β spectrin-dependent and domain specific mechanisms for Na+ channel clustering
Figures
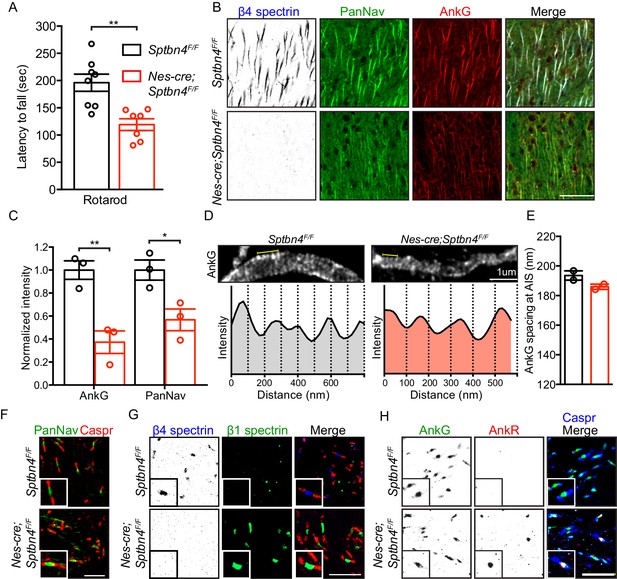
Mice lacking β4 spectrin in the central nervous system have impaired motor behavior and disrupted AIS, but intact nodal Nav clustering.
(A) Accelerating rotarod test performed on 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice. Sptbn4F/F, N = 8; Nes-cre;Sptbn4F/F, N = 7. Data are mean ± SEM, **p=0.0017. (B) Immunostaining of cortical brain sections from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice using antibodies against the β4 spectrin (blue), PanNav channels (green), and AnkG (red). Scale bar, 50 μm. (C) Normalized fluorescence intensity of AnkG and PanNav channel at AIS were measured from cortical brain sections of 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice. N = 3 mice with a total of 51–70 AIS were measured in each genotype. Data are mean ± SEM, For AnkG, **p=0.0079; PanNav channel, *p=0.0282. (D) STED images of AnkG at cortical AIS from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice. The regions between the yellow lines (as shown in images) were used to generate intensity profile as shown in lower panels. (E) Measurements of cortical AIS AnkG spacing by STED imaging from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice. Data are mean ± SEM. Sptbn4F/F, n = 107 spacings; Nes-cre;Sptbn4F/F, n = 96 spacings were measured from 2 mice of each genotype. (F) Immunostaining of corpus callosum from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice using antibodies against PanNav channel (green), and Caspr (red). Scale bar, 10 μm. (G) Immunostaining of corpus callosum from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice using antibodies against β4 spectrin (blue), β1 spectrin (green), and Caspr (red). Scale bar, 10 μm. (H) Immunostaining of corpus callosum from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice using antibodies against Caspr (blue), AnkG (green), and AnkR (red). Scale bar, 10 μm.
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/56629/elife-56629-fig1-data1-v1.xlsx
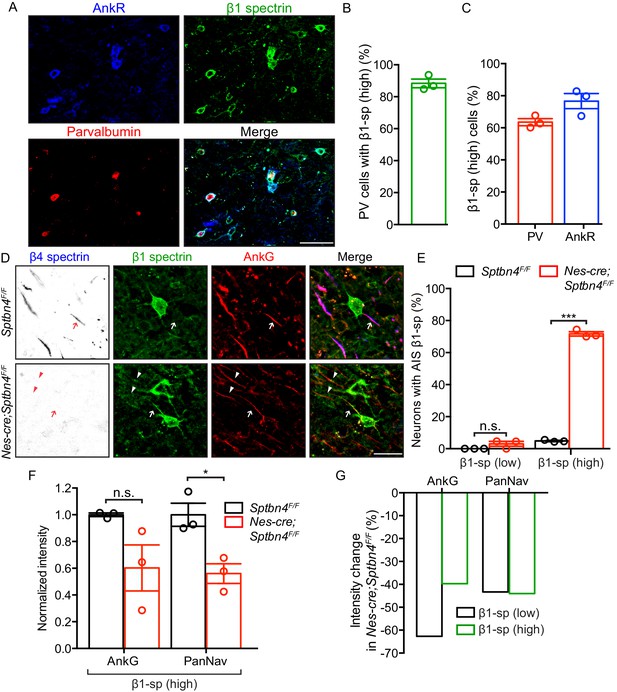
β1 spectrin is localized at the AIS of parvalbumin-positive neurons in β4 spectrin deficient mice.
(A) Immunostaining of brain cortical sections from 3 month-old SptbF/F mice using antibodies against the AnkR (blue), β1 spectrin (green), and parvalbumin (PV, red). Scale bar, 50 μm. (B) The percentage of PV-positive neurons labeled with high β1 spectrin in 3 month-old SptbF/F mice cortex. N = 3 animals, with total 166 neurons counted. (C) The percentage of high β1 spectrin signal in cortical neurons labeled with PV or AnkR in 3 month-old SptbF/F mice. N = 3 animals, with total 231 and 251 neurons counted, respectively. (D) Immunostaining of brain cortical sections from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice using antibodies against β4 spectrin (blue), β1 spectrin (green), and AnkG (red). AIS of high and low β1 spectrin expression neurons are indicated by arrows and arrowheads, respectively. Scale bar, 25 μm. (E) The percentage of neurons with AIS β1 spectrin in β1 spectrin low and β1 spectrin high 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice. N = 3 mice in each genotype. For Sptbn4F/F mice, 122 and 125 β1 spectrin low/high neurons were counted, respectively; for Nes-cre; Sptbn4F/F mice, 122 and 120 β1 spectrin low/high neurons were counted, respectively. For β1 spectrin (low) population, p=0.1162; for β1 spectrin (high) population, ***p=1.14E-06. (F) Normalized fluorescence intensity of AnkG and PanNav channel at AIS from β1 spectrin (high) cortical neurons measured in 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice. N = 3 mice with a total of 50–63 AIS were measured in each neuron type per genotype. Data are mean ± SEM, For AnkG, p=0.0829; PanNav channel, *p=0.018. (G) The average changes of percentage of fluorescence intensity of AnkG and PanNav channel in β1 spectrin low and β1 spectrin high neurons in 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice.
-
Figure 2—source data 1
Source data for Figure 2.
- https://cdn.elifesciences.org/articles/56629/elife-56629-fig2-data1-v1.xlsx
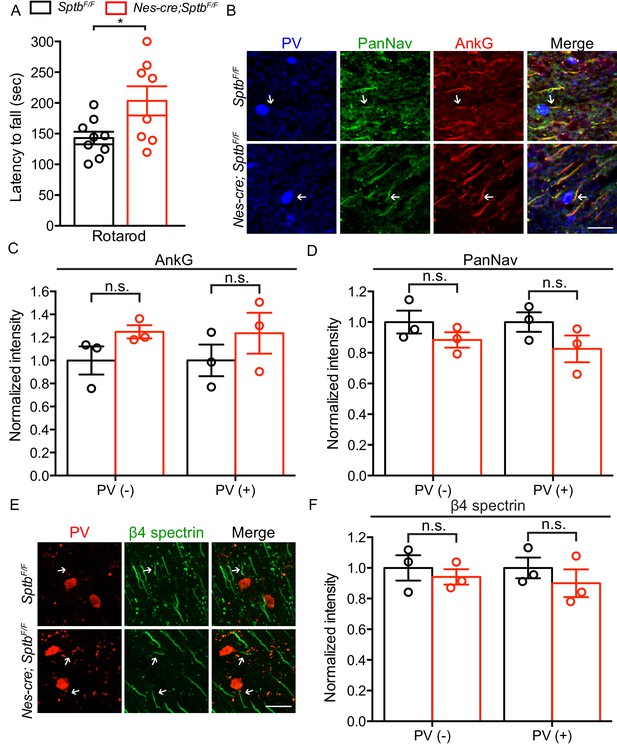
Mice lacking β1 spectrin in the central nervous system show normal motor performance and AIS structure.
(A) Accelerating rotarod test performed on 3 month-old SptbF/F and Nes-cre;SptbF/F mice. SptbF/F, N = 9; Nes-cre;Sptbn4F/F, N = 8. Data are mean ± SEM, *p=0.0276. (B) Immunostaining of brain cortical sections from 3 month-old SptbF/F and Nes-cre;SptbF/F mice using antibodies against the parvalbumin (PV, blue), PanNav channel (green) and AnkG (red). AIS of PV-expressing neurons are indicated by arrows. Scale bar, 25 μm. (C–D) Normalized fluorescence intensity of AnkG and PanNav at AIS in PV-positive (PV+) and negative (PV-) neurons were measured from cortical brain sections of 3 month-old SptbF/F and Nes-cre;SptbF/F mice. N = 3 mice with a total of 43–44 AIS were measured in each neuron type per genotype. Data are mean ± SEM. For AnkG in PV(-) and PV(+) neurons, p=0.1395 and p=0.3505, respectively; For PanNav in PV(-) and PV(+) neurons, p=0.1813 and p=0.1812, respectively. (E) Immunostaining of brain cortical sections from 3 month-old SptbF/F and Nes-cre;SptbF/F mice using antibodies against the parvalbumin (red) and β4 spectrin (green). AIS of PV-expressing neurons are indicated by arrows. Scale bar, 25 μm. (F) Normalized fluorescence intensity of β4 spectrin at AIS in PV-positive (PV+) and negative (PV-) neurons measured in cortical brain sections of 3 month-old SptbF/F and Nes-cre;SptbF/F mice. N = 3 mice in each genotype, with a total of 57–69 AIS were measured in each neuron type per genotype. Data are mean ± SEM. For β4 spectrin in PV(-) and PV(+) neurons, p=0.5775 and p=0.4268, respectively.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/56629/elife-56629-fig2-figsupp1-data1-v1.xlsx
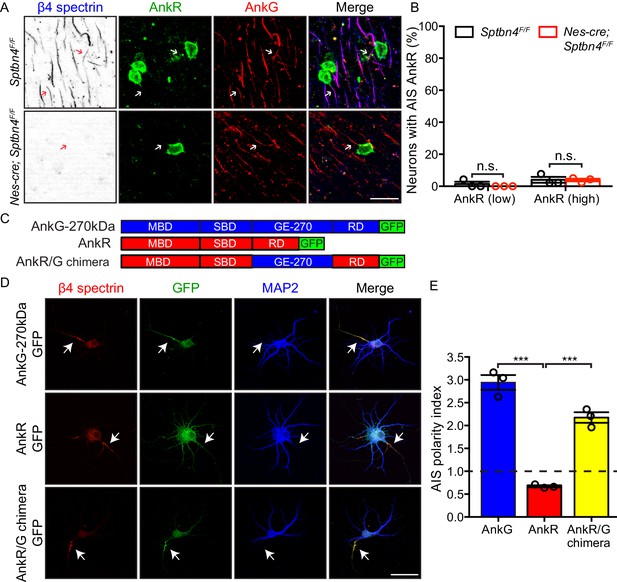
AnkyrinR fails to target to the AIS.
(A) Immunostaining of brain cortical sections from 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice using antibodies against β4 spectrin (blue), AnkR (green), and AnkG (red). AIS of high AnkR expression neurons are indicated by arrows. Scale bar, 25 μm. (B) The percentage of neurons with AnkR in the AIS in two subpopulations (AnkR low/high) of 3 month-old Sptbn4F/F and Nes-cre;Sptbn4F/F mice. N = 3 mice in each genotype. For Sptbn4F/F mice, 150 and 129 AnkR low/high neurons were counted; for Nes-cre; Sptbn4F/F mice, 127 and 128 AnkR low/high neurons were counted. For AnkR (low) population, p=0.3739; for AnkR (high), p=0.9497. (C) Domain structure and design of AnkG-270kDa-GFP, AnkR-GFP, and AnkR/G chimera-GFP expression constructs. (D) Immunostaining of cultured rat hippocampal neurons at DIV10 after transfected with AnkG-270kDa-GFP, AnkR-GFP, or AnkR/G chimera-GFP expression plasmids. Antibodies were used against β4 spectrin (red), GFP (green), and the somatodendritic marker MAP2 (blue). AIS are indicated by arrows. Scale bar, 50 μm. (E) Quantification of the ratio of GFP signal intensity at AIS versus proximal dendrite in AnkG-270kDa-GFP, AnkR-GFP, or AnkR/G chimera-GFP transfected cultured hippocampal neurons. N = 3 batches of cultured neurons for each transfected plasmids, with total 43–49 neurons were measured for each plasmid. Data are mean ± SEM. For AnkG-270kDa-GFP versus AnkR-GFP, ***p=0.0002; for AnkR-GFP versus AnkR/G chimera-GFP, ***p=0.0002.
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/56629/elife-56629-fig3-data1-v1.xlsx
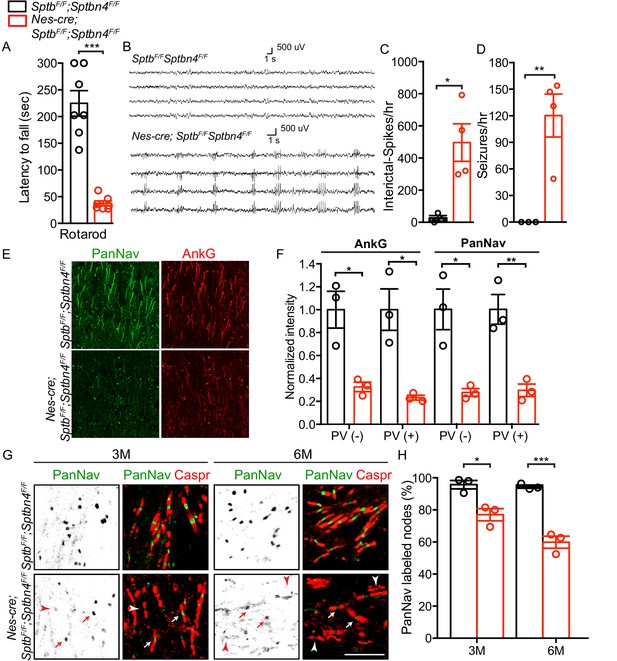
Mice lacking both β1 and β4 spectrin have severe motor impairment, epileptic activity, disrupted AIS, and gradual loss of nodal Nav channel clustering.
(A) Accelerating rotarod test performed on 3 month-old SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F mice, with N = 7 animals tested per genotype. Data are mean ± SEM, ***p=5.78E-06. (B) Video EEG monitoring of awake and behaving 3 month-old mice showed generalized seizure discharges in Nes-cre;SptbF/F; Sptbn4F/F mice that were not detected in SptbF/F; Sptbn4F/F littermates. (C–D) Quantification of interictal-spikes/hr and seizures/hr in SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F mice. N = 3 and 4 for SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F mice, respectively. Data are mean ± SEM. For interictal-spikes/hr, *p=0.0194; for seizures/hr, **p=0.0085. (E) Immunostaining of brain cortical sections from 3 month-old SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F mice using PanNav (green) and AnkG (red) antibodies. Scale bar, 50 μm. (F) Normalized fluorescence intensity for AnkG and PanNav at AIS in PV(-) and PV(+) neurons in cortex from 3 month old SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F mice. N = 3 mice with a total of 37–50 AIS measured in each neuron type per genotype. Data are mean ± SEM. For AnkG in PV(-) and PV(+) neurons, *p=0.0152 and *p=0.0135, respectively; for PanNav in PV(-) and PV(+) neurons, *p=0.0159 and **p=0.0073, respectively. (G) Immunostaining of corpus callosum from 3 and 6 month-old SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F mice using PanNav (green) and Caspr (red) antibodies. Arrows indicate intact nodal Nav clusters, whereas arrowheads indicate nodes devoid of Nav channels. Scale bar, 10 μm. (H) Quantification of the percentage of corpus callosum nodes labeled for Nav channels in SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F mice at the indicated ages. Data are mean ± SEM. N = 3 animals with a total of 345–377 nodes counted in each genotype per age. For 3 month-old, *p=0.0152; 6 month-old, ***p=0.0009.
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/56629/elife-56629-fig4-data1-v1.xlsx
Videos
6 month-old SptbF/F; Sptbn4F/F and Nes-cre;SptbF/F; Sptbn4F/F.
Mice lacking β1 and β4-spectrin in the central nervous system showed motor impairments.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Sptb | https://www.ncbi.nlm.nih.gov/gene/20741 | Gene ID: 20741 | |
| Gene (M. musculus) | Sptbn4 | https://www.ncbi.nlm.nih.gov/gene/80297 | Gene ID: 80297 | |
| Genetic reagent (M. musculus) | Nestin-cre | The Jackson Laboratory | Stock No:003771 | |
| Genetic reagent (M. musculus) | Sptbflox/flox | PMID:32052742 | ||
| Genetic reagent (M. musculus) | Sptbn4flox/flox | PMID:30226828 | ||
| Antibody | Anti-Ankyrin G (Mouse monoclonal) | Neuromab | Clone: N106/36; RRID:AB_10673030 | IF (1:500) |
| Antibody | Anti-Ankyrin G (Mouse monoclonal) | Neuromab | Clone N106/65; RRID:AB_10675130 | IF (1:100) for STED |
| Antibody | Anti-Ankyrin R (Mouse monoclonal) | Neuromab | Clone N380/A10; RRID:AB_2491109 | IF (1:500) |
| Antibody | Anti-Parvalbumin (Mouse monoclonal) | Neuromab | Clone L114/3; RRID:AB_2651167 | IF (1:500) |
| Antibody | Anti-PanNav (Mouse monoclonal) | Neuromab | Clone: N419/78; RRID:AB_2493099 | IF (1:300) |
| Antibody | Anti-PanNav (Mouse monoclonal) | Sigma-Aldrich | Clone: K58/35; RRID:AB_477552 | IF (1:300) |
| Antibody | Anti-β1 spectrin (Mouse monoclonal) | Neuromab | Clone: N385/21; RRID:AB_2315815 | IF (1:500) |
| Antibody | Anti-β4 spectrin SD antibody (Rabbit polyclonal) | PMID:28123356 | IF (1:500) WB (1:1000) | |
| Antibody | Anti-Ankyrin R (Rabbit polyclonal) | PMID:25362473 | IF (1:500) | |
| Antibody | Anti-Caspr (Rabbit polyclonal) | PMID:10460258 | RRID:AB_2572297 | IF (1:500) |
| Antibody | Anti-Pan Neurofascin (Chicken polyclonal) | R and D Systems | Cat.#: AF3235; RRID:AB_10890736 | IF (1:500) |
| Antibody | Anti-MAP2 (Chicken polyclonal) | Encor | Cat.#: CPCA-MAP2; RRID:AB_2138173 | IF (1:500) |
| Antibody | Anti-GFP (Rat monoclonal) | Biolegend | Cat.#: 338002; RRID:AB_1279414 | IF (1:500) |
| Antibody | Anti-Parvalbumin (Rabbit polyclonal) | Novus | RRID:AB_791498 | IF (1:500) |
| Antibody | Anti-active Caspase 3 (Rabbit polyclonal) | R and D Systems | RRID:AB_2243952 | IF (1:500) |
| Antibody | Anti-βAPP (Rabbit polyclonal) | Thermo Fisher Scientific | RRID:AB_2533902 | IF (1:1000) |
| Sequence-based reagent | Genotyping primer for Sptbn4flox/flox mouse (sense) | PMID:30226828 | 5’-GAGCTGCATAAGTTCTTCAGCGATGC-3’ | |
| Sequence-based reagent | Genotyping primer for Sptbn4flox/flox mouse (anti-sense) | PMID:30226828 | 5’-ACCCCATCTCAACTGGCTTTCTTGG-3’ | |
| Sequence-based reagent | Genotyping primer for Sptbflox/flox mouse (sense) | PMID:32052742 | 5’- ACAGAGACAGATGGCCGAAC-3‘ | |
| Sequence-based reagent | Genotyping primer for Sptbflox/flox mouse (anti-sense) | PMID:32052742 | 5’-CTCTGGTTCCCAGGAGAGC-3’ | |
| Sequence-based reagent | Genotyping primer for Avil-cre mouse (primer 1) | PMID:29038243 | 5’-CCCTGTTCACTGTGAGTAGG-3’ | |
| Sequence-based reagent | Genotyping primer for Avil-cre mouse (primer 2) | PMID:29038243 | 5’- AGTATCTGGTAGGTGCTTCCAG-3’ | |
| Sequence-based reagent | Genotyping primer for Avil-cre mouse (primer 3) | PMID:29038243 | 5’-GCGATCCCTGAACATGTCCATC-3’ | |
| Transfected construct (Rat) | pEGFP-N1-AnkG-270kDa | PMID:9744885 | Transfected construct (Rat) | |
| Transfected construct (Human) | pEGFP-N1-AnkR | This paper | Transfected construct (Human) Rasband laboratory | |
| Transfected construct (Human/Rat) | pEGFP-N1- AnkR/G chimera | This paper | Transfected construct (Human/Rat) Rasband laboratory | |
| Sequence-based reagent | AnkR-F | This paper | 5’-ATCTCGAGATGCCCTATTCTGTGG-3’ Rasband laboratory | |
| Sequence-based reagent | AnkR-R | This paper | 5’-AGCTTGAGGGGGTTGGGTGTCGA-3’ Rasband laboratory | |
| Sequence-based reagent | pEGFP-N1-F | This paper | 5’-CCAACCCCCTCAAGCTTCGAATTCTG-3’ Rasband laboratory | |
| Sequence-based reagent | pEGFP-N1-R | This paper | 5’-TAGGGCATCTCGAGATCTGAGTCC-3’ Rasband laboratory | |
| Sequence-based reagent | AnkR-SBD-F | This paper | 5’-CCCCTGGTACAGGCAACGTTCCCGGAGAATG-3’ Rasband laboratory | |
| Sequence-based reagent | AnkR-SBD-R | This paper | 5’-ACTGTTTTGTATCGCAGGGCCAG-3’ Rasband laboratory | |
| Sequence-based reagent | AnkG-RD-F | This paper | 5’-TGCGATACAAAACAGTTGAACGGAG-3’ Rasband laboratory | |
| Sequence-based reagent | AnkG-RD-R | This paper | 5’- GTACCGTCGACTGCAGAATTCGGTGGGCTTTCTTCTC-3’ Rasband laboratory | |
| Sequence-based reagent | AnkR-RD-F | This paper | 5’- TCCGATATCAGCATTCTCAGTGAGTCC-3’ Rasband laboratory | |
| Sequence-based reagent | AnkR-RD-R | This paper | 5’- TAGAATTCGGGGGTTGGGTGTCGAGGTG-3’ Rasband laboratory | |
| Commercial assay or kit | GeneArt Seamless Cloning and Assembly Kit | Thermo Fisher Scientific | Cat#: A13288 | |
| Software, algorithm | Zen | Carl Zeiss | RRID:SCR_013672 | |
| Software, algorithm | Labchart 8.0 | ADI Systems | RRID:SCR_017551 | |
| Software, algorithm | Leica Application Suite X | Leica | RRID:SCR_013673 | |
| Software, algorithm | Fiji | National Institutes of Health | RRID:SCR_002285 | |
| Software, algorithm | Prism | Graph Pad | RRID:SCR_002798 | Version 6 |





