Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes
Figures
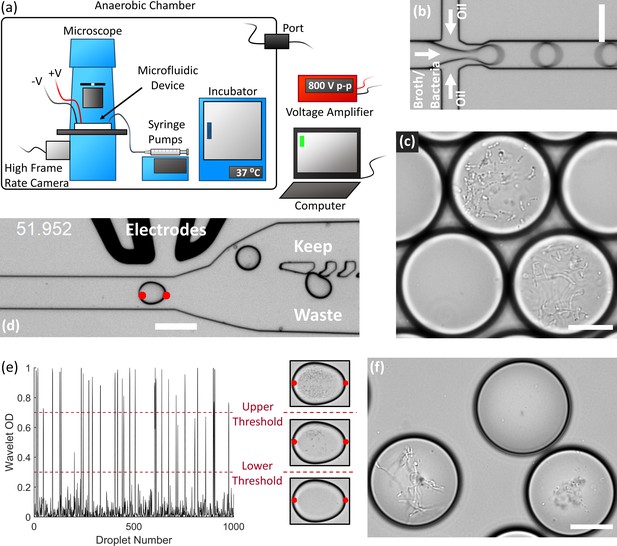
End-to-end system for efficient isolation and culture of gut anaerobes in microfluidic droplets.
(a) The experimental setup for isolating, culturing, and sorting anaerobic bacteria in microfluidic droplets consists of a microscope, microfluidic devices, a high frame rate camera, syringe pumps, an incubator, and electrodes all contained within an anaerobic chamber. The computer controls the syringe pumps, high frame rate camera, and electrodes (via a voltage amplifier). The equipment power and control wires are introduced to the anaerobic chamber through sealed rubber ports to strictly maintain the anaerobic conditions within the chamber. (b) Single bacteria cells are isolated in droplets containing anaerobic culture medium and the resulting emulsion is cultured inside the incubator. (c) An example of human gut bacteria isolated and cultivated inside droplets. (d) Droplets are sorted by optical detection and subsequent deflection via dielectrophoresis near a sorting junction. Specifically, droplets with bacterial colonies which meet a certain thresholding criteria were determined using image analysis (region between the red dots), and these droplets were deflected into the ‘keep’ path by actuating an on-chip electrode while sending the remaining droplets to waste. (e) The colony density measured by image analysis (Wavelet OD) for 1000 successive droplets. Droplets with a dense colony, a sparse colony, and empty droplets (no colony) are represented by a wavelet OD value above an upper threshold, between an upper and lower threshold, or below a lower threshold, respectively. (f) Two slow-growing human gut-associated bacteria colonies (bottom left and bottom right) grown in droplets after sorting and a false positive empty droplet (top). Scale bar in (b) and (d) is 100 μm and scale bar in (c) and (f) is 20 μm.
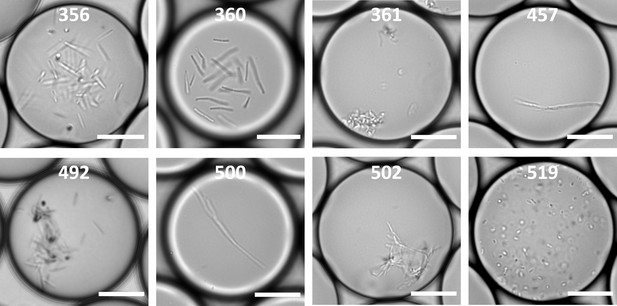
The Altered Schaedler’s Flora, ASF, is an important and widely studied gnotobiotic mouse model used for understanding microbiota-host dynamics in both health and disease.
The ASF community contains two facultative anaerobes, Lactobacillus intestinalis (ASF 360) and Lactobacillus murinus (ASF 361), two anaerobes, Mucispirillum schaedleri (ASF 457) and Parabacteroides goldsteinii (ASF 519), and four extremely oxygen sensitive anaerobes, Clostridium sp. (ASF 356), Eubacterium plexicaudatum (ASF 492), Pseudoflavonifactor sp. (ASF 500), and Clostridium sp. (ASF 502) (Wymore Brand et al., 2015). Pure ASF strains were cultured from stock solutions in liquid BHIS-ASF broth, then diluted and encapsulated in droplets using our microfluidic platform, and incubated at 37°C. After 17 to 20 hr, we observed that all 8 ASF strains were successfully cultured in the droplets. Scale bars are 20 μm.
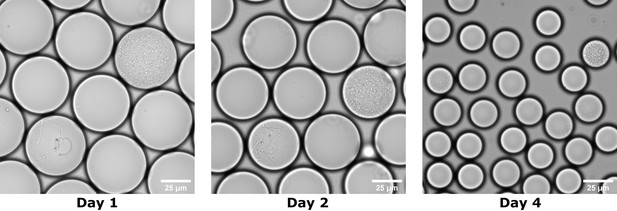
Droplet volume decreased during cultivation in the anaerobic chamber.
The figure shows cultivation of the donor FMT sample in BHIS droplets at 1 day, 2 days, and 4 days of cultivation time. The droplet volume decreased ~10 x from Day 1 to Day 4.
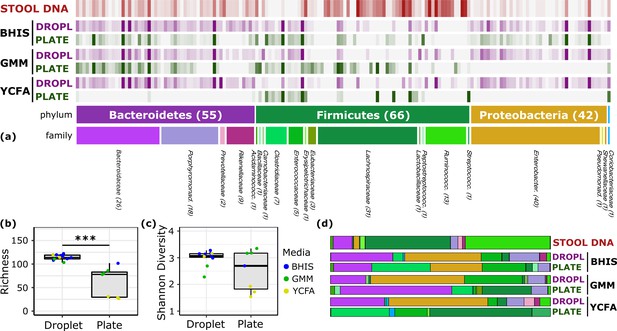
Comparison of human stool bacteria cultured on plates versus in droplets.
(a) The relative abundance for each ASV organized by phylum and family is plotted for the raw stool and for representative droplet and plate cultures in each medium. The dark bars indicate a higher relative abundance. (b) The community richness averaged over cultivation time and media is increased in droplets over plates (p<0.005) but (c) the Shannon diversity is not. (d) Family-level relative abundance for representative droplet and plate cultures. The community composition between droplet cultures of different media is more similar than between plates.
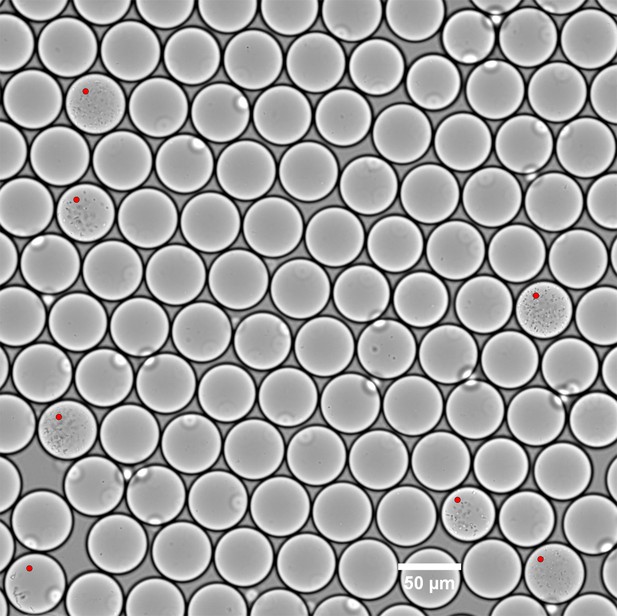
A one day cultivation of the donor FMT stool sample in GMM medium.
The seven red dots mark drops with growing colonies and correspond to 6% of the total drops within the image. For the initial live cell loading density of ~7.5 ± 5.0 x 105/mL and of ~65 pL droplets, the Poisson process of droplet loading suggests ~1.6–7.5% of droplets should contain a single live cell at time zero. The agreement between the expected single cell loading density at time zero and the measured number of colonies after 1 day of culture, coupled with uniform cell morphology in each droplet, suggests that the majority of colonies grew from a single cell.
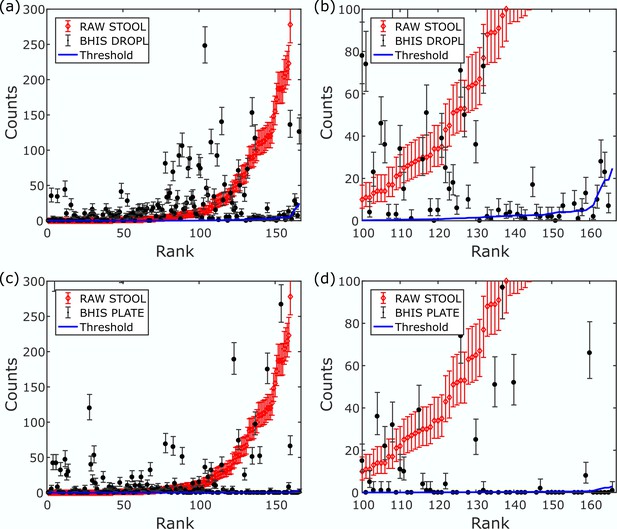
Amplicon sequence variant (ASV) filtering.
ASV counts for representative droplet (a–b) and plate (c–d) cultures are ordered by increasing rank of ASVs from the raw stool sample and a magnified region of (a) and (c) is shown in (b) and (d), respectively. The percentage of dead or nonviable DNA, pn.v., which is carried over from the initial innoculum into the collected DNA post culture is determined by fitting pn.v.*(RAW STOOL counts) to sample counts which are nonzero and at least 10x less than the raw stool counts. The best fit defines a threshold (blue line) for excluding reads. The error bars depict the 90% confidence interval of the true proportion of counts. Any ASV with a lower 90% confidence interval limit less than the threshold is discarded.
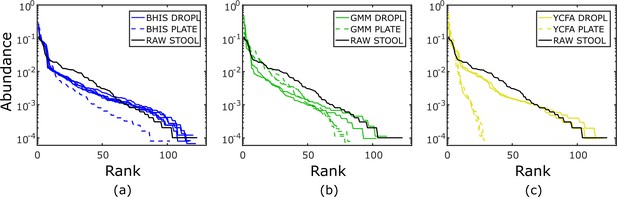
Rank-abundance curves for independent experiments in BHIS, GMM, and YCFA medium cultivated in droplets or on plates.
No two experiments within a given media and condition (i.e., BHIS droplet, GMM droplet, GMM plate, YCFA droplet, or YCFA plate) were found to reject the null hypothesis that the two data samples can be generated from the same distribution under the two-sample Kolmogorov–Smirnov test, indicating the reproducibility of cultivation between independent experiments.
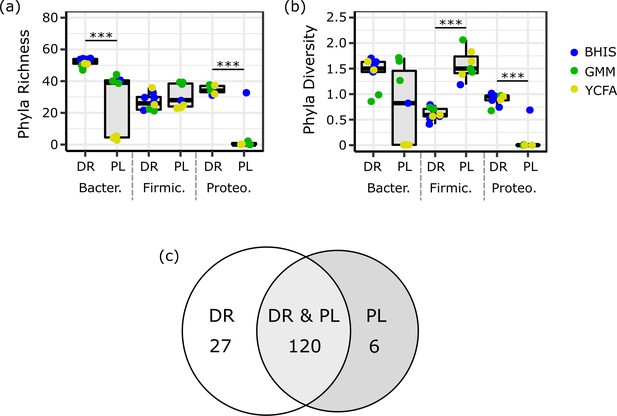
Intra-phyla (a) richness and (b) diversity for droplets (DR) and plates (PL) evaluated for Bacteroidetes (Bacter.), Firmicutes (Firmic.), and Proteobacteria (Proteo.).
The cultivated richness within Bacteroidetes and Proteobacteria on droplets is greater than plates across media (***, p<0.005, Mann-Whitney U test), while the diversity within Firmicutes is greater on plates than droplets (***, p<0.005, Mann-Whitney U test). Although our n is too low to statistically test within media, we note that very few Bacteroidetes were cultured on YCFA plates and similarly for Proteobacteria on GMM and YCFA plates. The intra-phyla richness of plates for each phyla is non-normally distributed as determined by a Shapiro-Wilk test for normality (R function shapiro.test, Plates – Bacteroidetes, p=0.0090, Plates – Firmicutes, p=0.035, Plates – Proteobacteria, p=1.7×10−5). The intra-phyla richness of droplets for each phyla was not found to be non-normally distributed. (c) The number of unique ASVs across all three rich media among both droplet and plate cultures (DR and PL), only plate cultures (PL), and only droplet cultures (DR). Despite a wide range of rich media, 6 ASVs could only be detected in droplets and 27 ASVs could only be detected in droplets, suggesting some organisms grow in the competitive plate environment while a larger majority prefer confined isolation in drops (likely due to either removal of competition or increase in quorum sensing).
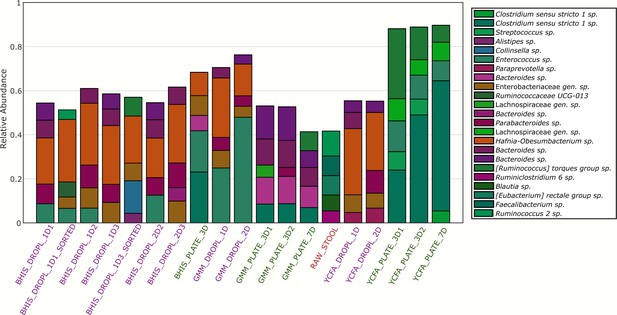
Relative abundance of the top five most abundant ASVs in each sample with taxonomic identification.
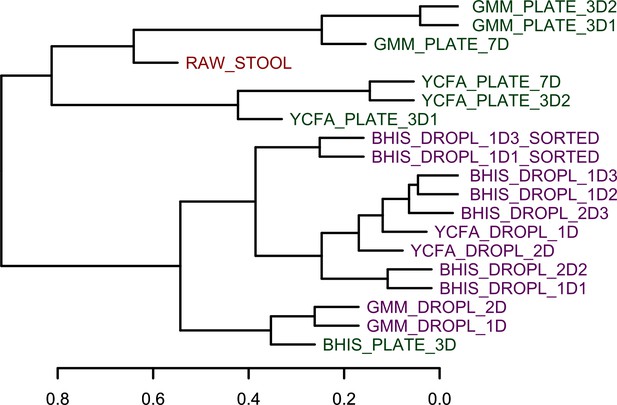
Bray-Curtis hierarchical clustering of the family-level composition for all samples.
The droplet-based culture reduces the variation in cultivated composition between media as compared to plate cultures.
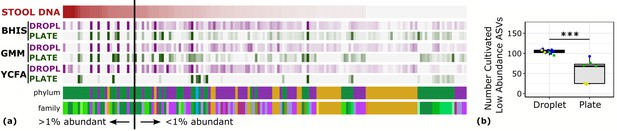
Droplet culture improves the cultivation of low abundant organisms.
(a) The relative abundance per ASV organized by relative abundance in raw stool. The phylum and family colors correspond to the labels shown in a. (b) The number of cultivated low abundant ASVs in the raw stool sample (<1%, total of 130 ASVs) averaged over cultivation time and three different media is increased in droplets over plates. The legend for the phylum and family color labels is depicted in Figure 2a.
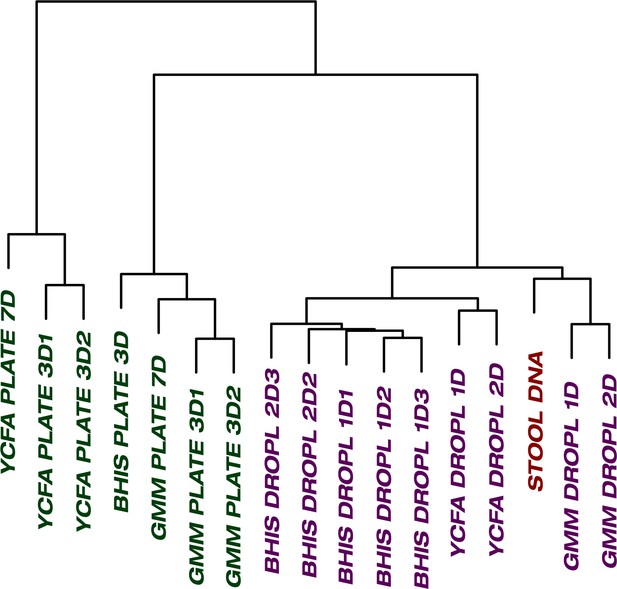
The culture of stool in droplets enables cultivation of clinically relevant Bacteroides spp.
which did not grow on plate cultures. Hierarchical clustering of Bacteroides oligotypes across seven plate cultures and nine droplet cultures reveals a stronger association of droplet cultures towards the raw stool as compared to plates.
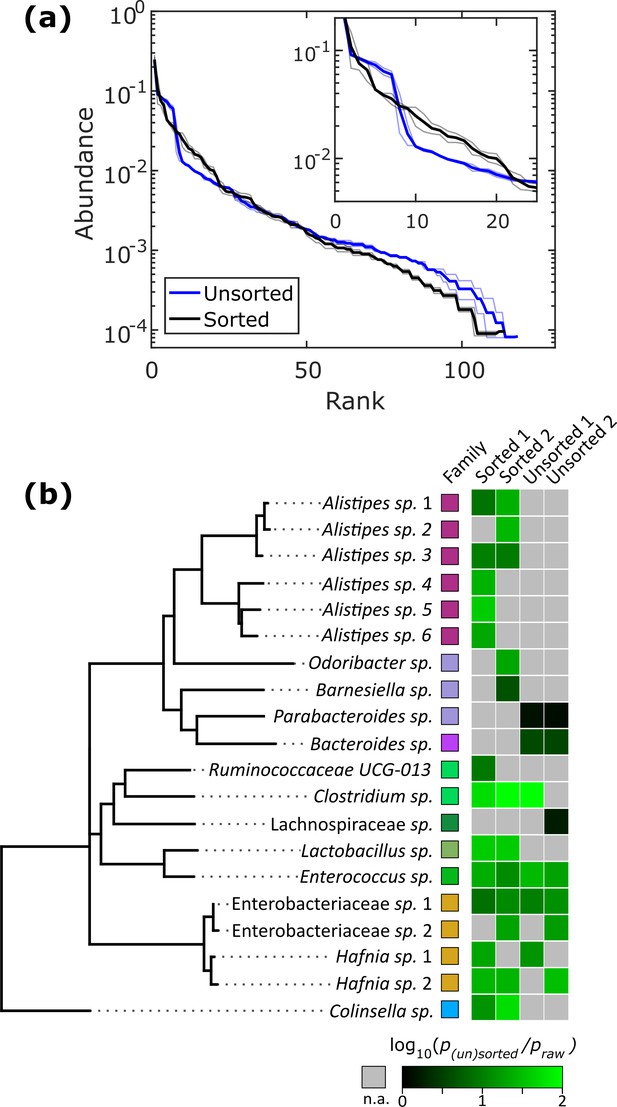
Isolation of slow-growing species from human stool microbiota.
(a) The rank-abundance curves show that sorting based on colony density changes the overall community composition. A zoomed in portion of the rank-abundance curve is shown in the inset. (b) Phylogenetic tree of ASVs which were <1% abundant in raw stool but were increased to >1% in at least one sorting experiment. The ratio of amplification for strains amplified above the 1% limit is depicted in the heat map. N.A. (shown in gray) indicates ASVs which were not amplified from <1% to>1%. The legend for the family color labels is depicted in Figure 2a.
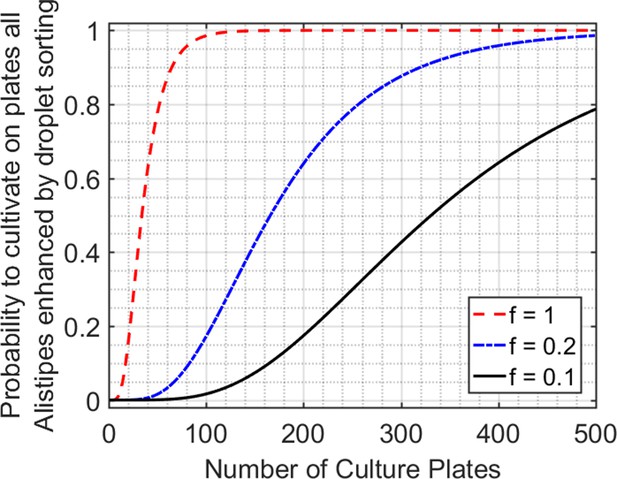
Comparison between droplets and plates for the isolation and cultivation of the 6 Alistipes populations enhanced by droplet sorting shown in Figure 5b.
The droplet technology enables high-throughput isolation and cultivation of organisms in discrete picoliter volumes with millions of individual drops per mL of droplet emulsion. In contrast, the conventional method of low-dilution plating achieves pure strain isolation by spreading the inoculum at low enough density on agar plates so that growing colonies do not overlap. In this Supplementary Figure, we calculate the equivalent number of culture plates needed to isolate and cultivate the 6 Alistipes populations enhanced by droplet sorting. For low-dilution plating aimed at isolating slow-growing organisms, we assume a plating density of 1 cell/cm2 on a standard 100 mm circular culture plate = 78 cells/plate. The probability for a given population, X1, to be present and grow follows from binomial random sampling and is P(X1 > 0) = 1 – P(X1 = 0) = 1 – (1 – f*p1)n, where n is the number of colonies, p1 is the abundance of X1 in the initial inoculum, and f is a constant which describes the fraction of X1 which will grow in the environmental conditions. If the number of colonies is much greater than the number of populations we are aiming to grow, then P(Isolate and cultivate all populations of interest) = P(X1 > 0) * P(X2 > 0) *. . .* P(Xm > 0), where m is the number of unique populations. The figure above plots P(Isolate and cultivate the 6 droplet sorting enhanced Alistipes populations) for varying f. Even in the most optimistic scenario, where every Alistipes cell is guaranteed to grow on the low-dilution culture plate, it would still require 65 culture plates to have a 90% chance at isolating and growing all 6 populations.
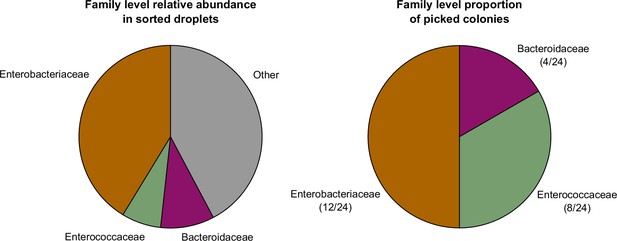
Bacteria can be cultured on traditional plates following cultivation in droplets.
(Left) The family level relative abundance of the pooled droplets from experiment BHIS_DROPL_1D1_SORTED. (Right) The family level taxonomic assignment of 24 randomly picked colonies on a BHIS plate streaked from the same sample shown on the left. For each family detected on plate, only one genera was observed within that family. In particular, we detected Hafnia (12/24), Enterococcus (8/24), and Bacteroides (4/24).
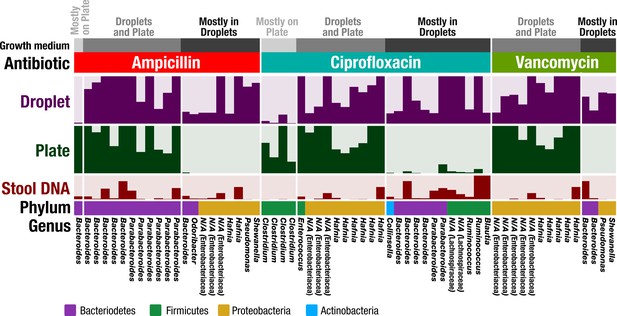
Antibiotic resistance screening of FMT donor stool in droplet cultures reveals antibiotic resistant members.
The relative abundance of ASVs detected from droplet and plate cultures (rows) is shown for the three separate antibiotics along with the relative abundance in the raw stool sample. Each row is a linear scale between 0% and 2.5% relative abundance. For each antibiotic, ASVs are grouped according to plate preferred growth (light grey), droplet preferred growth (dark grey), or growth in droplets and plates (grey). Dead and nonviable cell DNA was removed via droplet sorting (for droplets only) and ASV filtering so that the ASVs depicted represent only organisms which grew in the presence of the antibiotics during the culture period. N/A indicates taxa which are unassigned at the genus level.
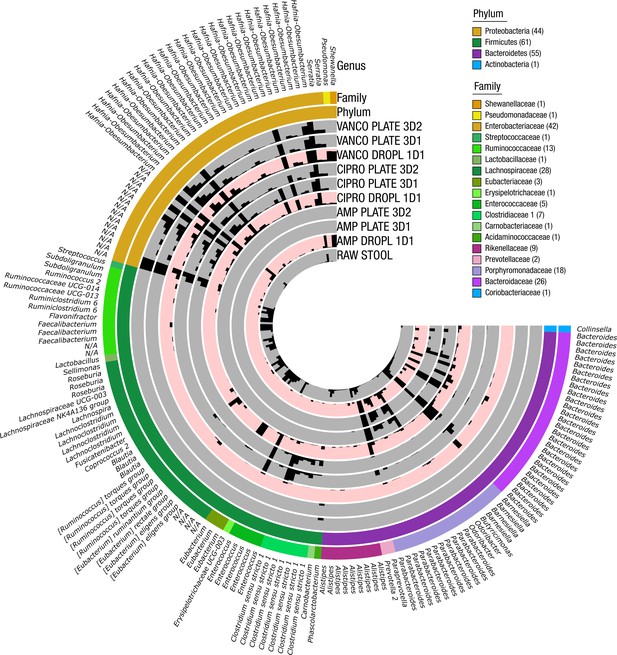
ASVs with taxonomic assignment in antibiotic droplet and plate experiments.
The FMT donor stool sample was grown in droplets for 1 day (DROPL) and plates for 3 days (PLATE) in the presence of ampicillin (AMP), ciprofloxacin (CIPRO), and vancomycin (VANCO). The relative abundance for each sample is shown with a linear scale from 0% to 2%. N/A indicates taxa unassigned at the genus level.
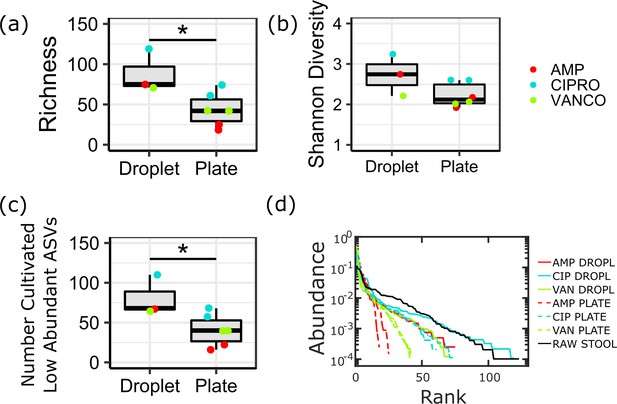
Ecological measures of the cultivated community composition on droplets and plates in the presence of ampicillin (AMP, red), ciprofloxacin (CIPRO, blue), and vancomycin (VANCO, green) for the (a) richness, (b) the Shannon diversity, (c) number of ASVs which are <1% in the raw stool sample, and (d) rank-abundance.
Videos
Human stool bacteria cultured in BHIS droplets for 1 day.
Droplet sorting in an anaerobic environment for human stool bacteria cultured in BHIS droplets for 1 day.
The top left time stamp is in seconds. For each droplet, the region between the 2 red dots is analyzed using the Wavelet OD. For empty droplets and droplets containing a dense colony, the Wavelet OD does not satisfy the thresholding criteria (the decision is labeled as false, ‘F’) and the droplets flow down the waste path. When the threshold criteria are met for a sparse colony (the decision is labeled as true, ‘T’), the electrodes are actuated sending the droplet to the ‘keep’ path. We note that the spots in the oil phase were observed even for droplets generated without bacteria.
Additional files
-
Source code 1
ASV filtering to remove nonviable or dead cell DNA from reads.
- https://cdn.elifesciences.org/articles/56998/elife-56998-code1-v2.m.zip
-
Source data 1
Experiment_Info.
The droplet generation and cultivation information is listed for each experiment. 2. ASV_Counts_Matrix. The counts matrix for each ASV determined by Minimum Entropy Decomposition (MED) for the raw human stool sample and the human stool cultured on plates and in droplets. 3. ASV_Filtered_Percent_Matrix. Percent matrix for each ASV obtained by filtering the reads using the Matlab analysis file ASV_Filtering.m. 4. Sequences_Taxonomy. Closest taxonomic assignment of each ASV from raw and cultivated human stool samples. 5. Sequences. Sequences from MED analysis for raw and cultivated human stool samples. 6. Ecological_Measures. Richness, Diversity, Dominance, and Number Rare measurements for each sample and the intra-phyla richness and diversity across all 4 detected phyla. 7. Oligotyping_Percent_Matrix. Percent matrix for Bacteroides oligotype assignment for each sample. 8. Oligotyping_Representatives. Oligotypes of Bacteroides spp. 9. Sorting_Enhancement_Ratio. Enhancement ratio for ASVs which were <1% abundant in the raw stool sample but >1% in the droplet sorting experiments. 10. Sanger_Consensus_Sequences. The consensus sequence for each of 24 randomly picked colonies from sorted droplets which were streaked onto a plate. For each sequence, the closest BLAST taxonomic assignment along with percent identity is also listed.
- https://cdn.elifesciences.org/articles/56998/elife-56998-data1-v2.xlsx
-
Supplementary file 1
CAD file for microfluidic droplet generating and sorting devices.
- https://cdn.elifesciences.org/articles/56998/elife-56998-supp1-v2.dwg.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56998/elife-56998-transrepform-v2.docx





