The Mla pathway in Acinetobacter baumannii has no demonstrable role in anterograde lipid transport
Figures
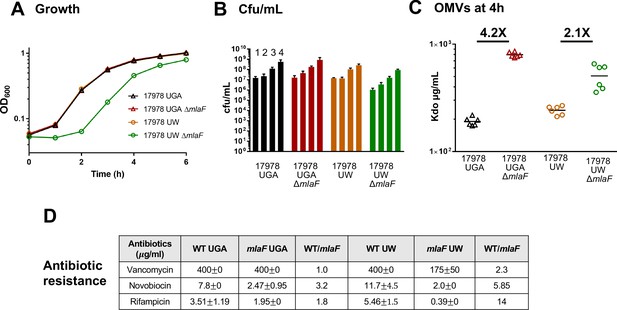
ΔmlaF mutant from the UW group exhibits significant growth defects.
(A) Growth curves of WT and ΔmlaF deletion mutants from the UGA lab (triangles) and UW lab (circles). Standard deviation error bars, if smaller than symbols, are not shown. (B) Colony-forming units (cfu) of cultures at hours 1–4. (C) Outer membrane vesicle (OMV) quantification of WT and ΔmlaF deletion mutants harvested at 4 hr and quantified by measuring 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) levels using the Purpald assay. Growth curves were performed in biological triplicate. OMV assays were performed in biological duplicate and quantified in technical triplicates. (D) MICs for vancomycin, novobiocin, and rifampicin for both UGA and UW strains. MICs were calculated as described in the Materials and methods.
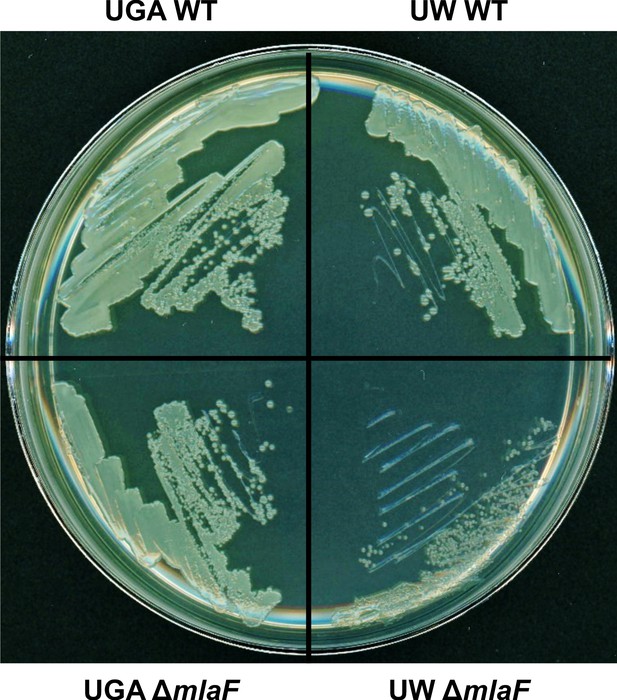
UW ΔmlaF has a distinct colony morphology.
Top left – UGA WT grown on LB + 1.5% agar. Bottom left – UGA ΔmlaF. Top right – UW WT. Bottom right – UW ΔmlaF.
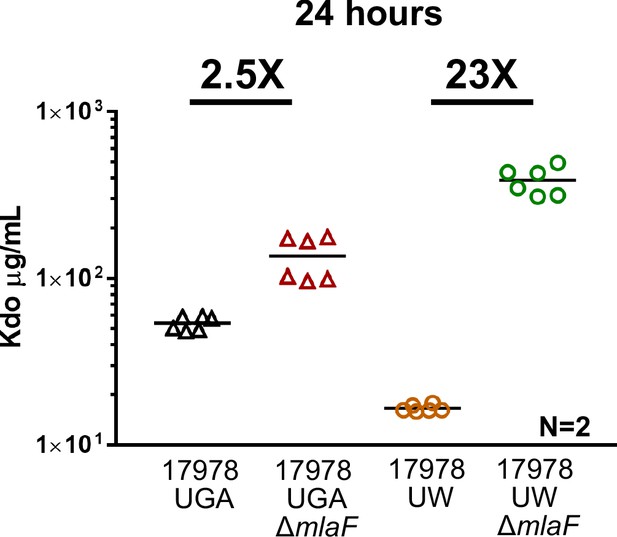
OMVs collected from stationary phase cultures at 24 hr.
The UW ΔmlaF strain exhibits lysis in stationary phase, artificially inflating OMVs with cellular debris when collected after 24 hr.
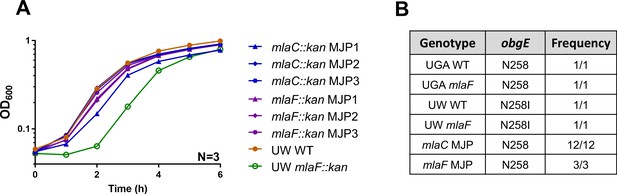
obgE* alleles are selected against in the absence of Mla.
(A) Growth curves of ΔmlaC (blue) or ΔmlaF (green) mutants generated in the UW WT. Any strains generated by our hands in this background are annotated with MJPX, with X indicating an independently generated clone. For comparison, the UW WT and UW ΔmlaF provided to us are displayed as black and orange circles respectively. Growth curves were performed in biological triplicate. (B) Table displaying frequency of obgE* to obgE reversion. Both UGA strains contain the WT N258 allele. Both UW strains contain the obgE* allele of N258I. When ΔmlaC strains were generated in the UW WT, obgE* reverted to obgE 12/12 times tested. When ΔmlaF strains were generated in the UW WT, obgE* reverted to obgE 3/3 times tested. SNP presence was determined by sanger sequencing.
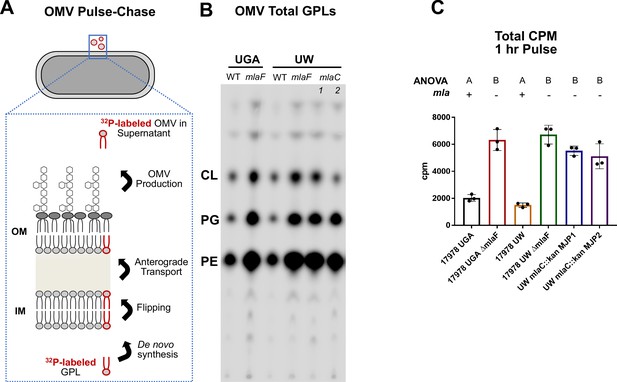
Mla mutants exhibit no defect in anterograde transport of GPLs.
(A) Schematic of the biological principle behind the OMV pulse chase assay. Cultures were pulsed with 32Pi, which is incorporated in de novo synthesized lipids (red outlined GPL). After synthesis, lipids are flipped across the IM and anterograde transported to the OM. Over the course of the experiment, a percentage of lipids from the OM will be shed as OMVs. Any 32P-labeled OMVs must have incorporated GPLs that underwent anterograde transport. (B) Representative TLC of supernatant GPLs. Total sample was loaded after LSC quantification. GPLs were separated in solvent system containing chloroform, methanol, and acetic acid (65:25:10, v/v/v). (C) LSC quantification of extracted GPLs. Circles represent individual replicates. Lettering above denotes significantly different clusters as determined by a one-way ANOVA.
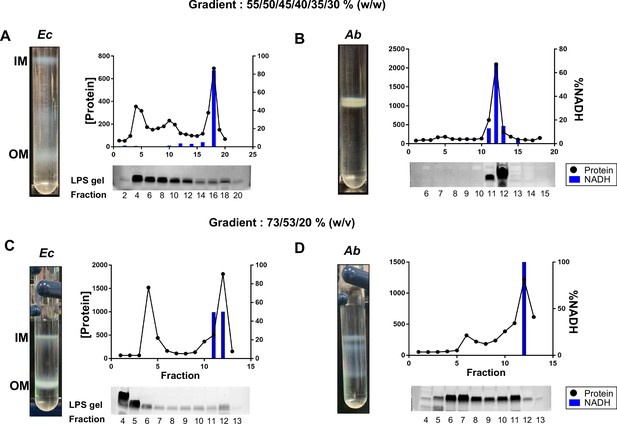
Membrane separations do not work in A. baumannii.
(A) Left – Representative image of IM and OM separations of E. coli membranes using the protocol as designed by Osborn’s group. Right – Marker analysis. Graph displays protein content on the left y-axis with black circles and %NADH activity (IM-marker) on the right y-axis in blue bars across fractions for the entire gradient (x-axis). An SDS-PAGE gel stained for LPS (OM marker) is displayed immediately below the respective graph and numbered by fraction. (B) Left – Representative image of IM and OM separations of A. baumannii membranes using the protocol as designed by Osborn’s group. Right – Marker analysis as described above. (C) Left – Representative image of IM and OM separations of E. coli membranes using the protocol as described by the UW group. Right – Marker analysis as described above. (D) Left – Representative image of IM and OM separations of A. baumannii membranes using the protocol as described by the UW group. Right – Marker analysis as described above.
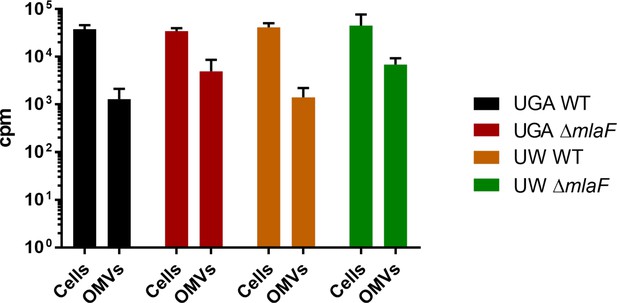
Liquid scintillation counts of whole cells and OMVs from pulse-chase assay.
After pulse-chase, total GPLs were isolated from cell pellets in addition to the isolation of OMVs for the assay. GPLs isolated in this manner were counted by liquid scintillation counting.

Validation of OMV pulse-chase assay.
(A) α-OmpA blot of either OMVs or total membranes (Mem) for each genotype tested. Total protein was normalized to 5 µg. Blot is representative of biological duplicates. (B) NADH oxidase activity of OMVs or total membranes over time. For each sample, protein was normalized to 3 µg. Error bars are standard deviation of technical triplicates and graph is representative of biological duplicates.
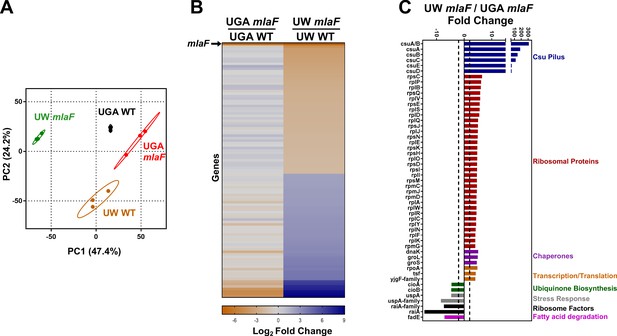
Transcriptomic profiling highlights substantial differences between ΔmlaF mutants.
(A) Principal components analysis (PCA) of total transcriptomic profiles of UGA WT (purple square) and ΔmlaF (blue square) and UW WT (green circle) and ΔmlaF (red circle). Each point represents a biological replicate. (B) Heat map comparing differentially regulated genes between UW ΔmlaF and UW WT. All genes in the right column are differentially expressed with a > |2| log2 fold change and a FDR p-value<0.05. For comparison, fold-change values of the same set of genes for the UGA ΔmlaF vs UGA WT are displayed in the left column. The row representing mlaF is denoted with an arrow. A full list of genes, fold-changes, and p-values for both comparisons are in Supplementary files 1 and 2. (C) Comparison of total transcriptomic profiles between UW ΔmlaF and UGA ΔmlaF mutants. Genes are clustered and colored by predicted or known function. Dashed lines denote +/- 2 fold-change. All transcriptomic data are presented as average RPKM values from three biological replicates.
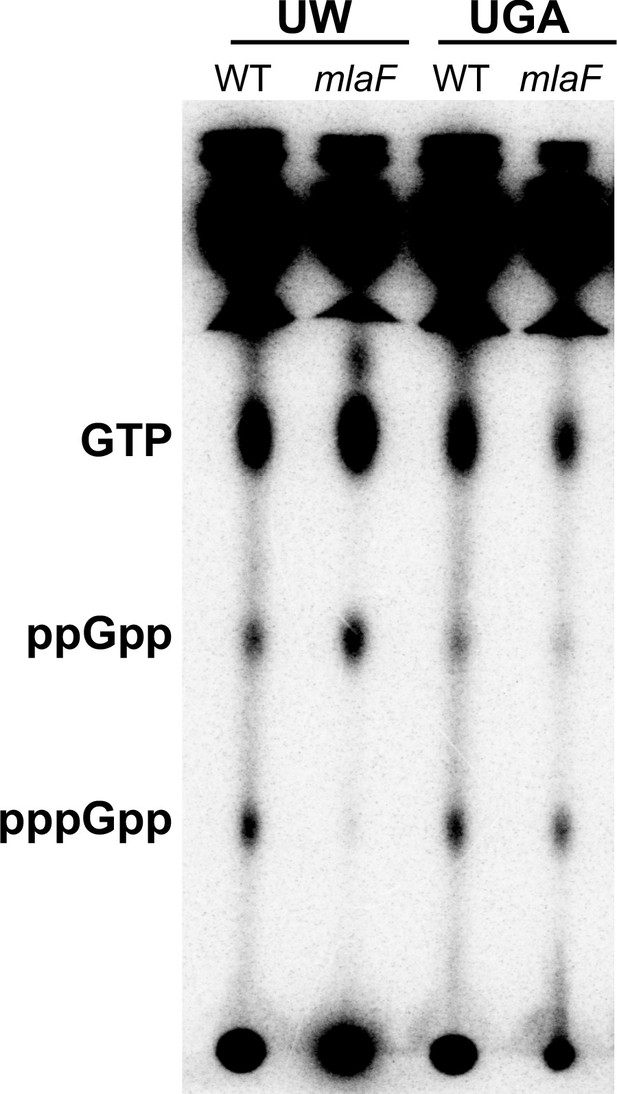
UW ΔmlaF mutant accumulates ppGpp.
(A) Representative TLC of 32Pi-labeled nucleotides extracted from whole cells separated on PEI-cellulose plates in 1.5 M KH2PO4 mobile phase. Positions of GTP, ppGpp, and pppGpp are denoted on the left. The image shown here is representative of three biological replicates. Additional replicate images are shown in Figure 5—figure supplement 1.
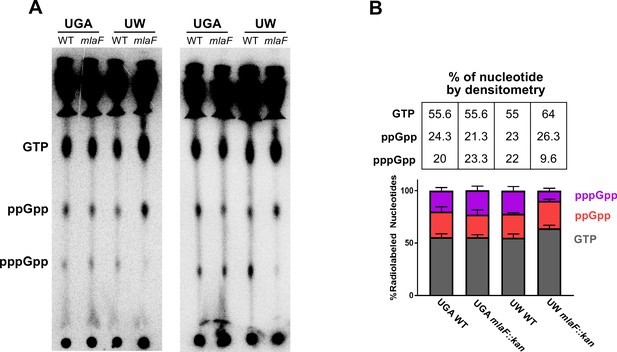
Nucleotide profile of UGA and UW strains.
(A) Additional replicates of TLC separation of radiolabeled nucleotides as described in Figure 5. Relative positions of pppGpp, ppGpp, and GTP are labeled. (B) Densitometry quantification of pppGpp, ppGpp, and GTP spots. Relative percentages were calculated within a given genotype. Error bars are standard deviation of three replicates. Distributions are visualized by bar chart and colored by nucleotide species. Average percentages of three biological replicates are displayed in the table.
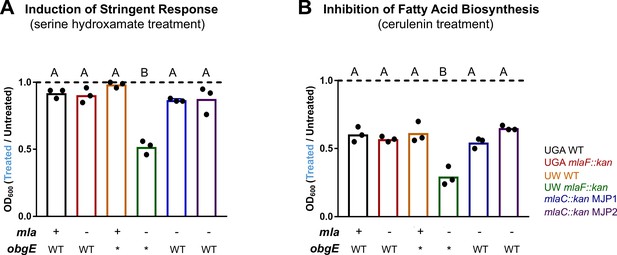
Δmla with obgE* is synthetically sensitive to chemical modulation of stringent response or fatty acid biosynthesis.
(A) Normalized growth of cultures treated with 100 µg/mL serine hydroxamate which induces stringent response. Graphs represent normalized OD600 readings of treated vs untreated. Dashed line at 1.0 indicates no difference in growth of treated culture. Values < 1.0 mean growth was decreased after treatment. Bars are colored by genotype with individual circles representing biological replicates. The mla and obgE alleles are indicated below for ease of comparison. Lettering above denotes significantly different clusters as determined by a one-way ANOVA. (B) Normalized growth of cultures treated with 100 µg/mL cerulenin which inhibits fatty acid biosynthesis. Graphs are displayed identically as panel (A).

Multiple sequence alignment of obgE across A. baumannii.
Portion displayed is from amino acid positions 210 to 269, with position 258 annotated by an arrow. Asterisks represent conservation across all samples. Top 10 rows are strains utilized during this study. Any strains derived from the UW WT are denoted. Bottom 14 rows are from published A. baumannii genomes. I258 is boxed in red.

Quantification of (p)ppGpp levels after serine hydroxamate treatment at 100 µg/mL.
Cultures were treated identically to that of Figure 6A except with the addition of 32P. Cultures were normalized by OD600 and equivalent OD units were harvested for each sample. Nucleotides were extracted and separated by TLC as described in the methods. Due to an overall lower OD600, UW ΔmlaF strains were loaded equivalently but less than other genotypes tested. (A) Densitometry was normalized by comparing treated versus untreated for both pppGpp and ppGpp and is biological triplicates. (B) Representative TLC image.
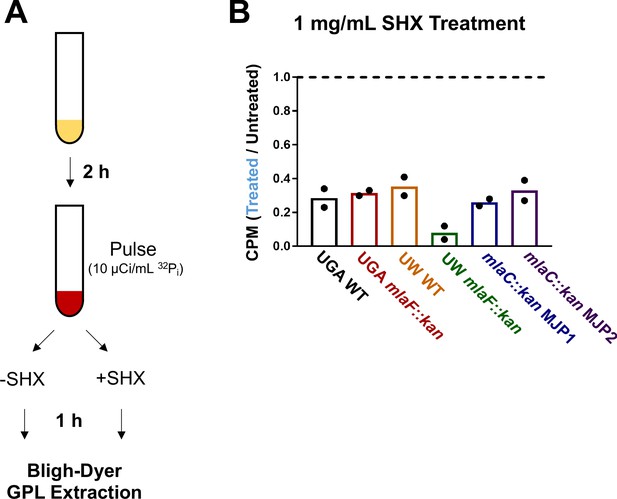
Increased stringent response inhibits de novo GPL biosynthesis.
(A) Schematic detailing serine hydroxamate (SHX) treatment. Cultures were grown for 2 hr and pulsed with 10 µCi/mL. This labeled culture was immediately split in half, and half was treated with 1 mg/mL SHX. Following an hour of growth, cells were pelleted and total GPLs extracted via Bligh-Dyer. (B) Isolated GPLs were quantified via LSC. Counts of the treated culture were normalized to that of untreated. A value = 1 indicates no change (dashed line) in GPL content and a value <1 indicates a decrease in GPLs under SHX treatment. Individual circles represent biological replicates.
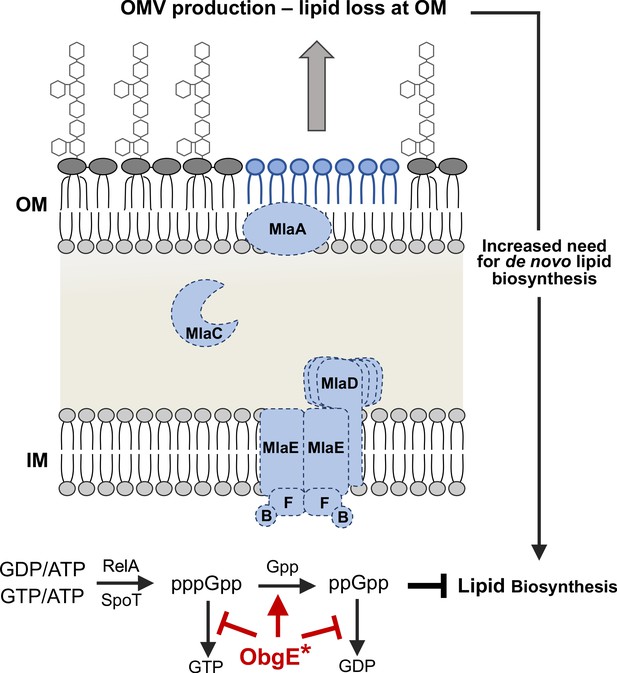
Model for the observed synthetic sick phenotype of Δmla with obgE*.
Mla mutants normally exhibit a 2-4X increase in OMV production, due in large part to the perturbed asymmetry of the OM (Roier et al., 2016). Additional evidence in E. coli suggests that fatty acids derived from OM phospholipase activity (PldA) stimulate lipid A biosynthesis (May and Silhavy, 2018). These factors result in an increased need for de novo fatty acid biosynthesis to contend with the rate of loss at the OM. Concurrently in the cytoplasm, the stringent response alarmone ppGpp has been shown to inhibit fatty acid biosynthesis (Heath et al., 1994). We know that Mla-null mutants (demonstrated here with dashed lines) with obgE* accumulate ppGpp at levels greater than that of WT (Figure 5, Figure 5—figure supplement 1). ObgE* could be acting through either partial inhibition of the hydrolysis of (p)ppGpp to GTP/GDP (red bars) or stimulating the hydrolysis of pppGpp to ppGpp (red arrow). The resultant accumulation of ppGpp would repress fatty acid biosynthesis despite the global need for increased biosynthesis due to OMV production.
Additional files
-
Supplementary file 1
Differentially regulated genes between UGA ΔmlaF and WT.
- https://cdn.elifesciences.org/articles/56571/elife-56571-supp1-v2.docx
-
Supplementary file 2
Differentially regulated genes between UW ΔmlaF and WT.
- https://cdn.elifesciences.org/articles/56571/elife-56571-supp2-v2.docx
-
Supplementary file 3
Unique mutations present in ΔmlaC, obgE::kan, pMMB67EH-obgE*.
- https://cdn.elifesciences.org/articles/56571/elife-56571-supp3-v2.docx
-
Supplementary file 4
Strains and plasmids used in this study.
- https://cdn.elifesciences.org/articles/56571/elife-56571-supp4-v2.docx
-
Supplementary file 5
Primers used in this study.
- https://cdn.elifesciences.org/articles/56571/elife-56571-supp5-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56571/elife-56571-transrepform-v2.docx





