H arylation with 4-bromo iodobenzene was performed on both racemic ((±)-S1) and enantioenriched (–)−5 piperidine amide substrates (Scheme 1). The racemic synthesis was performed on a 0.5 mmol scale according to our previously reported protocol, (Antermite et al., 2018) and afforded cis-arylated derivatives (±)-S2a in 34% (Scheme 1a). A minor trans-functionalized product (±)-S3a, formed via a trans-palladacycle, (Antermite et al., 2018) was also isolated in 14%. C–H Arylation of enantioenriched substrate (–)−5 proceed smoothly on a 4.0 mmol scale, and cis- and trans-piperidine products (+)−6a and (–)-S3a were isolated as single enantiomers in very similar yields (Scheme S1b). Subsequent treatment of enantiopure cis-derivative (+)−6a with DBU at 100°C afforded the trans-diastereomer as the right-handed enantiomer (+)−7a in 94% yield.
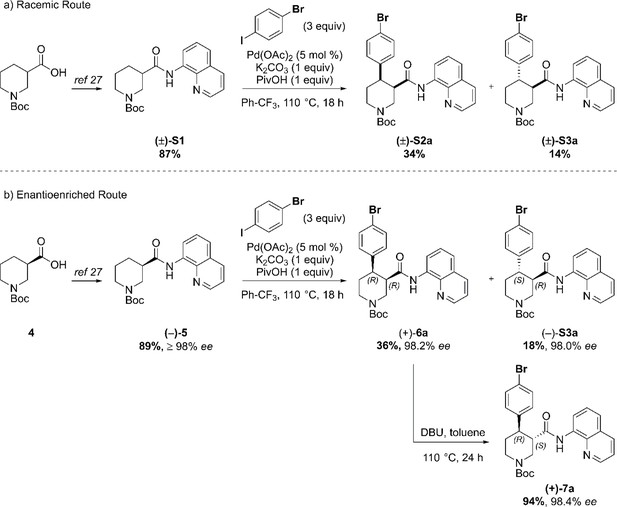
Synthetic sequence, including the Pd-catalyzed C(4)–H arylation step, to access racemic and enantioenriched cis- and trans-piperidine amide derivatives (±)-S2a, (±)-S3a, (+)−6a, (+)−7a and (–)-S3a.
(a) C–H Arylation conditions: (±)-S1 (0.5 mmol, one equiv) Ph-CF3 (500 μL, 1 M). (b) C–H Arylation conditions: (–)−5 (4.0 mmol, one equiv), Ph-CF3 (2.0 mL, 2 M). The enantiomeric excess of alcohol intermediates (+)-S4a and (–)−8a was evaluated after aminoquinoline removal on both enantiomeric trans-derivatives (–)-S3a and (+)−7a (Scheme 2). No undesired debromination was observed for the reductive aminoquinoline removal, and enantiopure alcohols (+)-S4a and (–)−8a were obtained in 70% and 77% yield, respectively. No erosion of enantiopurity should be expected after this step, given the literature precedents on the synthesis of (–)-paroxetine (Amat et al., 2000) and the absence of acidic protons in the substrate. Therefore, the synthesis was continued exclusively on alcohol derivative (–)−8a. O-Alkylation and Boc-deprotection with HCl finally afforded enantiopure Br-(–)-paroxetine analogue two as the corresponding hydrochloride salt in 12% yield over eight steps from commercial material.
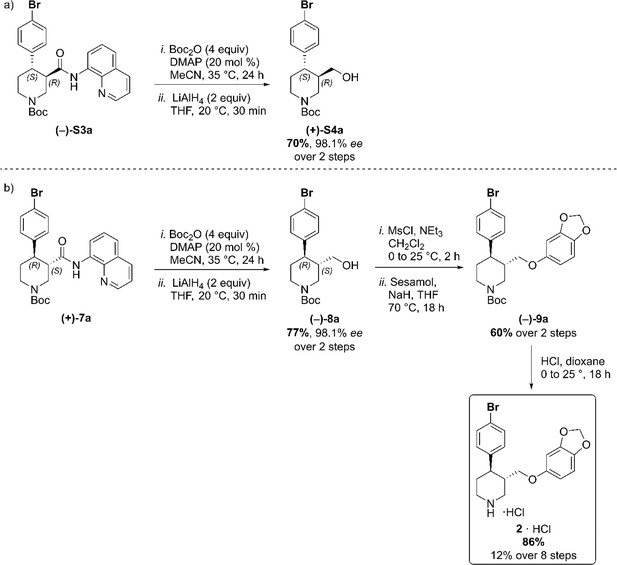
Reductive aminoquinoline removal and final steps in the synthesis of Br-(–)-paroxetine 2.
(a) AQ removal on enantiomerically pure trans-piperidine (–)-S3a (0.2 mmol, one equiv). (b) AQ removal on enantiomerically pure trans-piperidine (+)−7a (1.1 mmol, one equiv) and final steps in the synthesis of Br-(–)-paroxetine 2. Full Synthetic Route to Racemic and Enantioenriched I-Piperidine Derivatives (±)-S2b, (±)-S3b, (+)−6b, (+)−7b, (–)-S3b, (–)−8b, (+)-S4b, (–)−9b and I-(–)-paroxetine 3 Similarly to the Br-analogue, C–H arylation with 1,4-diiodobenzene was performed on both racemic ((±)-S1) and enantioenriched ((–)−5) piperidine amide substrates (Scheme 3). The reaction proceeded well on both substrates affording racemic cis- and trans-arylated products (±)-S2b and (±)-S3b in 35% and 19% yield, and enantioenriched cis- and trans-derivatives (+)−6b and (–)-S3b in 35% and 20% yield respectively.
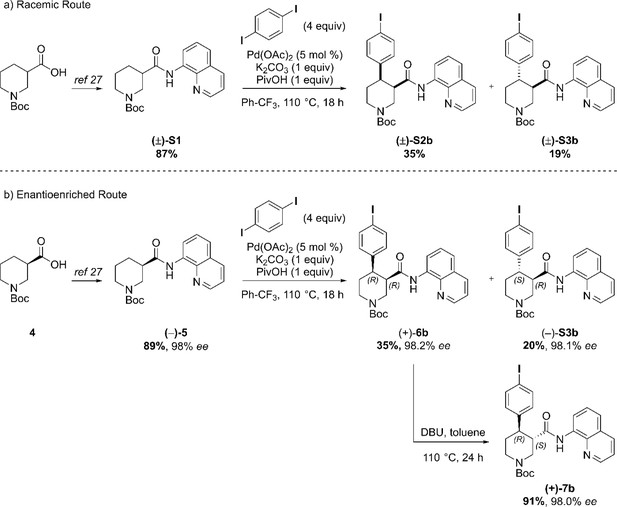
Synthetic sequence, including the Pd-catalyzed C(4)–H arylation step, to access racemic and enantioenriched cis- and trans-piperidine amide derivatives (±)-S2b, (±)-S3b, (+)−6b, (+)−7b and (–)-S3b.
(a) C–H Arylation conditions: (±)-S1 (0.5 mmol, one equiv) Ph-CF3 (500 μL, 1 M). (b) C–H Arylation conditions: (–)−5 (4.0 mmol, one equiv), Ph-CF3 (2.0 mL, 2 M). Reductive aminoquinoline cleavage was again performed to access enantiomeric trans-piperidine alcohols (+)-S4b and (–)−8b (Scheme 4). In both cases, a small degree of LiAlH4-mediated dehalogenation was observed, and an inseparable mixture of the desired product and 10–15% of deiodinated material was isolated. However, the contaminant could be effectively removed after O-Alkylation, affording the pure aryl ether derivative (–)−9b in 71% yield. Final HCl-mediated Boc deprotection formed the desired I-(–)-paroxetine three as the corresponding HCl salt in 81% yield (12% yield over eight steps from commercial material).
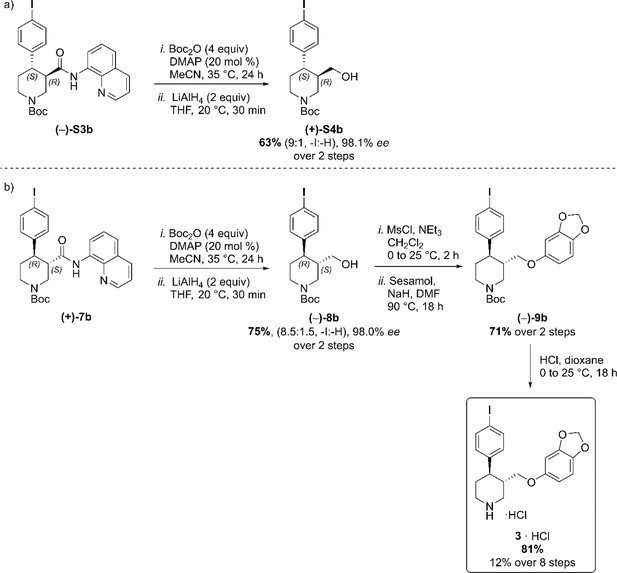
Reductive aminoquinoline removal and final steps in the synthesis of I-(–)-paroxetine 3.
(a) AQ removal on enantiomerically pure trans-piperidine (–)-S3b (0.2 mmol, one equiv). (b) AQ removal on enantiomerically pure trans-piperidine (+)−7b (1.0 mmol, one equiv) and final steps in the synthesis of I-(–)-paroxetine 3.
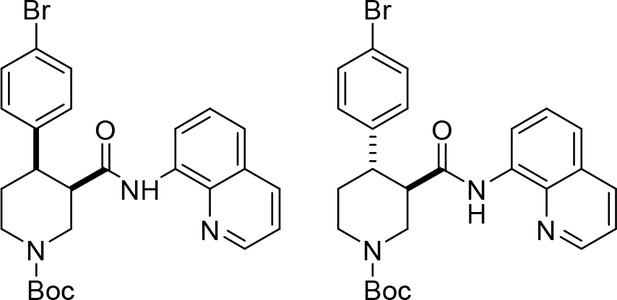
tert-Butyl cis-(±)−4-(4-bromophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((±)-S2a) and tert-butyl trans-(±)−4-(4-bromophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((±)-S3a).
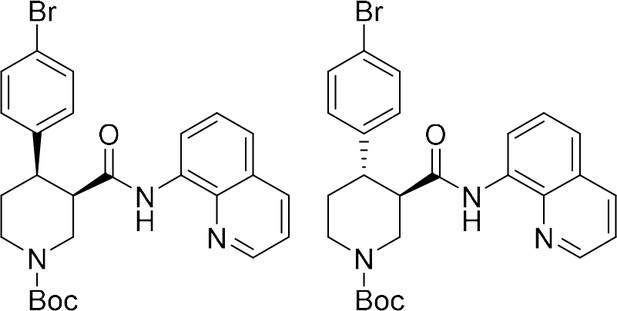
tert-Butyl (+)-(3R,4R)−4-(4-bromophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((+)−6a) and tert-butyl (–)-(3R,4S)−4-(4-bromophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((–)-S3a).
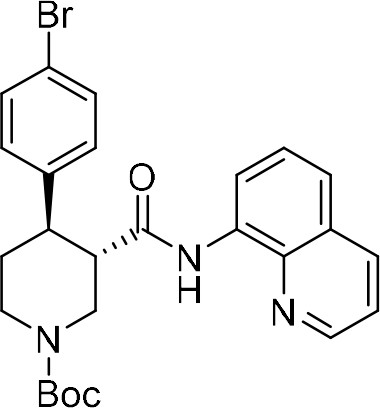
tert-Butyl (+)-(3S,4R)−4-(4-bromophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((+)−7a).

tert-Butyl (+)-(3R,4S)−4-(4-bromophenyl)−3-(hydroxymethyl)piperidine-1-carboxylate ((+)-S4a).
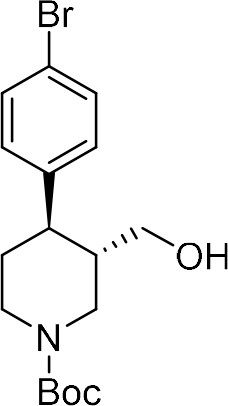
tert-Butyl (–)-(3S,4R)−4-(4-bromophenyl)−3-(hydroxymethyl)piperidine-1-carboxylate ((–)−8a).
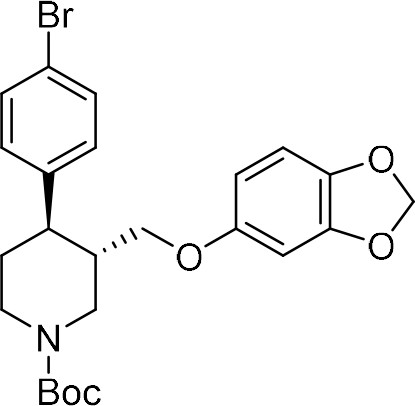
tert-Butyl (3S,4R)−3-((benzo[d][1,3]dioxol-5-yloxy)methyl)−4-(4-bromophenyl)piperidine-1-carboxylate ((–)−9a).

(3S,4R)−3-((Benzo[d][1,3]dioxol-5-yloxy)methyl)−4-(4-bromophenyl)piperidine-1-ium chloride (2 ∙ HCl).
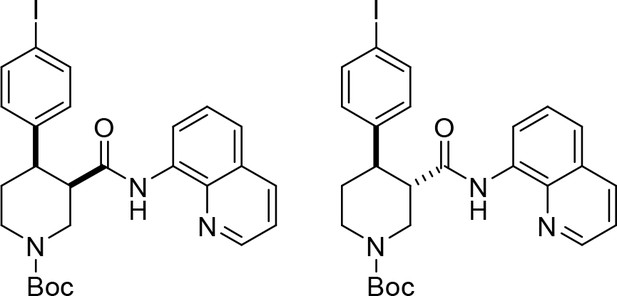
tert-Butyl cis-(±)−4-(4-iodophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((±)-S2b) and tert-butyl trans-(±)−4-(4-iodophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((±)-S3b).
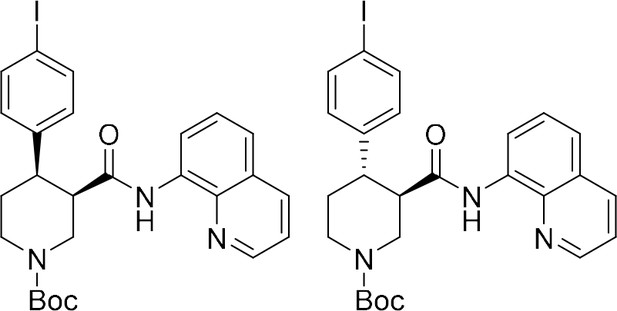
tert-Butyl (+)-(3R,4R)−4-(4-iodophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((+)−6b) and tert-butyl (–)-(3R,4S)−4-(4-iodophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((–)-S3b).

tert-Butyl (+)-(3S,4R)−4-(4-iodophenyl)−3-(quinolin-8-ylcarbamoyl)piperidine-1-carboxylate ((+)−7b).
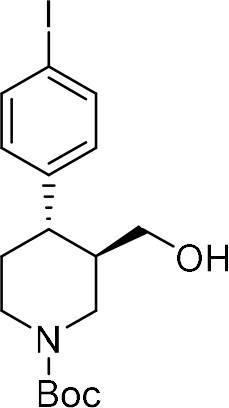
tert-Butyl (+)-(3R,4S)−4-(4-iodophenyl)−3-(hydroxymethyl)piperidine-1-carboxylate ((+)-S4b) .
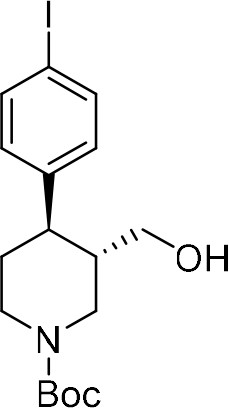
tert-Butyl (–)-(3S,4R)−4-(4-iodophenyl)−3-(hydroxymethyl)piperidine-1-carboxylate ((–)−8b) .
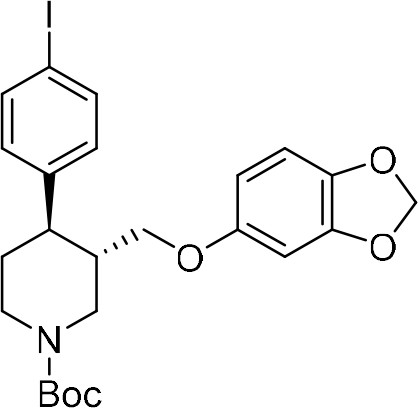
tert-Butyl (3S,4R)−3-((benzo[d][1,3]dioxol-5-yloxy)methyl)−4-(4-iodophenyl)piperidine-1-carboxylate ((–)−9b) Alcohol (–)−8b (203 mg, 0.49 mmol, one equiv) and triethylamine (96 μL, 0.69 mmol, 1.4 equiv) were added to a flame-dried round-bottom flask, dissolved in anhydrous CH2Cl2 (2.5 mL, 0.2 M) and cooled down to 0°C.
Methanesulfonyl chloride (49 μL, 0.64 mmol, 1.3 equiv) was then added by Gilson pipette. After stirring 5 min at 0°C, the reaction mixture was stirred at 25°C for 2 hr, then diluted with CH2Cl2 (5 mL) and sat. aq. NaHCO3 (5 mL). The phases were separated, and the aqueous layer was extracted with CH2Cl2 (3 × 10 mL). The combined organic extracts were dried over Na2SO4 and filtered. The solvent was removed under reduced pressure to afford the crude mesylated alcohol derivative.
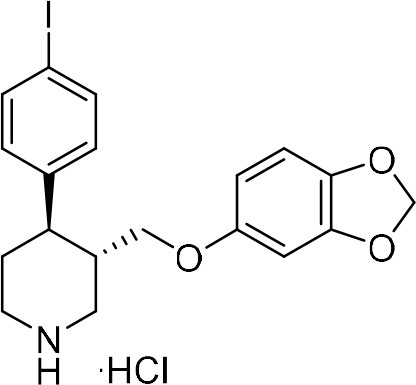
(3S,4R)−3-((Benzo[d][1,3]dioxol-5-yloxy)methyl)−4-(4-iodophenyl)piperidine-1-ium chloride.
(3 ∙ HCl) 4 N HCl in 1,4-dioxane (250 μL, 1.00 mmol, 10 equiv) was added to a solution of N-Boc protected piperidine (–)−9b (56.9 mg, 0.10 mmol) in 1,4-dioxane (250 μL, 0.4 M). At 0°C under air. The solution was stirred at 25°C for 18 hr, then an ice-cold 1:1 mixture of Et2O/pentane (1 mL) was added and formation of a solid precipitate was observed. This was filtered and washed with further ice-cold Et2O/pentane mixture (2 × 5 mL). The solid precipitate was dried under reduced pressure to afford (3S,4R)−3-((benzo[d][1,3]dioxol-5-yloxy)methyl)−4-(4-iodophenyl)piperidine-1-ium chloride 3 ∙ HCl (38.5 mg, 81%) as an off-white solid.
Tables
Expression constructs used in this study.
| Name | Expression construct | Experiment |
|---|---|---|
| Wild-type SERT | Full-length human SERT with a C-terminal thrombin-GFP-StrepII-His10 tag. | [3H] 5-HT transport assays |
| ∆N72/ ∆C13 SERT | Wild-type SERT modified by deletion of 72 residues on N-term and 13 residues on C-term | Cryo-electron microscopy |
| ts2-inactive | Full-length SERT with thrombin cleavage sites inserted after Gln76 and Thr618 and carrying the Tyr110Ala, Ile291Ala thermostabilizing mutations with additional mutations of surface-exposed cysteines Cys554, Cys580, and Cys622 to alanine | X-ray crystallography and [3H] citalopram binding assays |
| ts2-active | Full-length SERT with thrombin cleavage sites inserted after Gln76 and Thr618 and carrying the Ile291Ala, Thr439Ser thermostabilizing mutations with additional mutations of surface-exposed cysteines Cys554, Cys580, and Cys622 to alanine | [3H] citalopram binding assays |
| Asn177 mutants | Asn177 mutated to either Val, Thr, or Gln in ts2-active background | [3H] citalopram binding assays |
Inhibition of 5-HT transport by paroxetine and its derivatives.
| Ligand | IC50 |
|---|---|
| Paroxetine | 4 ± 1 nM |
| Br-paroxetine | 40 ± 20 nM |
| I-paroxetine | 0.18 ± 0.07 µM |
Binding of paroxetine and its derivatives to SERT variants used in this study.
| SERT variant | Ki (nM) | ||
|---|---|---|---|
| Paroxetine | Br-paroxetine | I-paroxetine | |
| ts2-inactive | 0.17 ± 0.02 | 0.94 ± 0.01 | 2.3 ± 0.1 |
| ts2-active | 0.31 ± 0.07 | 0.4 ± 0.2 | 1.7 ± 0.3 |
| Asn177Val | 1.11 ± 0.04 | 5 ± 1 | 7.3 ± 0.9 |
| Asn177Thr | 1.0 ± 0.1 | 5 ± 2 | 4.4 ± 0.4 |
| Asn177Gln | 0.58 ± 0.07 | 4 ± 1 | 3.6 ± 0.4 |
Cryo-EM data collection, refinement and validation statisticsa.
| #1 (EMDB-21368) (PDB 6VRH) (EMPIAR-10380) | #2 (EMDB-21369) (PDB 6VRK) | #3 (EMDB-21370) (PDB 6VRL) | |
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 77,160 | 77,160 | 77,160 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 54–60 | 54 | 54 |
| Defocus range (μm) | −0.8 to −2.2 | −0.8 to −2.2 | −0.8 to −2.2 |
| Pixel size (Å) | 0.648 | 0.648 | 0.648 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 4,147,084 | 4,545,318 | 4,470,768 |
| Final particle images (no.) | 420,373 | 503,993 | 414,091 |
| Map resolution (Å) FSC threshold | 3.3 0.143 | 4.1 0.143 | 3.8 0.143 |
| Map resolution range (Å)† | 4.25–3.25 | 5.75–3.75 | 5.50–3.50 |
| Refinement | |||
| Initial model used (PDB code) | 6AWN | 6VRH | 6VRH |
| Initial model CC Model resolution (Å)‡ FSC threshold | 0.64 3.7 0.5 | 0.70 4.3 0.5 | 0.71 4.1 0.5 |
| Model resolution range (Å) | 25.9–3.3 | 33.0–4.1 | 29.6–4.2 |
| Map sharpening B factor (Å2) | −85 | −174 | −161 |
| Model composition Non-hydrogen atoms Protein residues Ligands (atoms) | 6143 764 254 | 6142 764 254 | 6142 764 254 |
| B factors (Å2) Protein Ligand | 138 129 | 138 113 | 122 112 |
| R.m.s. deviations Bond lengths (Å) Bond angles (°) | 0.002 0.48 | 0.002 0.59 | 0.002 0.54 |
| Validation Refined model CC MolProbity score Clashscore Poor rotamers (%) | 0.73 1.86 9.67 0 | 0.74 1.96 10.26 0 | 0.75 1.88 10.59 0.00 |
| Ramachandran plot Favored (%) Allowed (%) Disallowed (%) | 94.84 5.16 0 | 93.54 6.46 0 | 95.12 4.88 0 |
-
aData set #1 is the paroxetine reconstruction, #2 is Br-paroxetine, #3 I-paroxetine.
†Local resolution range.
-
‡Resolution at which FSC between map and model is 0.5.
X-ray data collection statistics.
| Br-paroxetine (PDB 6W2B) | I-paroxetine (PDB 6W2C) | |
|---|---|---|
| Data collection | ||
| Space group | C2221 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 128.0, 161.9, 139.7 | 127.7, 161.9, 140.8 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 20.45–4.69 (4.82–4.69)* | 25.98–6.12 (6.30–6.12)* |
| Rmerge | 13.60 (339.3) | 7.9 (292.9) |
| I / σI CC1/2 | 5.51 (0.49) 99.9 (16.5) | 5.01 (0.32) 99.8 (20.0) |
| Completeness (%) | 99.2 (100.0) | 92.6 (89.7) |
| Redundancy | 6.8 (6.2) | 1.8 (1.7) |
-
*Values in parentheses are for highest-resolution shell.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | Human serotonin transporter | cDNA | NCBI Reference Sequence: NP_001036.1 | Dr. Randy D. Blakely (Florida Atlantic university brain institute) |
| Cell line (Homo sapiens) | HEK293S GnTI- | ATCC | Cat # ATCC CRL-3022 | Used for expression of SERT (PMID:27929454) |
| Cell line (Spodoptera frugiperda) | SF9 cells | ATCC | Cat # ATCC CRL-1711 | Used in production of baculovirus for transduction, and SERT antibodies (PMID:27929454) |
| Antibody | Mouse monoclonal. Isotype IgG2a, kappa | OHSU VGTI, Monoclonal Antibody Core | 8B6 | |
| Transfected construct (human) | pEG BacMam | Gouaux lab | PMID:25299155 | |
| Affinity chromatography resin | Strep-Tactin Superflow high capacity resin | Iba life sciences | Cat#2-1208-500 | |
| Chemical compound, drug | n-dodecyl-β-D-maltoside | Anatrace | Cat # D310 | Detergent |
| Chemical compound, drug | n-octyl β-D-maltoside | Anatrace | Cat # O310 | Detergent |
| Chemical compound, drug | fluorinated octyl-maltoside | Anatrace | Cat # O310F | Detergent |
| Chemical compound, drug | Cholesteryl Hemisuccinate | Anatrace | Cat # CH210 | Lipid |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine | Anatrace | Cat # P516 | Lipid |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine | Anatrace | Cat # P416 | Lipid |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol | Anatrace | Cat # P616 | Lipid |
| Chemical compound, drug | Paroxetine hydrochloride hemihydrate | Sigma | Cat # P9623 | Inhibitor |
| Chemical compound, drug | [3H]5-HT | PerkinElmer | Cat # NET1167250UC | Radiolabeled substrate |
| Chemical compound, drug | [3H]citalopram | PerkinElmer | Cat # NET1039250UC | Radiolabeled inhibitor |
| Software, algorithm | XDS | PMID:20124692 | RRID:SCR_015652 | http://xds.mpimf-heidelberg.mpg.de/ |
| Software, algorithm | Phaser | PMID:24189240 | RRID:SCR_014219 | https://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| Software, algorithm | Phenix | PMID:22505256 | RRID:SCR_014224 | https://www.phenix-online.org/ |
| Software, algorithm | SerialEM | PMID:16182563 | RRID:SCR_017293 | http://bio3d.colorado.edu/SerialEM |
| Software, algorithm | MotionCor2 | PMID:28250466 | RRID:SCR_016499 | http://msg.ucsf.edu/em/software/motioncor2.html |
| Software, algorithm | CTFFIND4 | PMID:26278980 | RRID:SCR_016732 | https://grigoriefflab.umassmed.edu/ctffind4 |
| Software, algorithm | DoG-Picker | PMID:19374019 | http://emg.nysbc.org/redmine/projects/software/wiki/DoGpicker | |
| Software, algorithm | cryoSPARC | PMID:28165473 | RRID:SCR_016501 | https://cryosparc.com/ |
| Software, algorithm | RELION | PMID:23000701 | RRID:SCR_016274 | http://www2.mrc-lmb.cam.ac.uk/relion |
| Software, algorithm | cisTEM | PMID:29513216 | RRID:SCR_016502 | https://cistem.org/ |
| Software, algorithm | UCSF-Chimera | PMID:15264254 | RRID:SCR_004097 | https://www.cgl.ucsf.edu/chimera/ |
| Software, algorithm | Coot | PMID:15572765 | RRID:SCR_014222 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| Software, algorithm | MolProbity | PMID:20057044 | RRID:SCR_014226 | http://molprobity.biochem.duke.edu/ |
| Other | R 2/2 200 mesh Au holey carbon grids | Electron Microscopy Sciences | Cat # Q2100AR2 | Cryo-EM grids |
| Other | Copper HIS-Tag YSI | PerkinElmer | Cat # RPNQ0096 | SPA beads |
Additional files
-
Supplementary file 1
HPLC Traces for Racemic, Scalemic and Enantioenriched Br-Piperidine Derivatives (±)-S2a, (±)-S3a, (+)-6a, (+)-7a, (–)-S3a, (–)-8a and (+)-S4a.
- https://cdn.elifesciences.org/articles/56427/elife-56427-supp1-v2.docx
-
Supplementary file 2
NMR Spectra for Novel Compounds.
- https://cdn.elifesciences.org/articles/56427/elife-56427-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56427/elife-56427-transrepform-v2.docx





