Modeling transfer of vaginal microbiota from mother to infant in early life
Figures
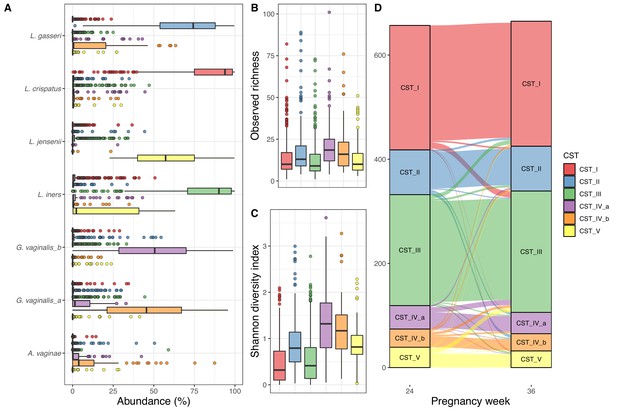
Vaginal community state type (CST).
(A) Boxplot of top amplicon sequence variant (ASV) abundance for each CST (including two most abundant ASVs from each CST), (B) boxplot of observed richness by CST, (C) boxplot of Shannon diversity index by CST, and (D) alluvial plot showing the CST at weeks 24–36 for each woman. All plots are colored by the CST.
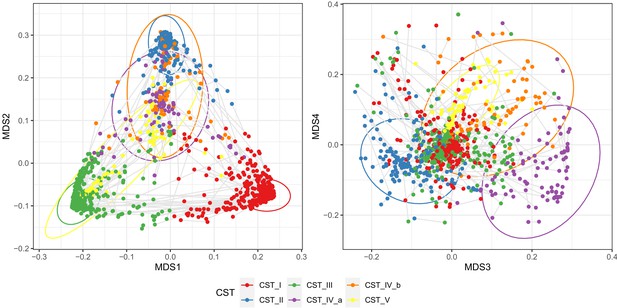
Non-metric multidimenionsal scaling plot based on Jensen–Shannon divergence; samples colored by community state type (CST), and gray lines connect samples from the individual women.
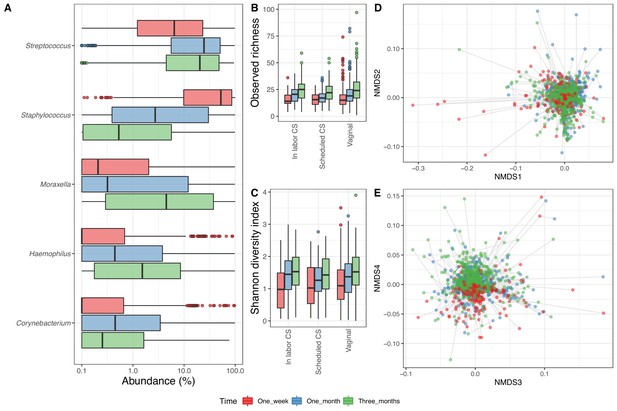
Airway microbiome.
(A) Boxplot of top genera with mean abundance of 5% at least one time point. Abundance is plotted on a log10 scale, and all abundances below 0.1% have been set to 0.1% in the plot. (B–C) Boxplot of alpha diversity, (B) observed richness, and (C) Shannon diversity index, by delivery mode. Three samples with observed richness above 100 were excluded from the plot. (D–E) Non-metric multidimensional scaling plots based on weighted UniFrac distances. Samples from the same individual are connected by gray lines. All plots are colored by time point.
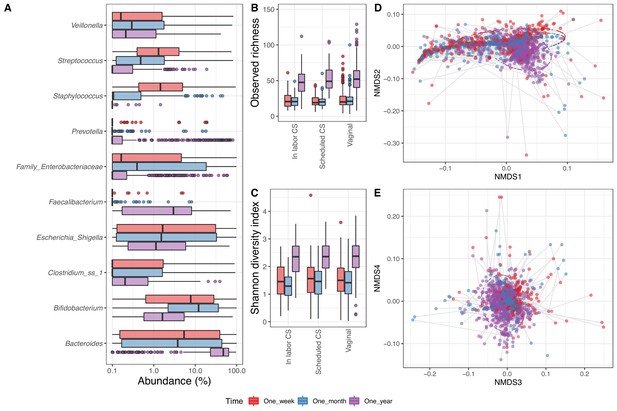
Fecal microbiome.
(A) Boxplot of top genera with mean abundance of 4% at least one time point. Abundance is plotted on a log10 scale, and all abundances below 0.1% have been set to 0.1% in the plot. (B–C) Boxplot of alpha diversity, (B) observed richness, and (C) Shannon diversity index, by delivery mode. One sample with observed richness of >150 was excluded from the plot. (D–E) Non-metric multi-dimensional scaling plot based on weighted UniFrac distances. Samples from the same individual are connected by gray lines. All plots are colored by time point.
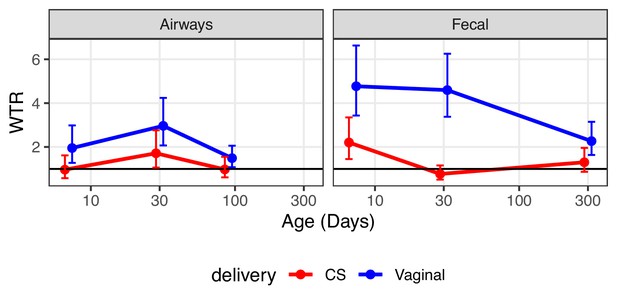
Weighted transfer ratios from vaginal week 36 to the fecal and airway compartments in first year of life stratified on mode of delivery (blue: vaginal birth, red: cesarean section).
A ratio above one indicates enrichment of microbial transfer. Error bars reflect standard errors.
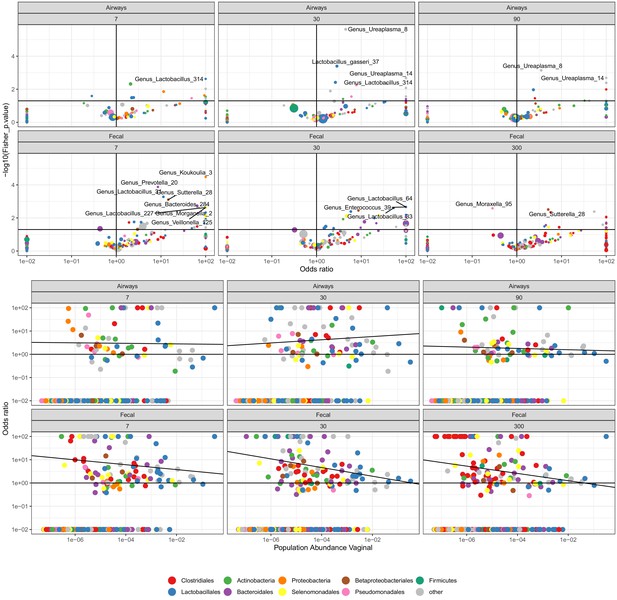
The odds for transfer between mother (week 36) and child for children delivered vaginally.
Top panel shows the odds ratio (OR) (x-axis) and the strength (p-value). Lower panel shows OR (y-axis) versus the population-wide vaginal abundance (x-axis).

The odds for transfer between mother (week 36) and child for children delivered by cesarean section (CS).
Top panel shows the OR (x-axis) and the strength (p-value). Lower panel shows OR (y-axis) versus the population-wide vaginal abundance (x-axis).

The odds for transfer between mother (week 36) and child for children delivered by in labor CS.
Top panel shows the OR (x-axis) and the strength (p-value). Lower panel shows OR (y-axis) versus the population-wide vaginal abundance (x-axis).
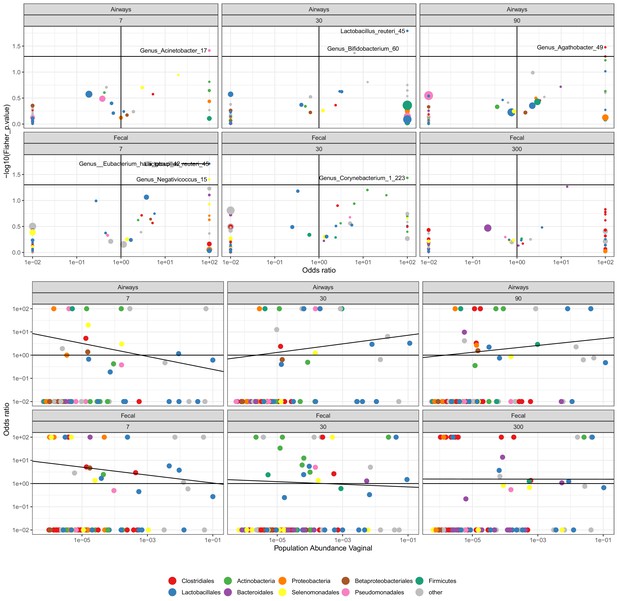
The odds for transfer between mother (week 36) and child for children delivered by scheduled CS.
Top panel shows the OR (x-axis) and the strength (p-value). Lower panel shows OR (y-axis) versus the population-wide vaginal abundance (x-axis).
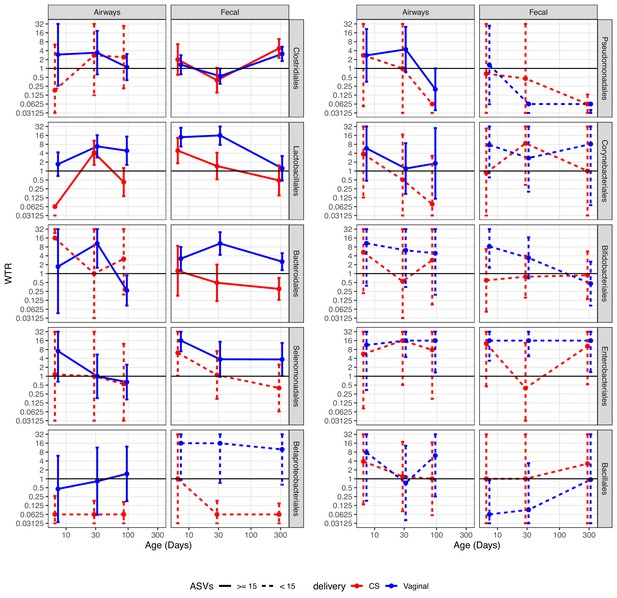
Weighted transfer ratios (WTRs) from vaginal week 36 to the fecal and airway compartments in the first year of life according to the mode of delivery (blue: vaginal birth, red: cesarean section) partitioned for the 10 most represented taxonomic classes at order level with upper left (Clostridiales) being the most represented order followed byLactobacillales and so forth.
Dashed lines represent analysis on less than 15 ASVs on average. Error bars reflect standard errors. WTR is truncated so values lower than 0.625 are plotted as 0.625 and values higher than 16 are plotted as 16.
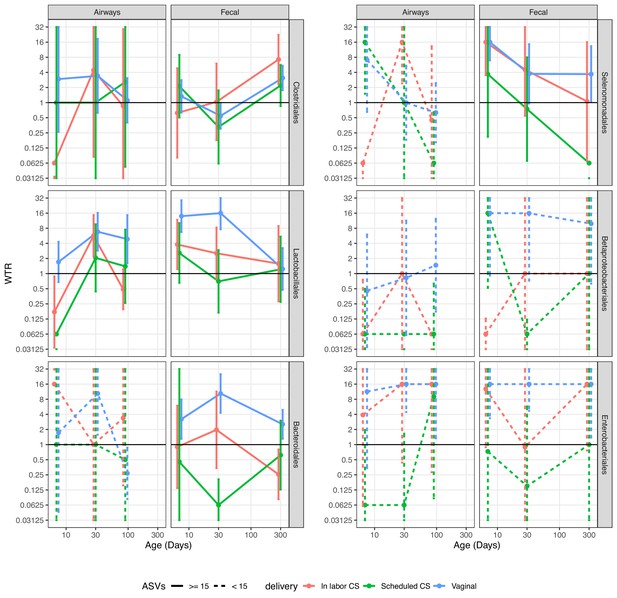
Weighted transfer ratios (WTRs) from vaginal week 36 to the fecal and airway compartments in the first year of life according to the mode of delivery (blue: vaginal birth, red: in labor CS, green: scheduled CS) partitioned for the 10 most represented taxonomic classes at order level with upper left (Clostridiales) being the most represented family followed by Lactobacillales and so forth.
Dashed lines represent analysis on less than 15 ASVs on average. Error bars reflect standard errors. WTR is truncated so values lower than 0.625 are plotted as 0.625 and values higher than 16 are plotted as 16.
Tables
Descriptives on testable Amplicon sequence variant (ASV) in terms of numbers of ASVs, vaginal, fecal, and airway total coverage, number of tests reaching nominal, and false discovery rate-corrected significance.
| Compartment | Delivery mode | Age (days) | Testable ASVs (n) | Vaginal relative abundance (%) | Child relative abundance (%) | Min (p)* (n) | Min (q)† (n) | p<0.01 (n) | p<0.05 (n) | q<0.05 (n) | q<0.10 (n) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Airways | CS | 7 | 104 | 31.0 | 36.5 | 0.058 | 0.990 | 0 | 0 | 0 | 0 |
| Fecal | CS | 7 | 160 | 31.6 | 73.3 | 0 | 0.008 | 3 | 5 | 1 | 1 |
| Airways | CS | 30 | 131 | 56.4 | 85.0 | 0.008 | 0.347 | 3 | 6 | 0 | 0 |
| Fecal | CS | 30 | 181 | 60.5 | 83.2 | 0.008 | 0.991 | 1 | 4 | 0 | 0 |
| Airways | CS | 90 | 152 | 56.8 | 84.4 | 0.033 | 0.992 | 0 | 3 | 0 | 0 |
| Fecal | CS | 300 | 161 | 61.7 | 59.3 | 0.018 | 0.991 | 0 | 2 | 0 | 0 |
| Airways | V | 7 | 293 | 64.0 | 46.0 | 0.002 | 0.691 | 3 | 13 | 0 | 0 |
| Fecal | V | 7 | 354 | 65.2 | 90.7 | 0 | 0.012 | 12 | 28 | 2 | 4 |
| Airways | V | 30 | 342 | 63.9 | 90.2 | 0 | 0.001 | 8 | 14 | 1 | 2 |
| Fecal | V | 30 | 395 | 65.8 | 92.4 | 0.002 | 0.312 | 11 | 28 | 0 | 0 |
| Airways | V | 90 | 364 | 62.2 | 87.7 | 0.001 | 0.260 | 3 | 9 | 0 | 0 |
| Fecal | V | 300 | 404 | 64.2 | 84.5 | 0.003 | 0.457 | 7 | 17 | 0 | 0 |
-
*Uncorrected p-values.
†False discovery rate corrected using the Benjamini-Hochberg procedure.
Additional files
-
Source code 1
Rmarkdown file (FullAnalysis.Rmd) with the entire analysis presented here as well as the scripts necessary to recreate the entire analysis (getTransferStats.R, transferFunctions.R, getWinnerStats.R, and inferenceTransferStat.R).
- https://cdn.elifesciences.org/articles/57051/elife-57051-code1-v1.zip
-
Source data 1
All tables produced in the analysis for this study.
- https://cdn.elifesciences.org/articles/57051/elife-57051-data1-v1.xlsx
-
Source data 2
Results of the BLAST analyses performed in this study.
- https://cdn.elifesciences.org/articles/57051/elife-57051-data2-v1.xlsx
-
Supplementary file 1
All results, including figures and tables, created as part of the analysis for this study.
- https://cdn.elifesciences.org/articles/57051/elife-57051-supp1-v1.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57051/elife-57051-transrepform-v1.docx





