Doa10 is a membrane protein retrotranslocase in ER-associated protein degradation
Figures
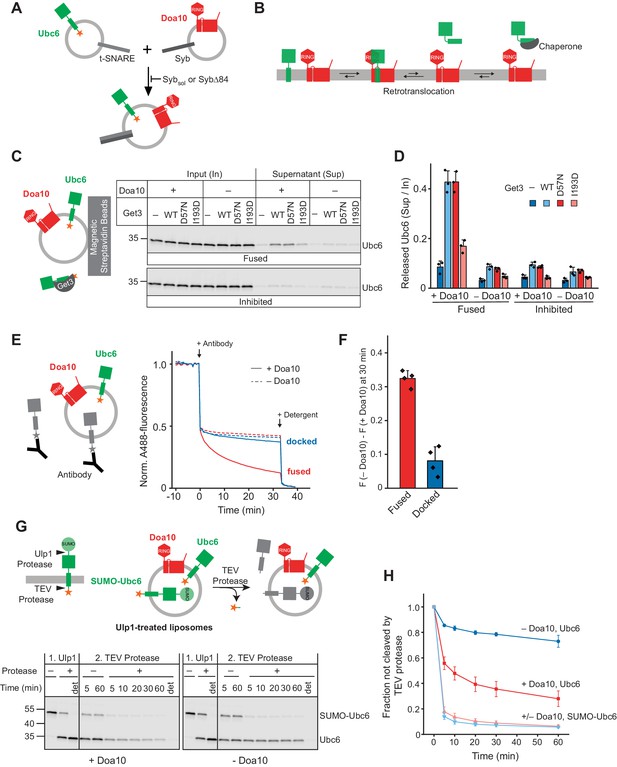
Retrotranslocation of Ubc6 by Doa10.
(A) SNARE-mediated co-reconstitution of Ubc6 and Doa10. Engineered versions of SNAREs involved in synaptic exocytosis were used, that is a Syntaxin 1A fragment, SNAP-25A, and Synaptobrevin 2 (Pobbati et al., 2006). Sybsol, a cytoplasmic fragment of Synaptobrevin (Syb). SybΔ84, Syb mutant that results in a docked state (Hernandez et al., 2012). See Figure 1—figure supplement 1, E to K for characterization of liposomes. (B) Working hypothesis for retrotranslocation by Doa10. (C) Membrane release of Ubc6 in the presence of Get3. Fluorescently labeled Ubc6 was co-reconstituted with Doa10 by SNARE-mediated fusion (+ Doa10), as shown in (A). Where indicated, Ubc6 liposomes were fused to liposomes lacking Doa10 (– Doa10), or fusion was inhibited with Sybsol (Inhibited). After incubation with the indicated Get3 variants or buffer, liposomes were immobilized (Figure 1—figure supplement 2B). Input and supernatant samples were analyzed by SDS-PAGE and fluorescence scanning. Final concentrations (f.c.): 0.1 µM Ubc6, 40 nM Doa10, 10 µM Get3. (D) Quantification (mean ± SD) of three independent experiments as in (C). (E) Retrotranslocation of Ubc6, measured as quenching of a C-terminal AlexaFluor488 (A488) label by an antibody. Liposomes were generated as shown in (A). Where indicated, liposomes lacked Doa10 (– Doa10), or co-reconstitution was inhibited by using SybΔ84 (docked). Arrows indicate addition of the quenching antibody or of solubilizing amounts of detergent (Triton X-100). F.c.: 0.2 µM Ubc6, 80 nM Doa10. (F) Quantification (mean ± SD) of four experiments as in (E). The fraction of accessible dye after 30 min was compared between conditions with and without Doa10. F, normalized fluorescence. (G) Retrotranslocation of Ubc6, measured by a protease protection assay. Ubc6 with an N-terminal SUMO tag (SUMO-Ubc6) and a TEV protease cleavage site between the C-terminus and the fluorescent dye was used. Arrow heads indicate cleavage sites for Ulp1 and TEV protease. SUMO-Ubc6 liposomes with or without Doa10 were incubated with Ulp1. Ulp1-treated liposomes were then incubated with buffer or TEV protease. Indicated reactions contained detergent to solubilize liposomes (det). Aliquots were taken at the indicated times and analyzed by SDS-PAGE and fluorescence scanning. F.c. during incubation with TEV protease: 0.1 µM Ubc6, 40 nM Doa10, 10 µM TEV protease. (H) Quantification (mean ± SD) of the fraction of Ubc6 and SUMO-Ubc6 inaccessible to TEV protease, from three experiments as in (G). Band intensities from samples treated with TEV protease were normalized to the corresponding band intensities of samples without TEV protease.
-
Figure 1—source data 1
This file contains the quantification of fluorescently labeled Ubc6 (Figure 1D) as well as of Rhodamine-labeled lipids (Figure 1—figure supplement 2B).
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig1-data1-v1.xlsx
-
Figure 1—source data 2
This file contains the quantification of the quenched fraction of Ubc6 in samples containing Doa10 compared to samples lacking Doa10, as shown in Figure 1F.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig1-data2-v1.xlsx
-
Figure 1—source data 3
This file contains the quantification of the TEV-protected fraction of SUMO-Ubc6 and Ubc6 shown in Figure 1H.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig1-data3-v1.xlsx
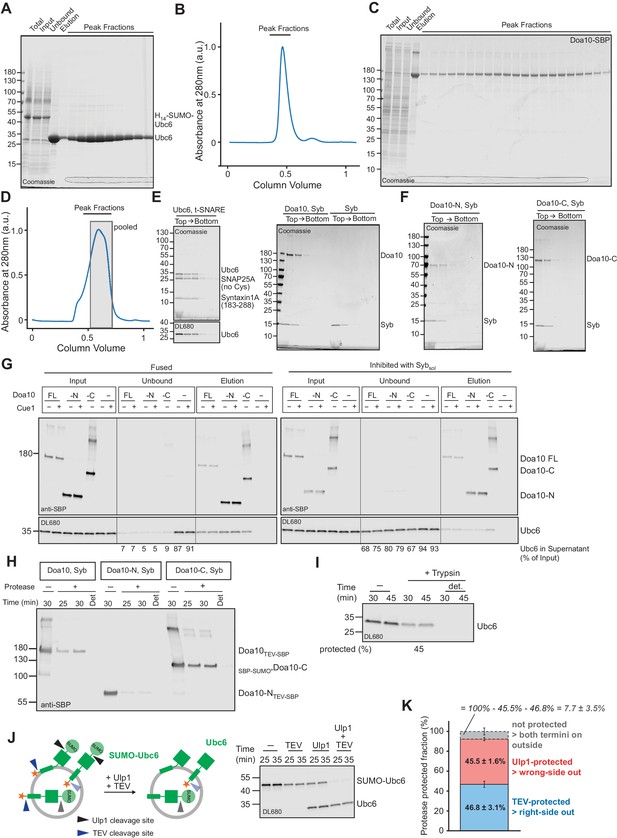
Quality control of liposomes.
(A) Coomassie Blue stained SDS-PAGE showing purification of Ubc6 from an E. coli membrane fraction by immobilized nickel ion chromatography and gel filtration. The protein was expressed with an N-terminal H14-SUMO tag that was cleaved with Ulp1 on beads. Samples of the membrane fraction before and after solubilization are denoted as Total and Input, respectively. (B) Chromatogram for gel filtration of Ubc6 on a Superdex 200 column. Peak fractions were analyzed by SDS PAGE, as shown in (A). (C) Coomassie Blue stained SDS-PAGE showing purification of SBP-tagged Doa10 from S. cerevisiae by streptavidin-affinity chromatography and gel filtration (Superose six column). SBP, streptavidin binding peptide. (D) Chromatogram for gel filtration of Doa10 purification shown in (C). Shaded area indicates fractions that were pooled. (E) Liposomes with co-reconstituted t-SNARE and Ubc6, fluorescently labeled with DyLight680 (DL680, left), and liposomes with co-reconstituted Doa10 and synaptobrevin (Syb, right) were subjected to a Nycodenz step gradient. After ultracentrifugation, the gradient was fractionated and analyzed by SDS-PAGE and Coomassie Blue staining (top) and fluorescence scanning (bottom). (F) Liposomes containing Syb and truncated versions of Doa10 used in Figure 4, Figure 5 and Figure 5—figure supplement 2 were analyzed as in (E). (G) Analysis of SNARE-dependent interaction of liposomes. Liposomes containing Syb and different SBP-tagged Doa10 versions were mixed with Ubc6 (fluorescently labeled)/t-SNARE liposomes in the absence (Fused) or presence (Inhibited) of Sybsol. Liposomes were then immobilized using streptavidin magnetic beads, washed, and eluted with biotin. Input, unbound, and elution fractions were analyzed by SDS-PAGE followed by Western blotting using an anti-SBP antibody to detect Doa10 variants (top), or fluorescence scanning to detect Ubc6 (bottom). Numbers below unbound fractions indicate the depletion of Ubc6 relative to input fractions. Doa10-N, and -C, refer to variants of Doa10 used in Figure 4, Figure 5 and Figure 5—figure supplement 2. Doa10-FL, full length Doa10. (H) Liposomes containing different Doa10 versions were subjected to either TEV protease (Doa10TEV-SBP and Doa10-NTEV-SBP) or a mutant version of Ulp1 (SBP-SUMO*Doa10-C Liu et al., 2008) for the indicated times in the absence or presence of Triton-X100 (det), and analyzed by SDS-PAGE and Western blotting using an anti-SBP antibody. Protease cleavage sites are only accessible in correctly oriented Doa10. (I) Liposomes containing fluorescently labeled Ubc6 and t-SNARE were subjected to a tryptic digest for the indicated times in the absence or presence of Triton-X100 (det). Samples were analyzed by SDS-PAGE and fluorescence scanning. The percentage of protected protein is indicated at the bottom. (J) Evaluation of Ubc6 reconstitution by protease protection. Liposomes containing Ubc6 with an N-terminal SUMO-moiety, and a TEV cleavage site between the TM anchor and the fluorescent dye (SUMO-Ubc6), were incubated with either TEV protease, Ulp1, or both proteases for the indicated times. Samples were analyzed by SDS-PAGE and fluorescence scanning. Ulp1 cleavage results in a size-shift in SDS-PAGE, TEV cleavage removes the dye, indicating correctly and wrongly oriented protein, respectively. Accessibility to both proteases indicates improper reconstitution. (K) Quantification (mean ± SD) of the fraction of non-cleaved Ubc6 from three experiments as in (J). 46.8 ± 3.1% of SUMO-Ubc6 was protected from TEV protease and is thus right-side out oriented. 45.5 ± 1.6% of SUMO-Ubc6 was protected from Ulp1 protease and is thus wrong-side out oriented. This indicates that a fraction of SUMO-Ubc6 (7.7 ± 3.5%) was not protected from either protease. In line with this, 26.9 ± 2.5% Ulp1-cleaved Ubc6 was also cleaved by TEV protease, corresponding to 14.7 ± 1.4% of total Ubc6. Thus, we estimate that about 10% of Ubc6 is not properly reconstituted.
-
Figure 1—figure supplement 1—source data 1
This file contains the quantification of the TEV- and Ulp1 cleaved fraction of SUMO-Ubc6, as well as of Ulp1-cleaved Ubc6 that is accessible to TEV protease, as shown in Figure 1—figure supplement 1K.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig1-figsupp1-data1-v1.xlsx
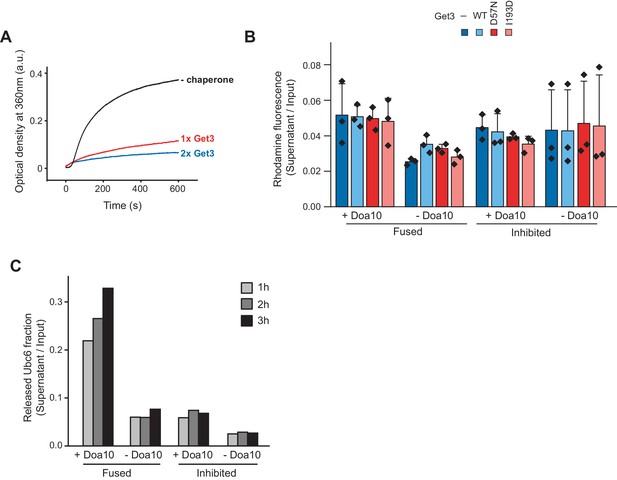
Retrotranslocation in the presence of Get3.
(A) Aggregation of Ubc6 in detergent-free buffer. Ubc6 in 0.03% (w/v) dodecyl maltoside was diluted 25-fold (f.c. 1.8 µM) into buffer containing no, 1.8 µM or 3.6 µM Get3. Optical density was measured at 360 nm. (B) Immobilization efficiency of liposomes used in Figure 1C,D was quantified (mean ± SD) via co-reconstituted fluorescent Rhodamine-phosphatidyl ethanolamine (PE). n = 3 independent experiments. (C) Release of Ubc6 in the presence of Get3, as in Figure 1C,D, but for shorter incubation times.
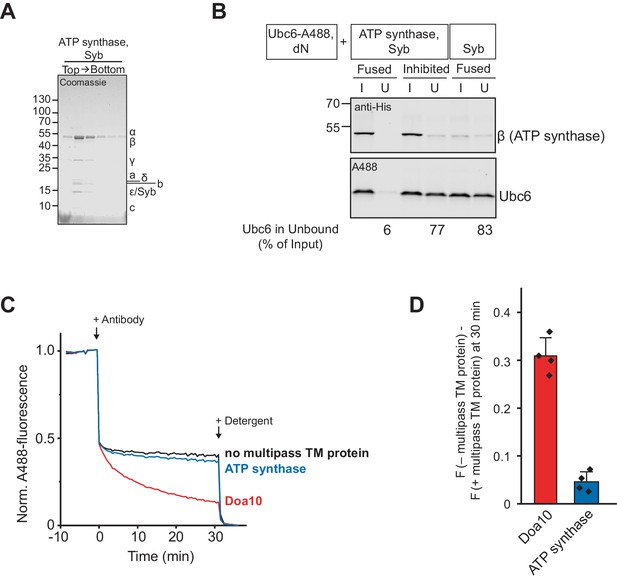
Co-reconstitution with ATP synthase.
(A) Liposomes with co-reconstituted ATP synthase and synaptobrevin (Syb) were subjected to a Nycodenz step gradient. After ultracentrifugation, the gradient was fractionated and analyzed by SDS-PAGE and Coomassie Blue staining. Bacillus PS3 ATP synthase is a multiprotein complex with a subunit composition of α3β3γδε ab2c20 (Guo et al., 2019). (B) Analysis of co-reconstitution efficiency of ATP synthase and Ubc6 upon SNARE-mediated fusion. Liposomes containing Syb and ATP synthase (tagged with His10-tag at β-subunit) were mixed with Ubc6 (fluorescently labeled)/t-SNARE liposomes in the absence (Fused) or presence (Inhibited) of Sybsol. Liposomes were then immobilized via the His-tag of ATP synthase with magnetic beads. Input (I) and unbound (U) fractions were analyzed by SDS-PAGE followed by Western blotting using an anti-His antibody to detect ATP synthase (top), or fluorescence scanning to detect Ubc6 (bottom). Numbers below unbound fractions indicate the depletion of Ubc6 relative to input fractions. (C) Retrotranslocation of Ubc6 in the presence of Doa10 or ATP synthase, measured as quenching of a C-terminal AlexaFluor488 (A488) label by an antibody. Where indicated, liposomes lacked Doa10 or ATP synthase (no multipass TM protein). Arrows indicate addition of the quenching antibody or of solubilizing amounts of detergent (Triton X-100). F.c.: 0.2 µM Ubc6, 80 nM Doa10/80 nM ATP synthase. (D) Quantification (mean ± SD) of four experiments as in (C). The fraction of accessible dye after 30 min was compared between conditions with and without a multipass TM protein. F, normalized fluorescence.
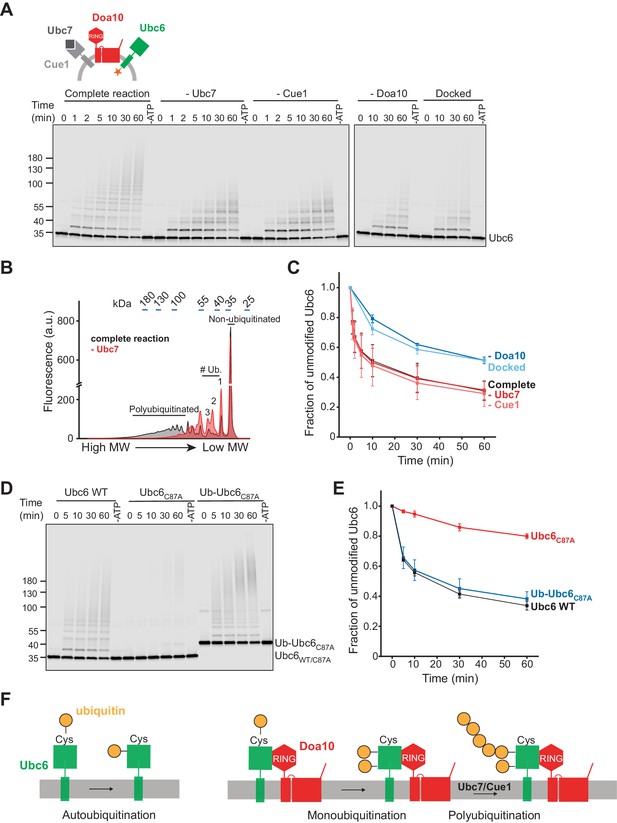
Ubiquitination of Ubc6.
(A) Time course of ubiquitination of Ubc6. Final concentrations in the complete reaction: 40 nM Doa10, 10 nM Cue1, 1 µM Ubc7, 100 nM Ubc6, 100 nM E1, 120 µM ubiquitin, and 2.5 mM ATP. Where indicated, individual components were omitted or co-reconstitution was inhibited by using SybΔ84 (Docked). For each reaction, a 60 min sample in the absence of ATP is shown. Samples were analyzed by SDS-PAGE and fluorescence scanning. (B) Analysis of ubiquitin-chain length on Ubc6 from an experiment as in (A). Line-scans were performed on fluorescence images for the complete reaction and in the absence of Ubc7 at t = 30 min. Approximate molecular weights are indicated on top. # ub. denotes number of ubiquitin moieties attached. (C) Quantification (mean ± SD) of the fraction of unmodified Ubc6 from three experiments as in (A). (D) Time course of ubiquitination of Ub-Ubc6C87A compared to Ubc6 WT and Ubc6C87A in the presence of Doa10, Cue1, and Ubc7. Concentrations and analysis as in (A). (E) Quantification (mean ± SD) of the fraction of unmodified Ubc6 variants from three experiments as in (D). (F) Model for ubiquitination of Ubc6. Ubc6 autoubiquitination activity results in transfer of ubiquitin from its active site cysteine to a non-cysteine residue (Weber et al., 2016). In the presence of Doa10, this activity is enhanced and Ubc6 is multi-monoubiquitinated. Ubc7/Cue1 are then required to form polyubiquitin chains on monoubiquitinated Ubc6.
-
Figure 2—source data 1
This file contains the quantification of the fraction of unmodified Ubc6 from three experiments as in Figure 2A, as shown in in Figure 2C.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig2-data1-v1.xlsx
-
Figure 2—source data 2
This file contains the quantification of the fraction of unmodified Ubc6 from three experiments as in Figure 2D, as shown in Figure 2E.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig2-data2-v1.xlsx
-
Figure 2—source data 3
This file contains the quantification of the fraction of unmodified Ubc6 from three experiments as in Figure 2—figure supplement 1C, as shown in Figure 2—figure supplement 1D.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig2-data3-v1.xlsx
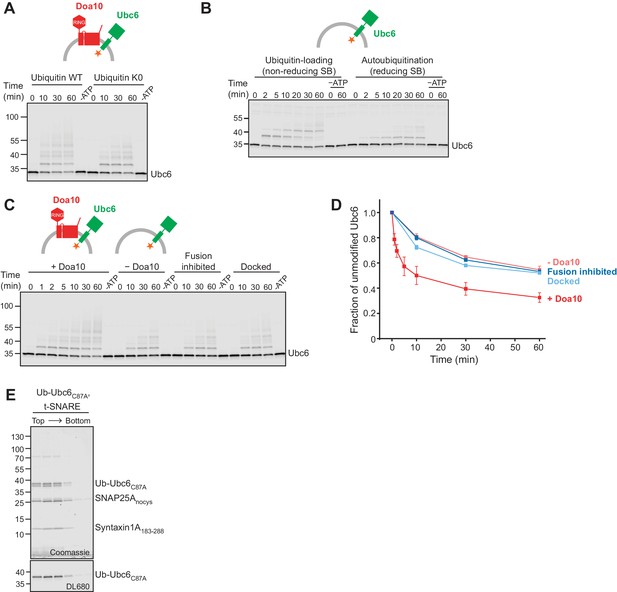
E3—independent and -dependent ubiquitination of Ubc6.
(A) Time course of ubiquitination of Ubc6 in the absence of Ubc7/Cue1 using either WT ubiquitin or a ubiquitin mutant with all Lys mutated to Arg (K0). 40 nM Doa10 and 0.1 µM Ubc6 in liposomes were incubated with 0.1 µM E1, 120 µM ubiquitin, and 2.5 mM ATP. A 60 min sample in the absence of ATP is shown for each reaction. Samples were analyzed by SDS-PAGE and fluorescence scanning. (B) Doa10-independent autoubiquitination of Ubc6. Liposomes containing fluorescently labeled Ubc6 were incubated with E1, ubiquitin, and ATP. Samples at indicated time points were analyzed by SDS-PAGE under non-reducing (left) and reducing conditions (right) and fluorescence scanning. Reactions lacking ATP are denoted as –ATP. Final concentrations: 0.1 µM Ubc6, 0.1 µM Uba1, 120 µM ubiquitin, 2.5 mM ATP. SB, SDS sample buffer. (C) Time course of ubiquitination of Ubc6 in the absence of Ubc7/Cue1. Ubc6 liposomes containing or lacking Doa10 were used. Where indicated, co-reconstitution was inhibited using either a soluble Syb fragment (Sybsol) or a Syb mutant (SybΔ84). Concentrations and analysis as in (A). (D) Quantification (mean ± SD) of the fraction of unmodified Ubc6 from three experiments as in (C). (E) Ub-Ubc6C87A/t-SNARE liposomes were subjected to a Nycodenz step gradient. After ultracentrifugation, the gradient was fractionated and analyzed by SDS-PAGE and Coomassie Blue staining (top) and fluorescence scanning (DL680, bottom).
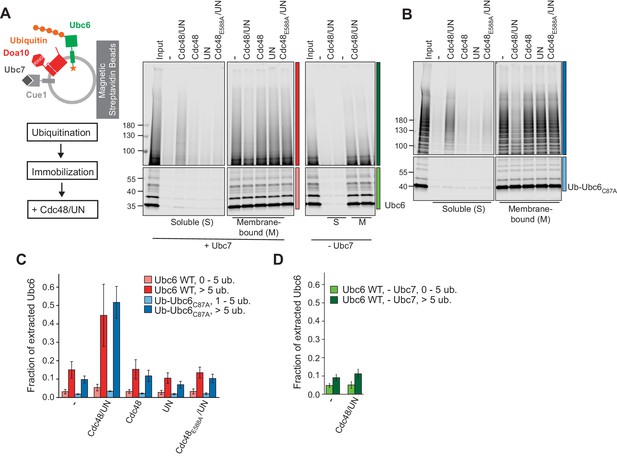
Cdc48-mediated Membrane Extraction of Ubc6.
(A) Extraction of Ubc6 by Cdc48 and Ufd1/Npl4 (UN). After ubiquitination, liposomes were immobilized (Figure 3—figure supplement 1,A to D). One bead equivalent was removed, and bound protein was eluted with SDS sample buffer (Input). Beads were then incubated with the indicated components. Soluble (S) and membrane-bound (M) material were analyzed by SDS-PAGE and fluorescence scanning. Colored bars indicate categorization of ubiquitin chain length as used for quantification in (C) and (D). For better visibility, bottom and top gel parts are scaled differently. See Figure 3—figure supplement 1E for uncut image. Final concentrations: 50 nM Ubc6, 20 nM Doa10, 0.1 µM Cdc48 hexamer, 0.1 µM Ufd1 and Npl4. (B) As in (A), but with Ub-Ubc6C87A instead of Ubc6. See Figure 3—figure supplement 1F for uncut image. (C) Quantification (mean ± SD) of three experiments as in (A) and (B). Ubiquitinated species were categorized according to ubiquitin chain length, as indicated in (A) and (B). The signal in the soluble fraction was normalized to that in the input. (D) Quantification (mean ± SD) of three experiments as in (A), when ubiquitination was performed in the absence of Ubc7.
-
Figure 3—source data 1
This file contains the quantification of the fraction of extracted Ubc6 from three experiments as in Figure 3A,B, as shown Figure 3C,D.
It also contains the quantification of the immobilization efficiency of Ubc6 (Figure 3—figure supplement 1C) and of Rhodamine-labeled lipids (Figure 3—figure supplement 1D), as well as the quantification of the extraction efficiency of Ubc6 modified with different ubiquitin chain lengths (Figure 3—figure supplement 1G).
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig3-data1-v1.xlsx
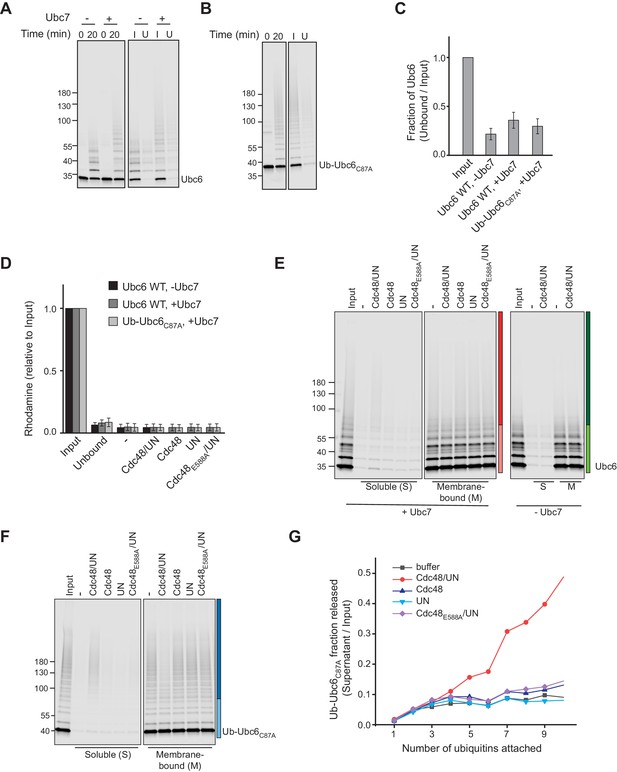
Cdc48-mediated extraction.
(A) Samples from ubiquitination and immobilization under conditions described in Figure 3A. Liposomes containing Ubc6, Doa10 and Cue1 were incubated with E1, ubiquitin, and ATP, with or without Ubc7. After 20 min, the ubiquitination reaction was stopped by adding EDTA. Liposomes were then immobilized to streptavidin magnetic beads. Input (I) and unbound (U) fractions were analyzed by SDS-PAGE and fluorescence scanning. (B) Samples from ubiquitination and immobilization under conditions described in Figure 3B. As in (A), but with Ub-Ubc6C87A instead of Ubc6 and ubiquitination in presence of Ubc7. (C) Quantification (mean ± SD) of protein immobilization efficiency from three experiments as in (A) and (B). (D) Quantification (mean ± SD) of the efficiency of liposome immobilization in experiments described in Figure 3A,B, measuring the fluorescence of Rhodamine-PE that was co-reconstituted into liposomes. Rhodamine content of Input and Unbound fraction from immobilization reaction (as in (A) and (B)) as well as of the soluble fraction after extraction (as in Figure 3A,B) was determined. n = 3 independent experiments. (E) Uncut image with uniform scaling of Figure 3A. (F) Uncut image with uniform scaling of Figure 3B. (G) Comparison of extraction efficiency of Ub-Ubc6C87A modified with ubiquitin chains of different length as shown in Figure 3B.
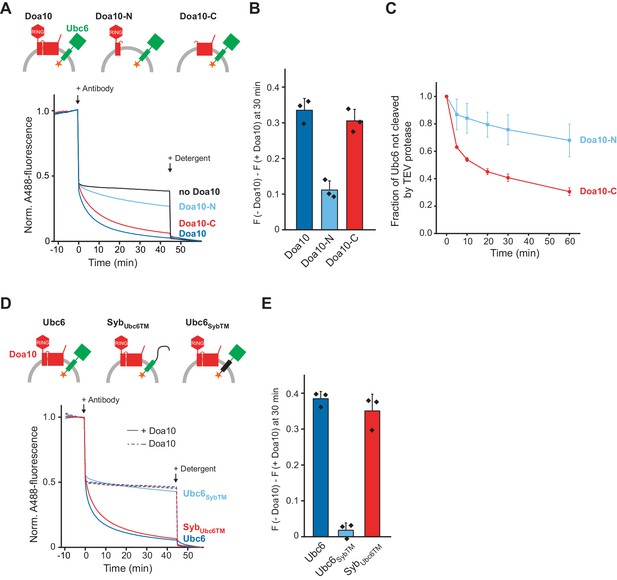
Structural Determinants for Retrotranslocation.
(A) Retrotranslocation of Ubc6 by Doa10 variants, as measured by accessibility of a fluorescence quenching antibody to a C-terminal A488 dye on Ubc6, as described in Figure 1E. Ubc6 liposomes containing the indicated Doa10 variants were used. Doa10-N, residues 1–468; Doa10-C, residues 434–1319. Arrows indicate addition of antibody or detergent. Final concentrations (f.c.): 0.2 µM Ubc6, 80 nM Doa10 variants. (B) Quantification (mean ± SD) of three experiments as in (A). The fraction of accessible dye after 30 min was compared between conditions with the indicated Doa10 variant and without Doa10. F, normalized fluorescence. (C) Retrotranslocation of Ubc6 by Doa10 variants, as measured by accessibility of TEV protease to the C-terminus of Ubc6, as described in Figure 1G. SUMO-Ubc6 liposomes with either Doa10-N or Doa10-C were treated with Ulp1 to identify right-side out oriented Ubc6. TEV protease was added and samples at different time points were analyzed by SDS-PAGE and fluorescence scanning. Quantification as in Figure 1H, but only for Ulp1-cleaved Ubc6. F.c. during incubation with TEV protease: 0.1 µM Ubc6, 40 nM Doa10 variants, 10 µM TEV protease. (D) Retrotranslocation of Ubc6 variants measured as in (A). A488-labeled Ubc6, Ubc6SybTM, or SybUbc6TM were directly co-reconstituted with Doa10 because SybUbc6TM was incompatible with SNARE-mediated co-reconstitution. Liposomes containing Doa10 were affinity-purified for this experiment (Figure 4—figure supplement 1A,B). (E) Quantification (mean ± SD) of three experiments as in (D). The fraction of accessible dye after 30 min was compared between conditions with and without Doa10.
-
Figure 4—source data 1
This file contains the quantification of the quenched fraction of Ubc6 in samples containing Doa10 or its variants compared to samples lacking Doa10 from three experiments as in Figure 4A, as shown in Figure 4B.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig4-data1-v1.xlsx
-
Figure 4—source data 2
This file contains the quantification of the TEV-protected fraction of Ubc6 from three experiments, as shown in Figure 4C.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig4-data2-v1.xlsx
-
Figure 4—source data 3
This file contains the quantification of the quenched fraction of Ubc6 or its variants in samples containing Doa10 compared to samples lacking Doa10 from three experiments as in Figure 4D, as shown in Figure 4E.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig4-data3-v1.xlsx
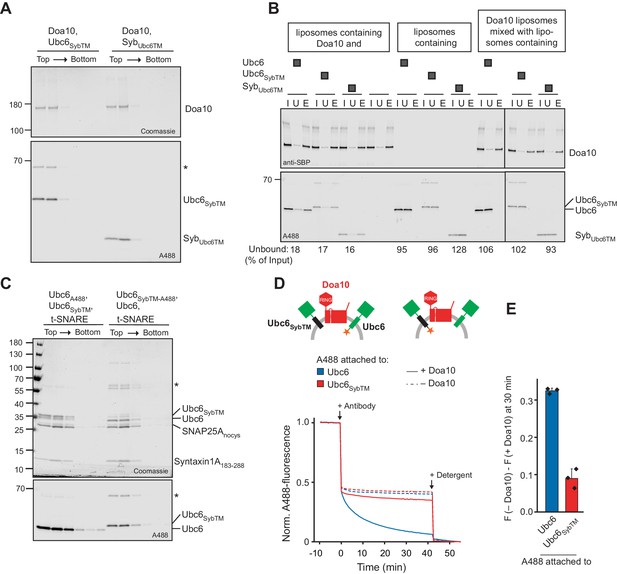
Antibody accessibility assay for Ubc6/Syb chimera.
(A) Doa10 liposomes directly co-reconstituted with fluorescently labeled Ubc6SybTM or SybUbc6TM were subjected to a Nycodenz step gradient. After ultracentrifugation, the gradient was fractionated and analyzed by SDS-PAGE and Coomassie Blue staining (top) and fluorescence scanning (bottom). The asterisk indicates a dimer of Ubc6SybTM that occurred in some sortase mediated labeling reactions of this construct. (B) Affinity-purification of Doa10-SBP liposomes for experiment in Figure 4D using streptavidin beads. Inputs (I), unbounds (U) and biotin-elutions (E) were analyzed by SDS-PAGE and by either Western Blot using an anti-SBP antibody (Doa10-SBP), or fluorescence scanning (Ubc6/Syb chimera). As an indication of the specificity of the pull-down, we also tested for binding of liposomes lacking Doa10, and for co-purification of Ubc6 liposomes, when Doa10-SBP and Ubc6 were reconstituted in separate liposome sets. Numbers at the bottom indicate the percentage of Ubc6 in the unbound fraction. Note that different intensities are due to different labeling efficiencies. (C) Liposomes containing t-SNARE, Ubc6SybTM and Ubc6, either fluorescently labeled on Ubc6SybTM or Ubc6, were subjected to a Nycodenz step gradient. After ultracentrifugation, the gradient was fractionated and analyzed by SDS-PAGE and Coomassie Blue staining (top) and fluorescence scanning (bottom). The asterisk indicates a dimer of Ubc6SybTM that occurred in some sortase mediated labeling reactions of this construct. (D) Antibody accessibility assay with liposomes containing both, Ubc6 and Ubc6SybTM, and with or without Doa10. In separate liposomes populations, A488 was either attached to Ubc6 or Ubc6SybTM. Arrows indicate addition of antibody or solubilizing detergent. Final concentrations: 0.2 µM for Ubc6 and Ubc6SybTM, 80 nM Doa10. (E) Quantification (mean ± SD) of three experiments as in (D). The fraction of accessible dye after 30 min was compared between conditions with and without Doa10. F, normalized fluorescence.
-
Figure 4—figure supplement 1—source data 1
This file contains numerical values for data shown in Figure 4—figure supplement 1E.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig4-figsupp1-data1-v1.xlsx
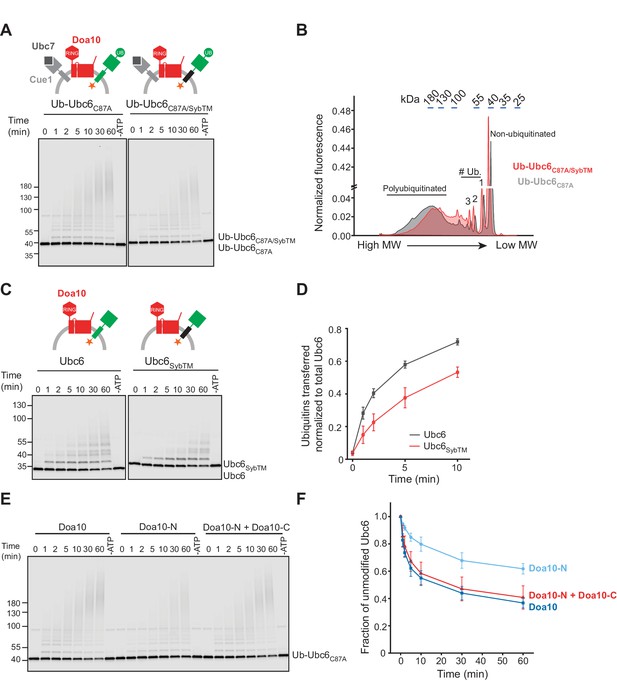
Structural Determinants for Ubiquitination.
(A) Time course of ubiquitination of Ub-Ubc6C87A or Ub-Ubc6C87A/SybTM by Doa10 in the presence of Cue1/Ubc7. For each reaction, a 60 min sample in the absence of ATP is shown. Samples were analyzed by SDS-PAGE and fluorescence scanning. Final concentrations: 40 nM Doa10, 10 nM Cue1, 1 µM Ubc7, 100 nM Ubc6 variants, 100 nM E1, 120 µM ubiquitin, and 2.5 mM ATP. See Figure 5—figure supplement 1A for quantification of unmodified Ubc6 variants. (B) Comparison of ubiquitin-chain length on Ub-Ubc6C87A or Ub-Ubc6C87A/SybTM. Line-scans were performed on fluorescence images of two representative gel samples (30 min timepoint) as in (A). Approximate molecular weights are indicated on top. # Ub., number of ubiquitin moieties attached. (C) Time-course of Ubc6 WT or Ubc6SybTM ubiquitination in the absence of Ubc7/Cue1. Analysis and concentrations as in (A). See Figure 5—figure supplement 1C for quantification of unmodified Ubc6 variants. (D) Quantification (mean ± SD) of total ubiquitin-transfer to Ubc6 or Ubc6SybTM from three experiments as in (C). Intensities of Ubc6 variants with one to four ubiquitin moieties attached were determined as described in Figure 5—figure supplement 1D, summed up for each time point and normalized to total Ubc6 in the reaction. (E) Time course of ubiquitination of Ub-Ubc6C87A by Doa10 variants in the presence of Cue1/Ubc7. Liposomes contained Ub-Ubc6C87A and either full-length Doa10, only Doa10-N, or both Doa10-N and -C. Analysis and concentrations as in (A). (F) Quantification (mean ± SD) of unmodified Ub-Ubc6C87A from three experiments as in (E).
-
Figure 5—source data 1
This file contains the quantification of the number of ubiquitins (n) transferred per Ubc6 or Ubc6SybTM (Figure 5—figure supplement 1D) as well as the quantification of the number of total ubiquitin transferred from three experiments as in Figure 5C, as shown in Figure 5D.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig5-data1-v1.xlsx
-
Figure 5—source data 2
This file contains the quantification of the fraction of unmodified Ubc6 from three experiments as in Figure 5E, as shown in Figure 5F.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig5-data2-v1.xlsx
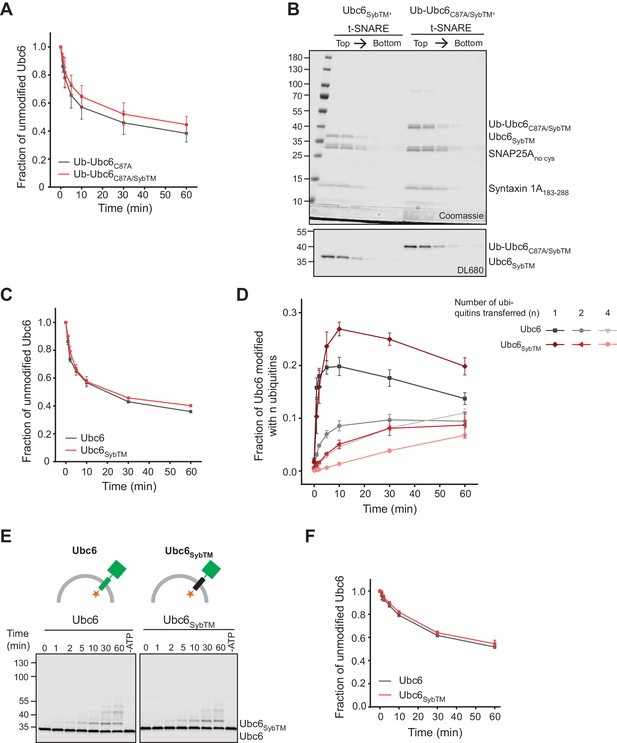
Ubiquitination of Ubc6/Syb chimera.
(A) Quantification (mean ± SD) of the fraction of unmodified Ub-Ubc6C87A or Ub-Ubc6C87A/SybTM from three experiments as in Figure 5A. (B) Liposomes with t-SNARE co-reconstituted with either Ubc6SybTM or Ub-Ubc6C87A/SybTM (fluorescently labeled) were subjected to a Nycodenz step gradient. After ultracentrifugation, the gradient was fractionated and analyzed by SDS-PAGE and Coomassie Blue staining (top) and fluorescence scanning (bottom). (C) Quantification (mean ± SD) of the fraction of unmodified Ubc6 or Ubc6SybTM from three experiments as in Figure 5C. (D) Quantification (mean ± SD) of mono-, di-, and tetra-ubiquitinated Ubc6 species relative to total Ubc6 from three experiments as in Figure 5C. (E) Time course of E3-independent autoubiquitination of Ubc6 and Ubc6SybTM. Liposomes containing the indicated Ubc6 variants (100 nM) were incubated with 100 nM E1, 120 µM ubiquitin, and 2.5 mM ATP. A 60 min sample in the absence of ATP is shown for each reaction. Samples were analyzed by SDS-PAGE and fluorescence scanning. (F) Quantification (mean ± SD) of the fraction of unmodified Ubc6 or Ubc6SybTM from three experiments as in (E).
-
Figure 5—figure supplement 1—source data 1
This file contains the quantification of the fraction of unmodified Ubc6 as shown in Figure 5—figure supplement 1A, C and F.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig5-figsupp1-data1-v1.xlsx
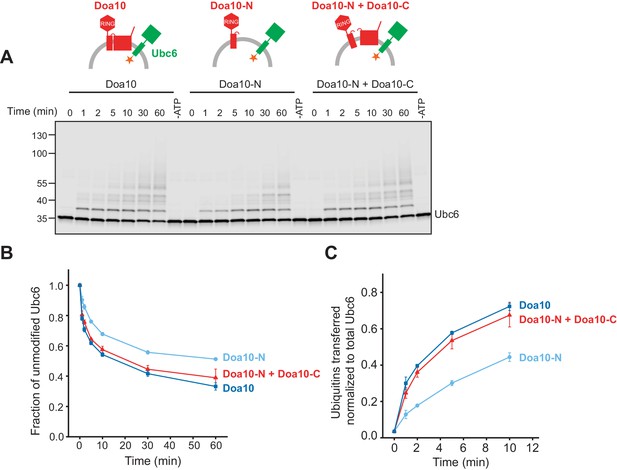
Ubc6 ubiquitination by Doa10 variants.
(A) Time course of Ubc6 ubiquitination in the presence of different Doa10 variants in the absence of Ubc7/Cue1. Indicated liposomes were incubated with 100 nM E1, 120 µM ubiquitin, and 2.5 mM ATP (f.c. 100 nM Ubc6, 10 nM Cue1, and 40 nM for Doa10-variants). A 60 min sample in the absence of ATP is shown for each reaction. Samples were analyzed by SDS-PAGE and fluorescence scanning. (B) Quantification (mean ± SD) of the fraction of unmodified Ubc6 from three experiments as in (A). (C) Quantification (mean ± SD) of total ubiquitin-transfer relative to Ubc6 from three experiments as in (A). Intensities of Ubc6 with one to four ubiquitin moieties attached were summed up for each time point and normalized to total Ubc6 in the reaction, as described in Figure 5D and Figure 5—figure supplement 1D.
-
Figure 5—figure supplement 2—source data 1
This file contains the quantification of the fraction of unmodified Ubc6 as shown in Figure 5—figure supplement 2B.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig5-figsupp2-data1-v1.xlsx
-
Figure 5—figure supplement 2—source data 2
This file contains the quantification of the number of total ubiquitin transferred in presence of different Doa10 variants from three experiments, as shown in Figure 5—figure supplement 2C.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig5-figsupp2-data2-v1.xlsx
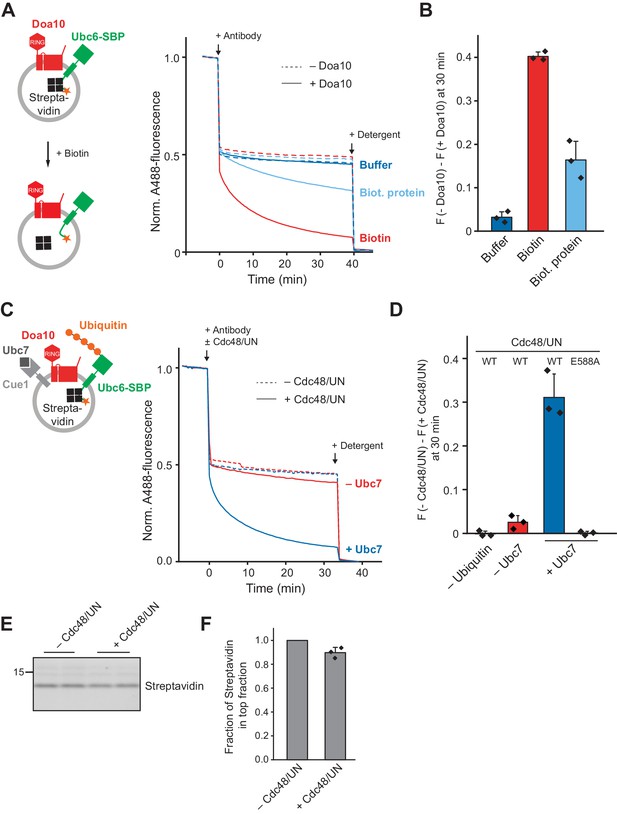
Effect of a Luminal Domain on Retrotranslocation.
(A) Retrotranslocation of Ubc6 with a C-terminal streptavidin binding peptide (SBP), reconstituted in complex with streptavidin, was measured by accessibility of a fluorescence quenching antibody to a C-terminal A488 dye on Ubc6, as described Figure 1E. Liposomes were incubated with buffer, biotin or a biotinylated protein prior to addition of the antibody. Final concentrations: 0.2 µM Ubc6, 80 nM Doa10, 0.25 µM streptavidin, 1.5 µM biotin or biotinylated protein. (B) Quantification (mean ± SD) of three experiments as in (A). The fraction of accessible dye after 30 min was compared between conditions with and without Doa10. F, normalized fluorescence. (C) Effect of Cdc48 and Ufd1/Npl4 (UN) on retrotranslocation of Ubc6-SBP in complex with streptavidin, measured using the antibody accessibility assay as in (A). Prior to the fluorescence measurement, liposomes were incubated with ubiquitination mix with or without Ubc7. Arrows indicate when antibody, with or without Cdc48/UN, or detergent were added. Final concentrations: 0.17 µM Ubc6-SBP, 68 nM Doa10, 0.17 µM hexameric Cdc48, Ufd1, and Npl4. See Figure 6—figure supplement 1F for gel samples of ubiquitination reaction. (D) Quantification (mean ± SD) of three experiments as in (C). The fraction of accessible dye after 30 min was compared between conditions with and without Cdc48/UN. In addition, experiments lacking ubiquitin or with the Cdc48 mutant E588A were quantified. (E) Determination of liposome-encapsulated streptavidin after extraction. Samples from experiments as in (D) were taken at t = 30 min. Biotin was added, and liposomes floated in a Nycodenz gradient. Co-floating streptavidin was detected in SDS-PAGE using stain-free technology. Two replicates are shown for each condition. (F) Quantification (mean ± SD) of the relative amount of streptavidin co-floating from three experiments as in (E). Each data point represents the mean of two replicates as shown in (E).
-
Figure 6—source data 1
This file contains the quantification of the quenched fraction of Ubc6 in samples containing Doa10 compared to samples lacking Doa10 from three experiments as in Figure 6A, as shown in Figure 6B.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig6-data1-v1.xlsx
-
Figure 6—source data 2
This file contains the quantification of the quenched fraction of Ubc6 in samples containing Cdc48/UN compared to samples lacking Cdc48/UN from three experiments as in Figure 6C, as shown in Figure 6D.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig6-data2-v1.xlsx
-
Figure 6—source data 3
This file contains the quantification of streptavidin in the top flotation fraction from three experiments as in Figure 6E, as shown in Figure 6F.
- https://cdn.elifesciences.org/articles/56945/elife-56945-fig6-data3-v1.xlsx
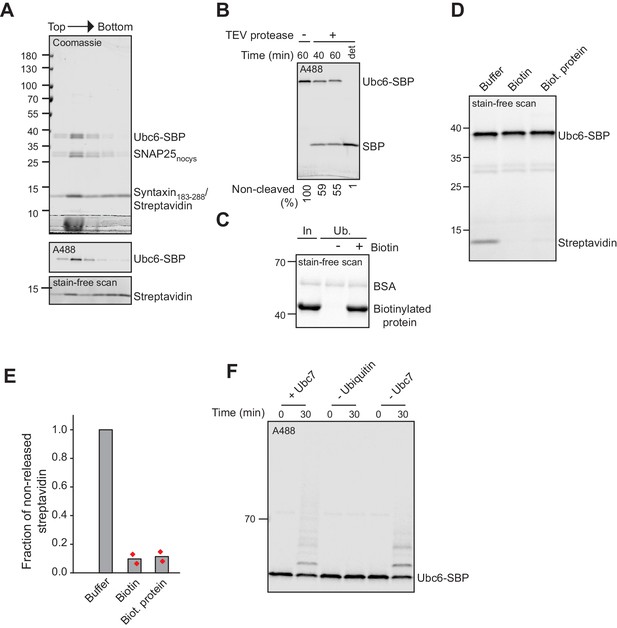
Liposomes with Ubc6-SBP and streptavidin.
(A) Liposomes containing t-SNARE and fluorescently labeled Ubc6-SBP in complex with streptavidin were subjected to a Nycodenz step gradient. After ultracentrifugation, the gradient was fractionated and analyzed by SDS-PAGE and Coomassie Blue staining (top), fluorescence scanning (middle), and using stain-free dye technology (bottom) to distinguish between streptavidin and Syntaxin, which is not visible with this technique as it lacks tryptophan residues. (B) The orientation of Ubc6-SBP in liposomes was assessed by testing for accessibility of TEV-protease to the TEV-cleavage site between the Ubc6 TM anchor and the SBP-tag. Liposomes were incubated with buffer or TEV protease with or without TX-100 (det) for the indicated times. Samples were analyzed by SDS-PAGE and fluorescence scanning. Numbers at the bottom indicate the fraction of protein right-side out oriented. (C) Streptavidin affinity pulldown of biotinylated protein in the presence and absence of biotin showing complete biotinylation of the protein. Samples of Input (In) and unbound (Ub) fractions were analyzed by SDS-PAGE and scanning of the gel using stain-free dye technology. (D) Streptavidin was added to liposomes containing fluorescently labeled Ubc6-SBP. After addition of buffer, biotin, or biotinylated protein, liposomes were floated in a sucrose step gradient. The top fraction of the gradient was analyzed by SDS-PAGE and the gel scanned using stain-free dye technology to visualize streptavidin. Ubc6 gives a strong signal here, because the fluorescent dye is also detected with the scanner used. (E) Quantification of co-floating streptavidin from two experiments as in (D). Streptavidin signal was first normalized to Ubc6-SBP signal and then normalized to the signal in buffer only control. (F) Ubiquitination of fluorescently labeled Ubc6-SBP. Samples of the ubiquitination reaction from experiments as in Figure 6C were analyzed by SDS-PAGE and fluorescence scanning.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (S. cerevisiae) | DOA10 | YIL030C | Amplified from BY4741 | |
| Gene (S. cerevisiae) | UBC6 | YER100W | Amplified from BY4741 | |
| Gene (S. cerevisiae) | UBA1 | YKL210W | Amplified from BY4741 | |
| Gene (S. cerevisiae) | UBC7 | YMR022W | Amplified from BY4741 | |
| Gene (S. cerevisiae) | CUE1 | YMR264W | Amplified from BY4741 | |
| Gene (S. cerevisiae) | CDC48 | YDL126C | Amplified from BY4741 | |
| Gene (S. cerevisiae) | UFD1 | YGR048W | Amplified from BY4741 | |
| Gene (S. cerevisiae) | NPL4 | YBR170C | Amplified from BY4741 | |
| Gene (S. cerevisiae) | GET3 | YDL100C | Amplified from BY4741 | |
| Gene (Rattus norvegicus) | Synaptobrevin 2 | NP_036795 | ||
| Strain, strain background (S. cerevisiae) | BY4741 | GE Dharmacon | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | |
| Strain, strain background (S. cerevisiae) | Δdoa10 | GE Dharmacon | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 doa10::kanR | |
| Strain, strain background (E. coli) | BL21 (DE3) | New England Biolabs | C2527I | Competent Cells |
| Strain, strain background (E. coli) | BL21-CodonPlus (DE3)-RIPL | Agilent | # 230280 | Competent Cells |
| Antibody | Anti-SBP (clone 20), mouse monoclonal | Merck | Cat#: MAB10764 | (1:2500) diluted in 5% milk TBS-T |
| Antibody | Anti-His6(Clone13/45/31‐2), mouse monoclonal | Dianova | Cat#: DIA-900 | (1:500) diluted in 2% BSA PBS-T |
| Antibody | Goat polyclonal anti-mouse IgG secondary antibody (IRDye 800 CW) | Li-Cor Biosciences | Cat#: 926–32210 RRID:AB_2687825 | (1:15000) |
| Antibody | Goat polyclonal anti-mouse IgG secondary antibody (IRDye 680 RD) | Li-Cor Biosciences | Cat# 926–68070, RRID:AB_10956588 | (1:15000) |
| Antibody | Rabbit polyclonal anti- Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11094f, RRID:AB_221544 | (1:15) diluted |
| Peptide, recombinant protein | Streptavidin | New England Biolabs | N7021S | |
| Peptide, recombinant protein | Gly-Gly-Gly-Cys peptide | Thermo Fisher Scientific | for Sortase-mediated labeling | |
| Commercial assay or kit | Gibson Assembly Master Mix | New England Biolabs | E2611S | |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | New England Biolabs | E0554S | |
| Commercial assay or kit | MasterPure Yeast DNA Purification Kit | Epicentre (Lucigen) | MPY80200 | |
| Chemical compound, drug | Decyl Maltose Neopentyl Glycol (DMNG) | Anatrace | NG322 | |
| Chemical compound, drug | GDN | Anatrace | GDN101 | |
| Chemical compound, drug | n-Octyl β-D-glucopyranoside (OG) | Glycon Biochemicals | D97001 | |
| Chemical compound, drug | n-Decyl β-D-Maltopyranoside (DM) | Glycon Biochemicals | D99003 | |
| Chemical compound, drug | Dodecyl-β-D-maltoside (DDM) | Carl Roth | CN26.5 | |
| Chemical compound, drug | Anapoe-X-100 (Triton X-100) | Anatrace | APX100 | |
| Chemical compound, drug | Sodium cholate hydrate | Sigma | C1254 | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-glycero3-phospho-choline (POPC) | Avanti Polar Lipids | 850457P | |
| Chemical compound, drug | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) | Avanti Polar Lipids | 850725P | |
| Chemical compound, drug | 1,2,-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) | Avanti Polar Lipids | 840035P | |
| Chemical compound, drug | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) | Avanti Polar Lipids | 870282P | |
| Chemical compound, drug | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhd-PE) | Avanti Polar Lipids | 810150P | |
| Chemical compound, drug | Ergosterol (>95%, HPLC) | Sigma-Aldrich | 45480 | |
| Chemical compound, drug | ATP | PanReac AppliChem | A1348 | |
| Chemical compound, drug | AlexaFluor488 maleimide | Thermo Fisher Scientific | A10254 | |
| Chemical compound, drug | DyLight 680 maleimide | Thermo Fisher Scientific | 46618 | |
| Chemical compound, drug | Pierce Detergent removal spin columns | Thermo Fisher Scientific | 87777 | |
| Chemical compound, drug | Ubiquitin (WT), yeast | Boston Biochem | U-100Sc | |
| Chemical compound, drug | Ubiquitin (K0) | LifeSensors | SI209 | |
| Chemical compound, drug | YEP broth | Formedium | CCM0410 | |
| Chemical compound, drug | Yeast Nitrogen Base (YNB) | US Biological Life Sciences | C19032801 | |
| Chemical compound, drug | CSM,-Ura | Formedium | DCS0161 | |
| Chemical compound, drug | D-(+)-Galactose | PanReac AppliChem | A1131 | |
| Other | HisPur NiNTA resin | Thermo Fisher Scientific | 88223 | |
| Other | Pierce High Capacity Streptavidin Agarose | Thermo Fisher Scientific | 20361 | |
| Other | Pierce Streptavidin Magnetic Beads | Thermo Fisher Scientific | 88817 | |
| Other | Novex DYNAL Dynabeads His-tag Isolation and Pulldown | Thermo Fisher Scientific | 10103D |





