Notch and TLR signaling coordinate monocyte cell fate and inflammation
Figures
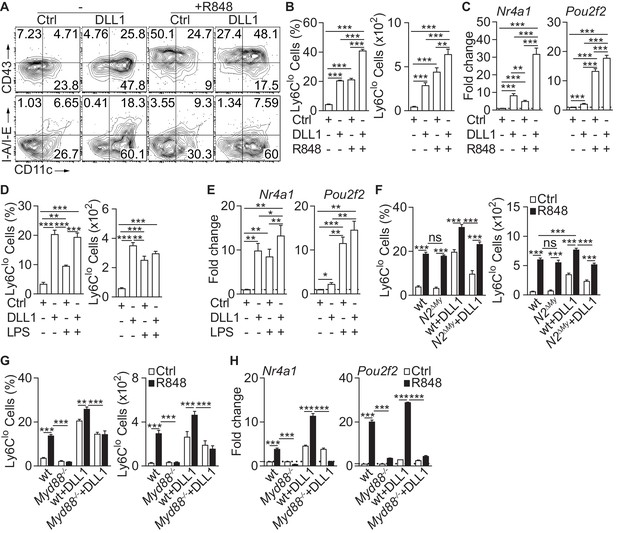
Inflammatory conditions enhance monocyte conversion in vitro.
(A–F) Monocyte conversion in the presence of DLL1 and TLR agonists in vitro: (A) Representative flow cytometry plot, (B) relative frequency of Ly6Clo monocyte-like cells in live CD11b+GFP+ cells (left) or absolute numbers of Ly6Clo monocyte-like cells recovered from each well (right) are shown (representative of 3 experiments, n = 3). (C) Bar graphs showing expression of Ly6Clo monocyte hallmark genes, Nr4a1 and Pou2f2 from in vitro cultures treated with R848 (pooled from four experiments, n = 8–12). (D) Relative frequency (in live CD11b+GFP+ monocytes) or absolute numbers of Ly6Clo monocyte-like cells (from three experiments, n = 3) and (E) expression of Nr4a1 and Pou2f2 (from four experiments, n = 4–6) in the presence of LPS in vitro are shown. (F) wt or N2ΔMy Ly6Chi monocyte conversion in the presence of DLL1 and R848 in vitro: relative frequency (in live CD11b+GFP+ monocytes) or absolute numbers of Ly6Clo monocyte-like cells (from three experiments, n = 4) is shown. (B, D, F) Absolute frequency of monocytes for Ctrl and DLL1 (in B, (D), and wt (Ctrl), wt+DLL1 (Ctrl) in (F) conditions are derived from the same experiments but are depicted as a three separate graphs for simplicity.(G, H) R848-enhanced conversion is Myd88 dependent in vitro. Relative frequency (in live CD11b+CX3CR1+ monocytes) or absolute numbers of Ly6Clo monocyte-like cells (G) and gene expression analysis in vitro (H) are shown (data are from two independent experiments, n = 3). (B, D, F–H) *p<0.05, **p<0.01, ***p<0.001; two-way ANOVA with Bonferroni’s multiple comparison test. (C, E) *p<0.05, **p<0.01, ***p<0.001; paired one-way ANOVA with Geisser-Greenhouse’s correction and Bonferroni’s multiple comparison test.
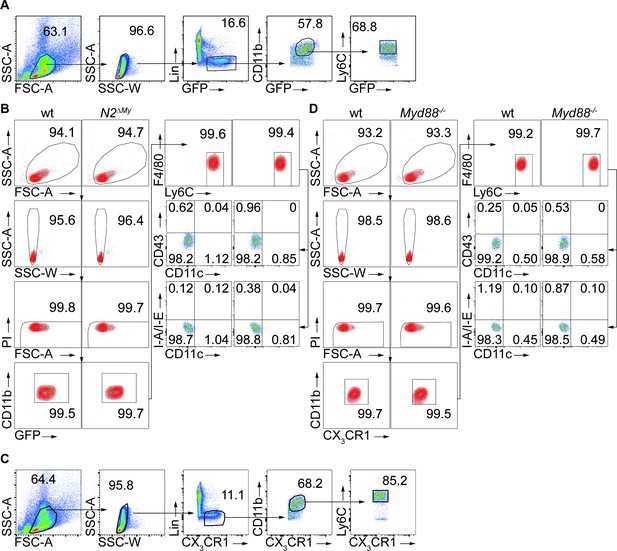
Strategy of monocyte isolation from mouse bone marrow.
(A) Sorting strategy of BM Ly6Chi monocytes from CX3CR1-GFP expressing (GFP+) wt or N2ΔMy mice used for in vitro culture experiments in Figure 1, or for adoptive transfer in Figure 2 and Figure 5. (B) Expression of CD11b, Ly6C, CX3CR1, F4/80, CD11c, CD43 and I-A/I-E, and purity of the cells sorted for in vitro culture (Figure 1) or adoptive transfer experiments Figure 2 and Figure 5, respectively. (C) Sorting strategy for isolation of BM Ly6Chi monocytes from wt or Myd88-/- mice used for in vitro culture (Figure 1) or adoptive transfer experiments (Figure 2). CX3CR1 depicts staining with anti-CX3CR1 antibody. (D) Expression of CD11b, Ly6C, CX3CR1, F4/80, CD11c, CD43 and I-A/I-E, and purity of wt or Myd88-/- cells sorted for in vitro culture (Figure 1) or adoptive transfer experiments Figure 2 and Figure 5 respectively.
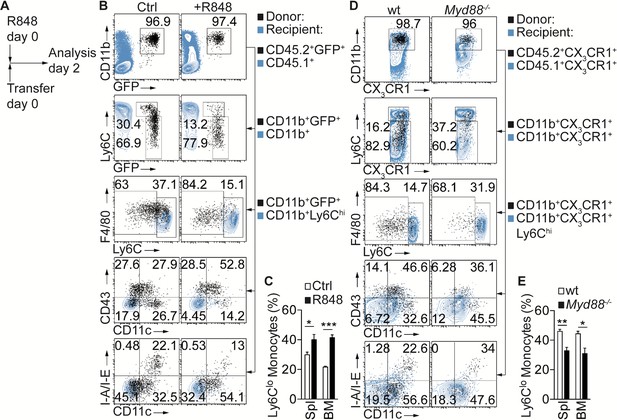
Inflammatory conditions enhance monocyte conversion in vivo.
(A–E) Adoptive transfer and flow cytometry analysis of BM CD45.2+ Ly6Chi monocytes in control or R848 injected CD45.1+ congenic recipients: (A) Experimental setup is depicted; (B) Flow cytometry plots showing the progeny of transferred CD45.2+CD11b+Ly6ChiCX3CR1-GFP+ (GFP+) cells in black and recipient CD45.1+ (1st row), CD45.1+CD11b+ (2nd row) or CD45.1+CD11b+Ly6Chi cells (3rd −5th rows) in blue; (C) Frequency of donor-derived Ly6Clo monocytes pooled from two independent experiments (n = 5). (D, E) R848-enhanced conversion is Myd88 dependent in vivo. (D) Flow cytometry plots showing transferred CD45.2+CX3CR1+ wt or Myd88-/- cells in black and recipient CD45.1+CX3CR1+ (1st row), CD45.1+CX3CR1+CD11b+ (2nd row) or CD45.1+CX3CR1+CD11b+Ly6Chi cells (3rd −5th rows) in blue. All recipient mice which received wt or Myd88-/- donor cells were treated with R848; (E) Frequency of donor-derived Ly6Clo monocytes pooled from two independent experiments are shown (n = 4/5). (C, E) *p<0.05, **p<0.01, ***p<0.001; Student’s t-test.
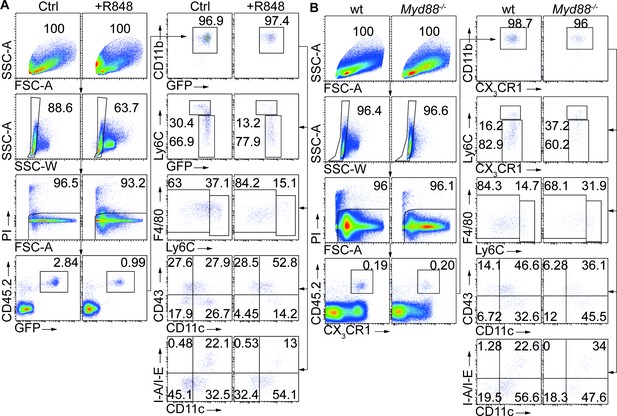
Inflammatory conditions enhance monocyte conversion in vivo.
(A, B) Full gating path of adoptively transferred donor CD45.2+ monocytes recovered from CD45.1+ recipients corresponding to Figure 2B and D respectively.
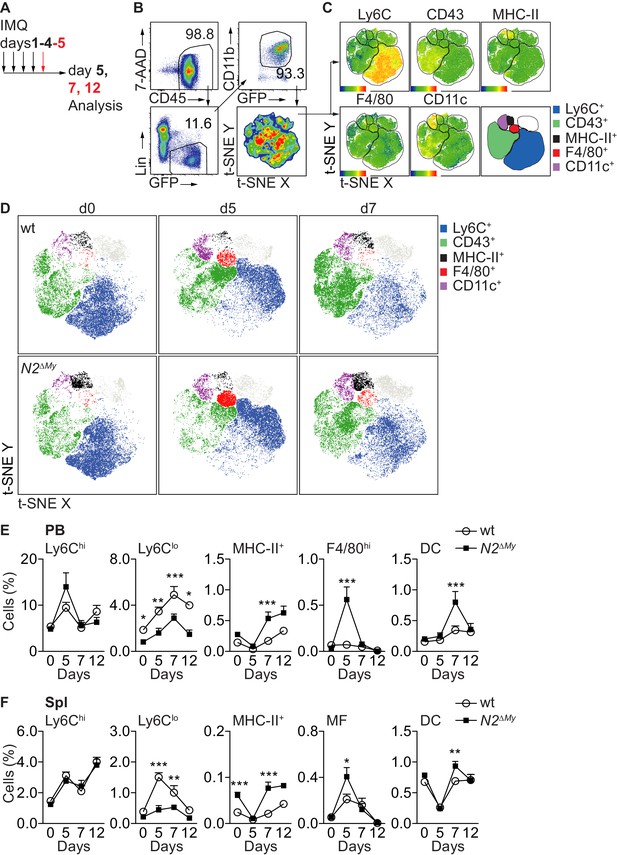
Acute inflammation triggers altered myeloid cell response in N2ΔMy mice.
(A) Experimental set-up for IMQ treatment and analysis of mice. (B, C) Gating strategy for t-SNE analysis and definition of cell subsets based on expression of surface markers are shown. t-SNE was performed on live CD45+Lin-CD11b+GFP+ cells concatenated from 48 PB samples from four independent experiments. (D) Unsupervised t-SNE analysis showing composition and distribution of cellular subsets from PB of wt or N2∆My IMQ-treated or untreated mice at different time points defined in B, C) (n = 8 mice are pooled for each condition). (E, F) Relative frequency of different myeloid subpopulations in PB and Spl of untreated or IMQ-treated mice are shown (data are pooled from six experiments n = 7–18). (E, F) *p<0.05, **p<0.01, ***p<0.001; two-way ANOVA with Bonferroni’s multiple comparison test.
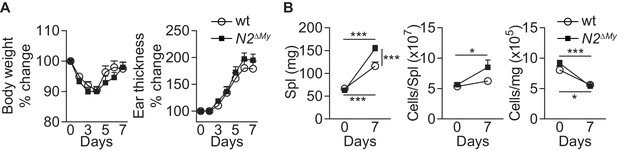
IMQ treatment induces systemic inflammation in mice.
(A) Body weight (left) and ear thickness (right) of IMQ-treated wt or N2ΔMy mice (data are from three experiments, n = 11/13). (B) Spleen weight and cell number of IMQ-treated wt or N2ΔMy mice (data are from three independent experiments, n = 11/12). (B) *p<0.05, ***p<0.001; two-way ANOVA with Bonferroni’s multiple comparison test.
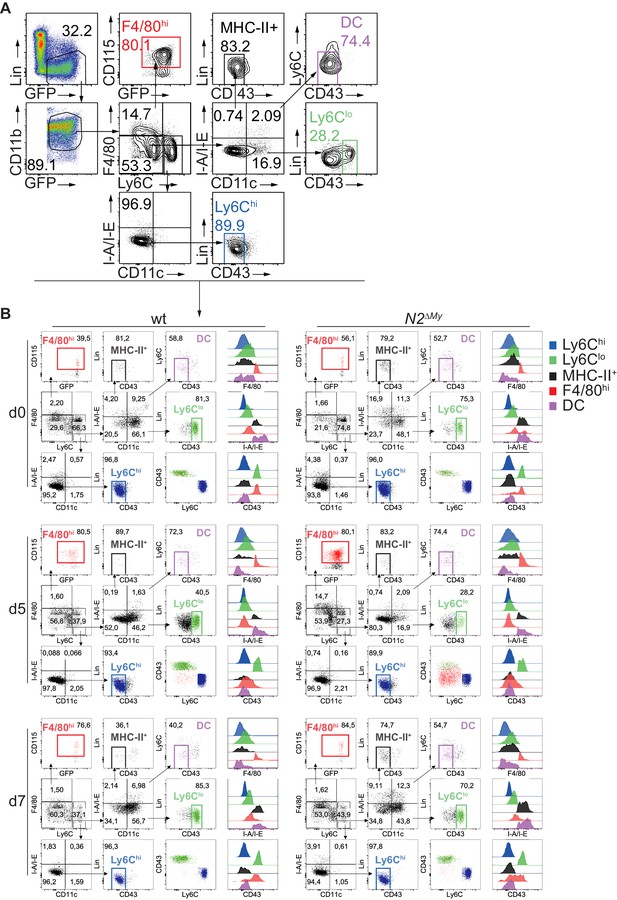
Identification of myeloid cell subsets in IMQ-treated mice.
(A) Gating strategy for definition and quantification of myeloid subsets from live CD45+Lin-CD11b+GFP+ cells by flow cytometry. Defined populations are color-coded and used for subsequent quantification. (B) Representative gating of color-coded myeloid subsets in mouse peripheral blood for each experimental condition corresponding to Figure 3. Overlay dot plots show CD43 vs Ly6C expression in Ly6Chi, Ly6Clo or FM+ monocytes. Histograms show intensity of F4/80, CD43 and I-A/I-E for all defined populations.
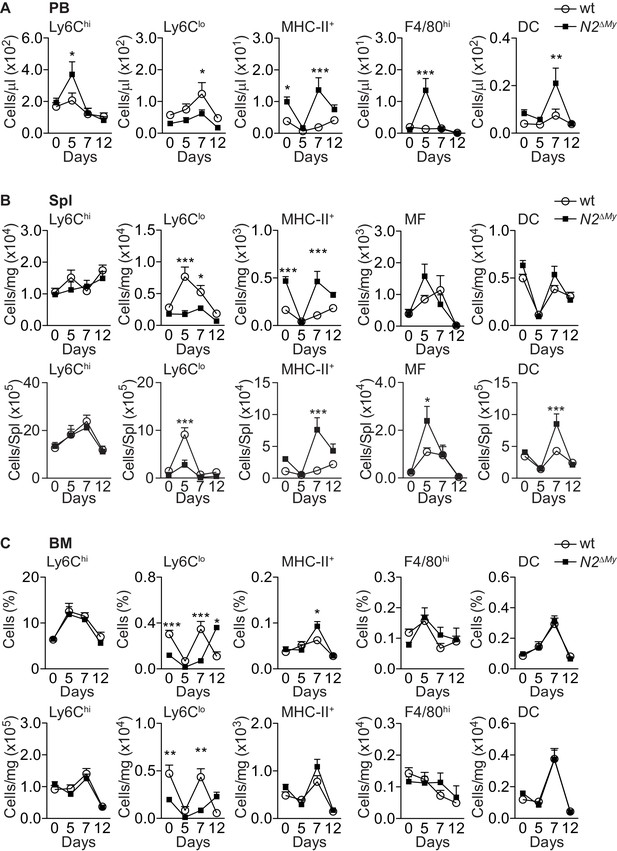
Flow cytometry analysis of myeloid cells in IMQ-driven inflammation.
(A) Absolute frequency of different myeloid subpopulations in PB. (B) Absolute frequency of different myeloid subpopulations normalized per mg tissue (top) or per spleen (bottom). (C) Relative (top) and absolute (bottom) frequency of different myeloid subpopulations in the bone marrow of Sham- or IMQ-treated mice are shown. (A–C) data are pooled from six experiments (n = 7–18). *p<0.05, **p<0.01, ***p<0.001; two-way ANOVA with Bonferroni’s multiple comparison test.
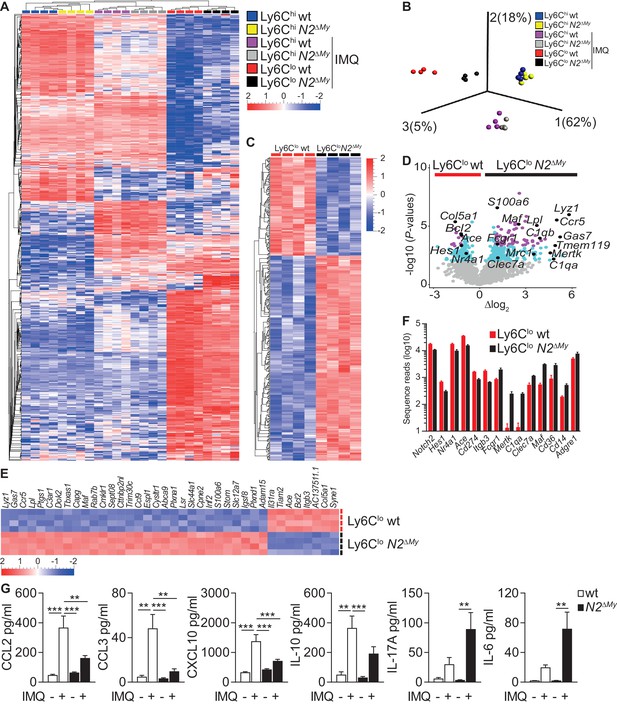
Enhanced macrophage gene expression signatures in monocytes and altered inflammatory response in N2ΔMy mice.
(A, B) Hierarchical clustering of 600 ANOVA-selected DEGs (A) and PCA of PB monocyte subsets (B) after IMQ treatment (n = 4) is shown (Variance filtering 0.295, ANOVA followed by the B-H correction (p<0.0076, FDR ≤ 0.01)). (C) Hierarchical clustering of 373 DEGs from IMQ-treated wt and N2ΔMy Ly6Clo monocyte subsets (Variance filtering 0.117,–0.6≥Δlog2 ≥ 0.6, Student’s t-test with B-H correction (p<0.01, FDR ≤ 0.05)). (D) Volcano plot showing 379 DEGs between wt and N2ΔMy Ly6Clo cells (-log10(p-value) ≥ 2, FDR ≤ 0.05, light blue); and 87 DEGs (−1≥Δlog2 ≥ 1 (FDR ≤ 0.01) purple); genes of interest are marked black (Student’s t-test with B-H correction). (E) Heat map of top 38 DEGs from (D) log10(p-value) ≥ 4,–1≥Δlog2 ≥ 1, Student’s t-test with B-H correction. (F) Bar graph showing mean number and SEM of sequence reads for selected genes from IMQ-treated wt and N2ΔMy Ly6Clo cell subsets. (G) Analysis of cytokine and chemokine profiles in the serum of IMQ-treated mice. n = 5–10, pooled from four independent experiments. (G) *p<0.05, **p<0.01, ***p<0.001; 2way ANOVA with Bonferroni’s multiple comparison test.
-
Figure 4—source data 1
List of 600 DEGs for hierarchical clustering and PCA (Figure 4A and B) from Ly6Chi and Ly6Clo subpopulations isolated from sham-or IMQ(Aldara)-treated wt or N2ΔMy mice.
- https://cdn.elifesciences.org/articles/57007/elife-57007-fig4-data1-v2.xlsx
-
Figure 4—source data 2
List of 373 DEGs between Ly6Clo cells isolated from IMQ(Aldara)-treated wt or N2ΔMy mice and used for the analysis in Figure 4C and D, Figure 5A and Supplementary file 2–5.
- https://cdn.elifesciences.org/articles/57007/elife-57007-fig4-data2-v2.xlsx
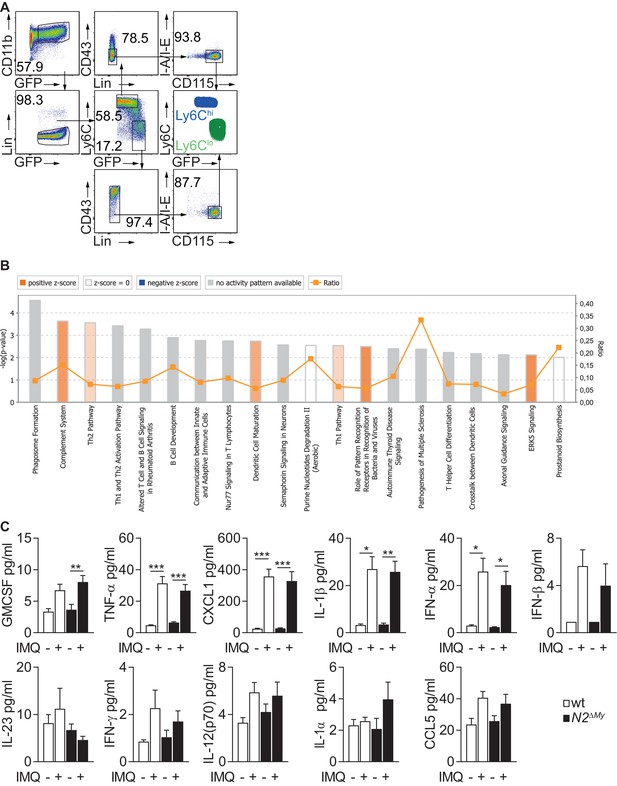
Gating strategy for cell sorting, IPA and cytokine and chemokine analysis in IMQ-treated mice.
(A) Sorting strategy of monocyte subsets for RNA-seq analysis. Lin-CD11b+GFP+CD115+Ly6ChiCD43-MHC-II- (Ly6Chi) and Lin-CD11b+GFP+CD115+Ly6Clo/-MHC-II- (Ly6Clo) monocytes were sorted from naive or IMQ-treated wt or N2ΔMy mouse PB and used for RNA-seq. (B) top 20 Ingenuity canonical pathways enriched in mutant Ly6Clo cells as compared to wt Ly6Clo subset from IMQ-treated mice. (C) Analysis of chemokine and cytokine levels in untreated or IMQ-treated wt or N2ΔMy mouse serum. Data are pooled from four independent experiments (n = 5–10).* p<0.05, **p<0.01, ***p<0.001; two-way ANOVA, Bonferroni’s post-test.
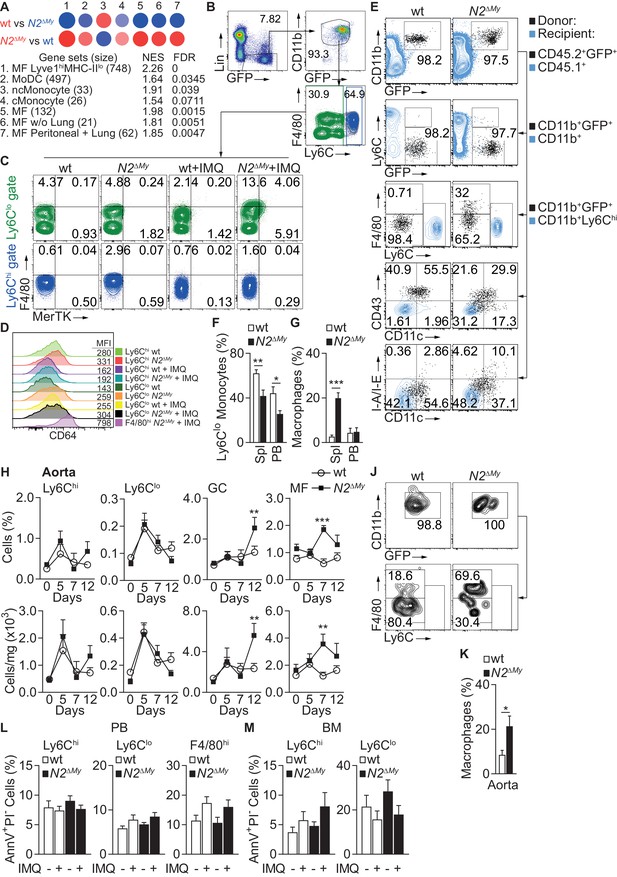
Notch2-deficient Ly6Clo cells show enhanced macrophage maturation during acute inflammation.
(A) GSEA based on 373 DEGs between IMQ-treated wt and N2ΔMy Ly6Clo subsets in PB. Red – positive-, and blue - negative enrichment in corresponding color-coded wt or N2ΔMy cells. Size of the circle corresponds to NES and intensity of the color to FDR. (B, C) Representative flow cytometry plots showing expression of F4/80 and MerTK in gated Lin-CD45+CD11b+GFP+Ly6Chi and Lin-CD45+CD11b+GFP+Ly6Clo cells from PB of Sham- or IMQ-treated wt or N2ΔMy mice. (D) Representative flow cytometry histograms with corresponding mean fluorescence intensities (MFI) showing expression of CD64 on myeloid cells in PB of sham or IMQ-treated mice. (E) Flow cytometry analysis 3 days after adoptive transfer of wt or N2∆My BM CD45.2+ Ly6Chi monocytes in IMQ-treated CD45.1+ recipients. Transferred cells are shown in black and recipient CD45.1+ (1st row), CD45.1+CD11b+ (2nd row) or CD45.1+CD11b+Ly6Chi cells (3rd −5th rows) are depicted in blue. (F, G) Frequency of donor-derived Ly6Clo monocytes (G) or macrophages (H) in CD45.2+CD11b+GFP+ donor cells after adoptive transfer of wt or N2∆My Ly6Chi monocytes is shown. Data are pooled from three independent experiments (n = 6/9). (H) Relative (top) and absolute number (bottom) of myeloid subpopulations in aortas of untreated or IMQ-treated mice. Data are pooled from six experiments (n = 7–18). (J, K) Representative flow cytometry plot of donor CD11b+GFP+ cells (J) and relative frequency of donor-derived macrophages (K) within CD45.2+CD11b+GFP+ cells recovered from aortas after adoptive transfer of wt or N2∆My Ly6Chi monocytes. (K) Data are pooled from three independent experiments (n = 9). (L, M) Relative frequency of apoptotic (AnnexinV+PIneg) cells in each myeloid subpopulation isolated from PB or BM of Sham- or IMQ-treated mice is shown (Data are from two independent experiments (n = 6–7)). (F, G, K) *p<0.05, **p<0.01, ***p<0.001; Student’s t-test. (H, L, M) **p<0.01, ***p<0.001; two-way ANOVA with Bonferroni’s multiple comparison test.
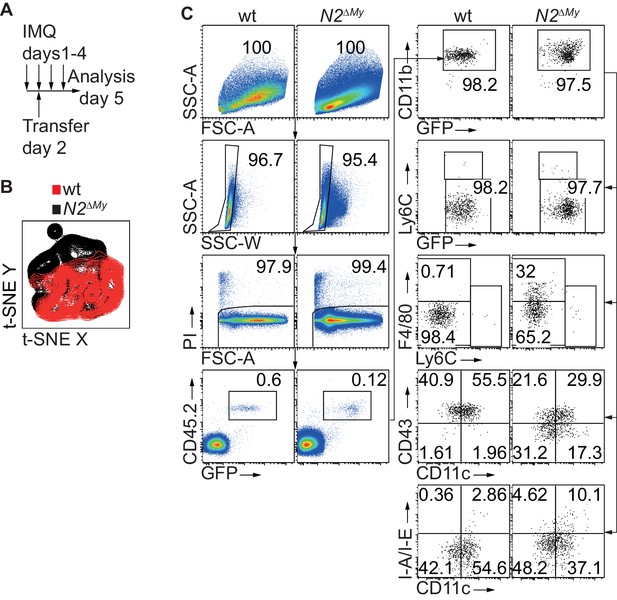
Characterization of F4/80hi monocytes in IMQ-treated mice.
(A) Experimental set-up for IMQ treatment, adoptive transfer and analysis of mice is depicted. (B) t-SNE analysis of donor CD45.2+CD11b+GFP+ cells extracted from Spl of IMQ-treated mice. Overlay of wt and N2∆My cells are depicted from concatenated samples. (C) Full gating path of adoptively transferred wt or N2ΔMy donor CD45.2+ monocytes recovered from IMQ-treated CD45.1+ recipients corresponding to Figure 5E.
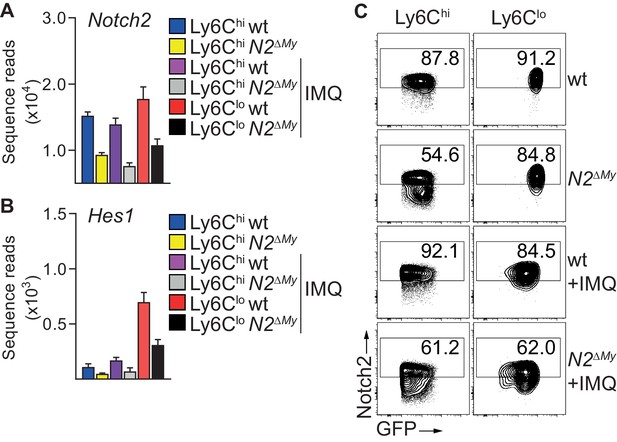
Expression of Notch2 and Notch-regulated gene in monocytes of IMQ-treated mice.
(A, B) Bar graph showing mean + SEM of Notch2 (A) or Hes1 (B) sequence reads from IMQ-treated wt and N2ΔMy PB monocyte subsets. (C) Representative flow cytometry plots showing expression of Notch2 in monocyte subsets from Sham-, or IMQ-treated wt or N2ΔMy mice.
Additional files
-
Supplementary file 1
Surface phenotype signatures for identification of distinct myeloid populations in vivo.
Lin: CD3, CD45R/B220, CD19, NK1.1, Ly6G, Ter119.
- https://cdn.elifesciences.org/articles/57007/elife-57007-supp1-v2.doc
-
Supplementary file 2
IPA of top five immunological diseases enriched in Ly6Clo cells from IMQ-treated N2ΔMy mice.
- https://cdn.elifesciences.org/articles/57007/elife-57007-supp2-v2.doc
-
Supplementary file 3
Top 20 gene sets involved in GO biological processes enriched in Ly6Clo cells from IMQ-treated N2ΔMy mice.
- https://cdn.elifesciences.org/articles/57007/elife-57007-supp3-v2.doc
-
Supplementary file 4
Parameters and the results of GSEA performed on 373 DEGs for Figure 5A.
- https://cdn.elifesciences.org/articles/57007/elife-57007-supp4-v2.xlsx
-
Supplementary file 5
List of the genes enriched in Lyve1hiMHC-IIlo MF gene set from Figure 5A.
- https://cdn.elifesciences.org/articles/57007/elife-57007-supp5-v2.xlsx
-
Supplementary file 6
List of antibodies and fluorescence dyes for flow cytometry used in the study.
- https://cdn.elifesciences.org/articles/57007/elife-57007-supp6-v2.doc
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57007/elife-57007-transrepform-v2.pdf





