Deep evolutionary origin of gamete-directed zygote activation by KNOX/BELL transcription factors in green plants
Figures
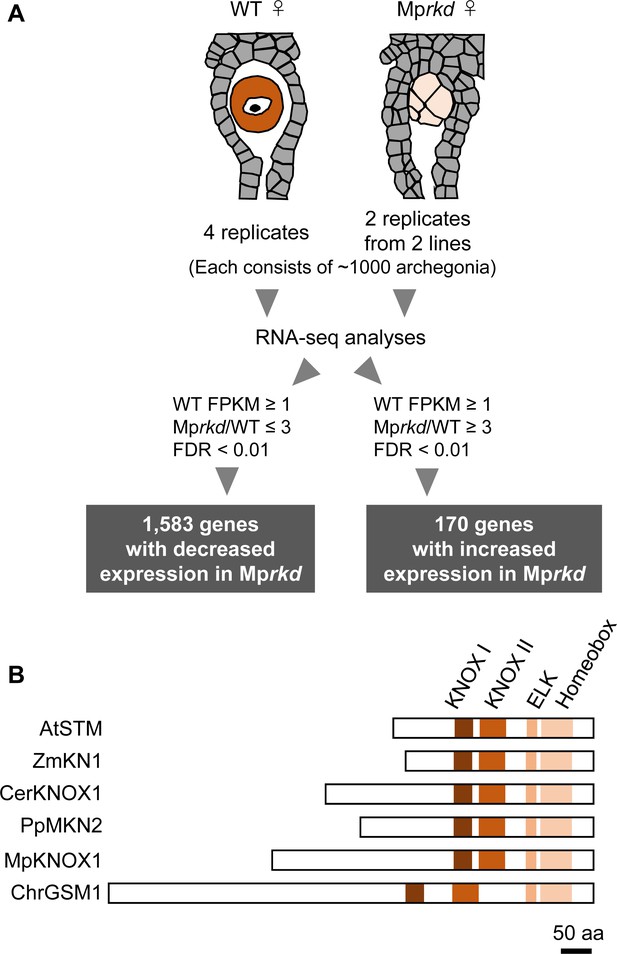
Comparative transcriptome analysis of wild-type and Mprkd archegonia and identification of MpKNOX1 as an egg-specific gene.
(A) Schematic illustration of RNA-seq analysis comparing the archegonia transcriptomes from wild-type females and egg-deficient Mprkd mutant females. About 4000 and 2000 archegonia were collected from wild-type and each of the two Mprkd mutant lines, and randomly allocated into four and two replicates, respectively. (B) Comparison of the domain arrangements of MpKNOX1 vs. representative class I KNOX proteins from Arabidopsis thaliana (AtSTM; GenBank accession number AEE33958.1), Zea mays (ZmKN1; AAP21616.1), Ceratopteris richardii (CerKNOX1, BAB18582.1), Physcomitrium patens (PpMKN2; AAK61308.2), and Chlamydomonas reinhardtii (ChrGSM1; ABJ15867.1).
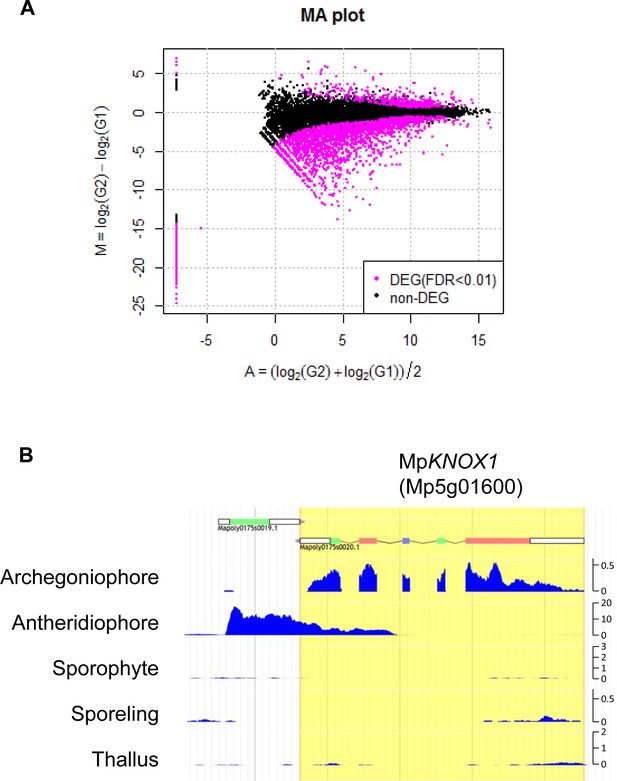
Comparative transcriptome analysis of wild-type and Mprkd archegonia and identification of MpKNOX1 as an egg-specific gene in M. polymorpha.
(A) M-A plot of differential expression analysis between wild-type and Mprkd archegonia. log2 differences between wild-type and Mprkd (Y-axis) are plotted against log2 average between wild-type and Mprkd (X-axis). Differentially expressed genes (DEGs) are plotted in magenta. (B) Captured Genome Browser image showing the expression levels along the MpKNOX1 locus in the indicated tissue types (archegoniophore: SRX301555; antheridiophore: SRX301553; sporophyte: SRX301556; sporeling: SRX301559; thallus: SRX301557). MpKNOX1 is specifically expressed in archegoniophores. The unit for Y axis is a read count normalized against 1× genome coverage.
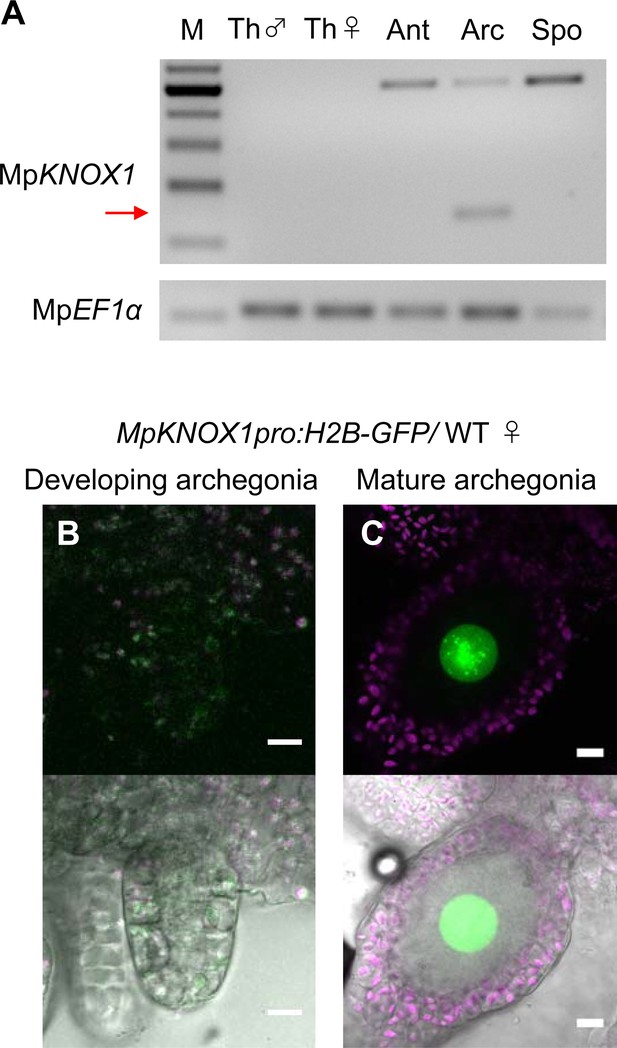
MpKNOX1 is specifically expressed in egg cells.
(A) RT-PCR analysis of MpKNOX1. Lanes are labeled as follows: M: size markers; Th ♂: male thalli; Th ♀: female thalli; Ant: antheridiophores; Arc: archegoniophores; Spo: sporophytes of 3-week-old plants. Constitutively expressed MpEF1α was used as a control. Red arrow indicates the expected size of PCR products from spliced MpKNOX1 mRNA. Bands at the top of the gel likely correspond to unspliced MpKNOX1 transcripts. Shown is a representative result from the experiments using three independently collected plant samples each with two technical replicates (two PCRs from each cDNA pool). See Figure 4—figure supplement 1A for the primer position. (B, C) Expression of the MpKNOX1 transcriptional reporter. Magenta: chlorophyll autofluorescence; green: GFP fluorescence. Lower panels are merged photographs of fluorescence and bright-field images. Bars, 10 μm.
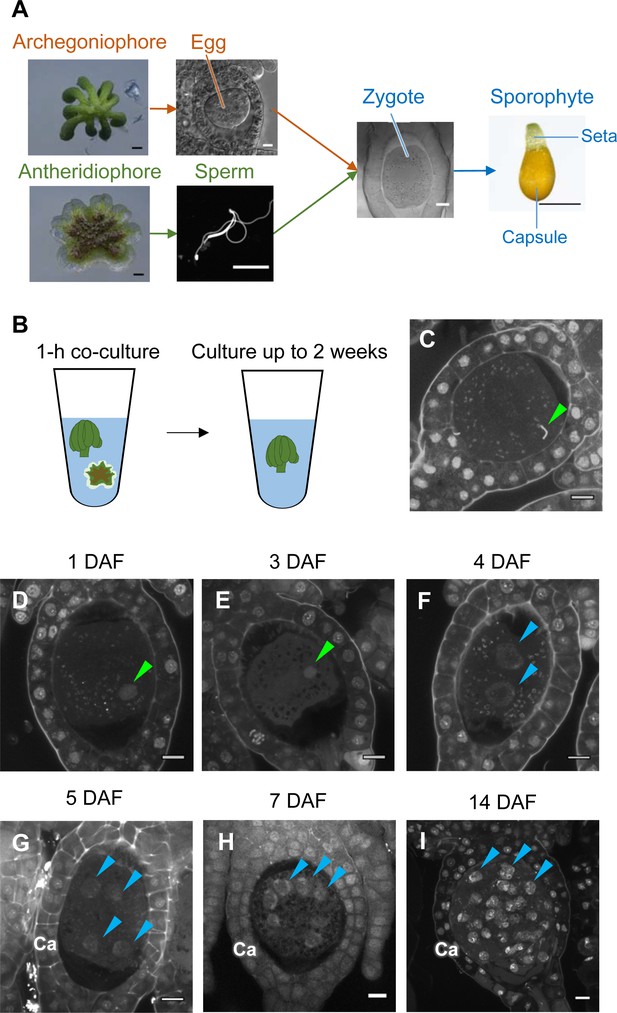
Time-course observation of subcellular dynamics during M. polymorpha fertilization.
(A) Schematic representation of sexual reproduction in M. polymorpha. Female plants develop umbrella-shaped sexual branches termed archegoniophores that form egg-containing archegonia. Male plants develop disc-shaped sexual branches termed antheridiophores that form antheridia, which produce numerous motile sperm cells. Upon soaking antheridiophores in water, the sperm cells are released from the antheridia and swim to egg cells in the archegonia. After fertilization, each zygote undergoes embryogenesis by dividing and differentiating into a sporophyte body consisting of a capsule containing haploid spores and a short supportive stalk called the seta. Black bars, 1 mm. White bars, 10 µm. (B) Illustration of the in vitro fertilization method used in this study. Excised archegoniophores and antheridiophores were co-cultured in water for 1 hr to allow fertilization to take place. The archegoniophores were transferred to a fresh tube containing water for further culturing. The tube lids were left open to allow gas exchange to occur. Archegoniophores containing sporophytes were cultured for up to 2 weeks. (C) A DAPI-stained zygote after 1 hr of co-culture. Most zygotes contained sperm nuclei at this time (green arrowhead). Bars, 10 µm. (D–I) DAPI-stained zygotes and sporophytes at the indicated days after fertilization (DAF). Male pronuclei (green arrowheads) were visible at 1–3 DAF (D, E) in wild-type fertilized eggs. In most zygotes, karyogamy was completed, and cells were cleaved at 4 DAF (F). Sporophyte cells continued to divide at 5–14 DAF (G–I), as visualized by the presence of multiple nuclei (blue arrowheads; not all nuclei are labeled in H and I). Ca: calyptra. Bars, 10 µm.
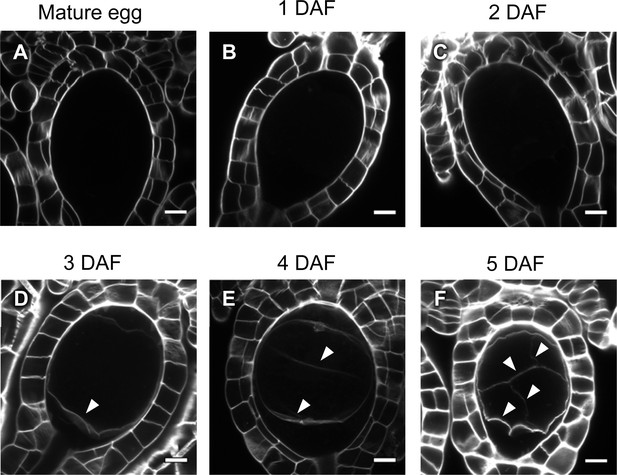
Cell wall regeneration during zygote development.
(A–F) Confocal images of archegonia containing an egg cell, zygotes, or embryos. Cell walls stained with SCRI Renaissance 2200 were detected only after embryogenesis (arrowheads), but not in mature eggs or zygotes at 2 days after fertilization or earlier. Bars, 10 µm.
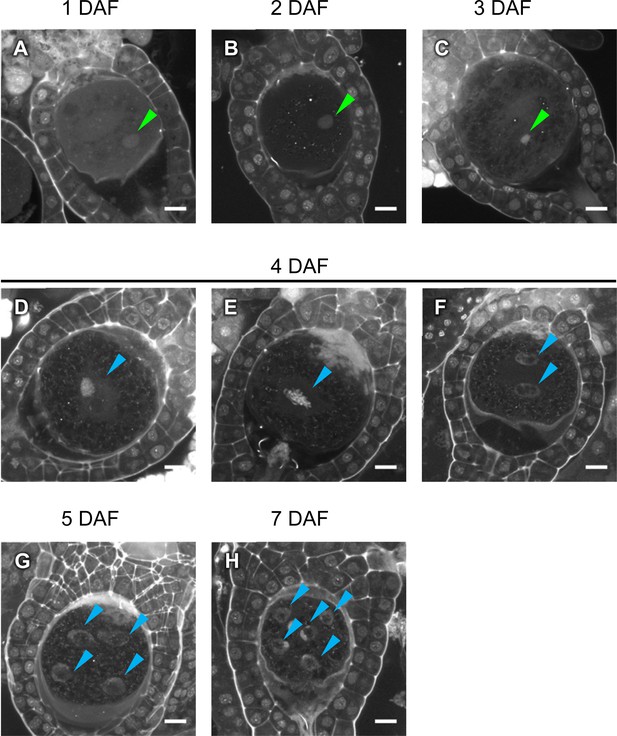
Cellular dynamics of zygotes and embryos generated by in planta crossing.
(A–H) DAPI-stained zygotes and embryos derived from in planta crosses at the indicated stages, showing that the timing of subcellular dynamics was equivalent to that generated by the in vitro fertilization method shown in Figure 3. At 4 days after fertilization, most zygotes completed karyogamy (D), whereas some showed an M-phase image (E) or completed the first division (F). Green arrowheads: male pronuclei; blue arrowheads: sporophytic nuclei. Bars, 10 µm.
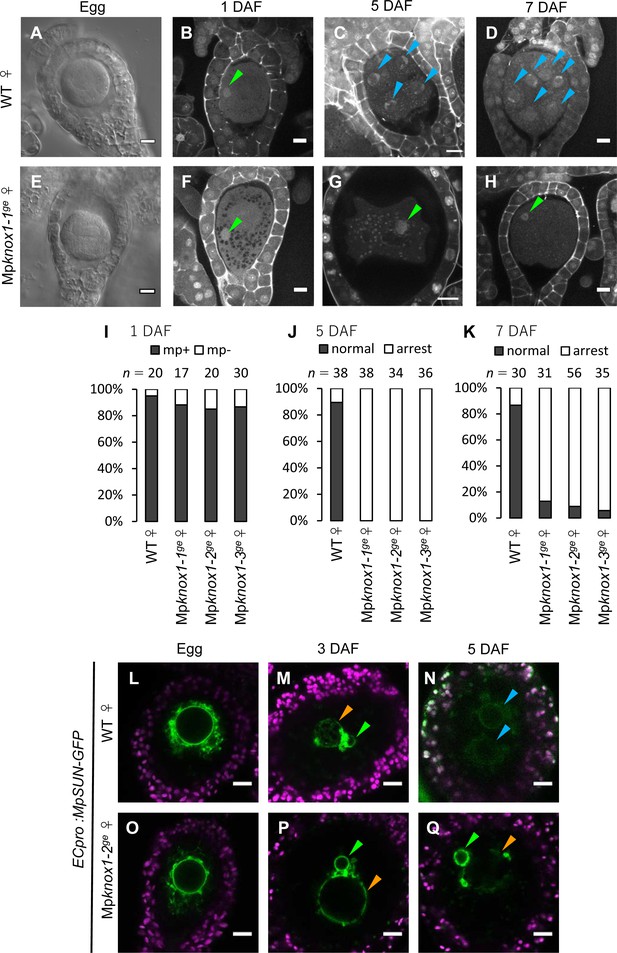
Maternally inherited MpKNOX1 is required for nuclear fusion.
(A, E) Bright-field images of wild-type (WT; A) and Mpknox1-1ge (E) archegonia. (B–D and F–H). 1 day after fertilization (DAF) (B, F), 5 DAF (C, G), and 7 DAF (D, H) zygotes from a cross between WT female and male plants (B–D), and a cross between Mpknox1-1ge female and WT male plants (F–H), indicating that maternal MpKNOX1 is dispensable for fertilization (B, F) but is required for embryogenesis (C, D, G, H). (I–K) Bar graphs showing the ratios of zygotes containing male pronuclei (mp+) vs. those not containing male pronuclei (mp-) at 1 DAF (I), and developed vs. arrested zygotes at 5 DAF (J) and 7 DAF (K) derived from the crosses of either WT or Mpknox1 females with WT males. Number of observed zygotes is shown above each bar. (L–Q) Egg cells of MpSUN-GFP marker lines in the WT (L) or Mpknox1-2ge (O) female background were crossed with WT males. At 3 DAF, male and female pronuclei were in contact with each other in both WT (M) and Mpknox1 (P) eggs. At 5 DAF, zygotes derived from a WT egg started to divide (N), while those from an Mpknox1 female (Q) were arrested without nuclear membrane fusion. Green arrowhead: male pronucleus; orange arrowhead: female pronucleus; blue arrowhead: embryo nucleus. Bars, 10 µm.
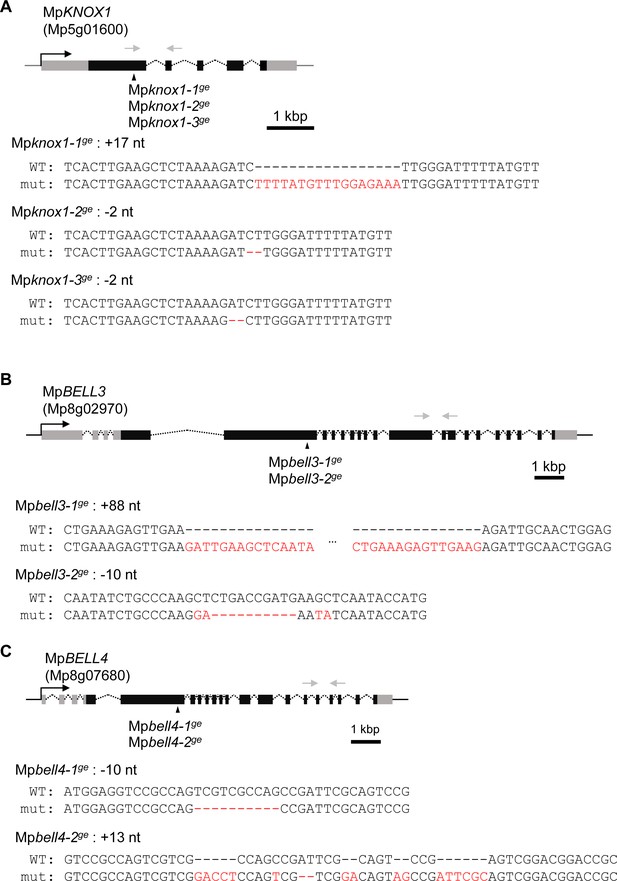
Generation of loss-of-function mutant lines by CRISPR/Cas9.
Gene organization and the locations of CRISPR/Cas9-introduced mutations in MpKNOX1 (A), MpBELL3 (B), and MpBELL4 (C) are indicated by the following symbols: gray line: 5′- and 3′-flanking sequences; gray box: 5′- and 3′-UTR; black box: coding regions; arrowhead: mutation positions; black arrow: transcriptional direction; dotted line: splice pattern; gray arrow: primers used in RT-PCR shown in Figures 2A and 5A. Sequence alignments of wild-type and mutant alleles are shown below each gene model. Mismatched nucleotides and gaps are shown in red.
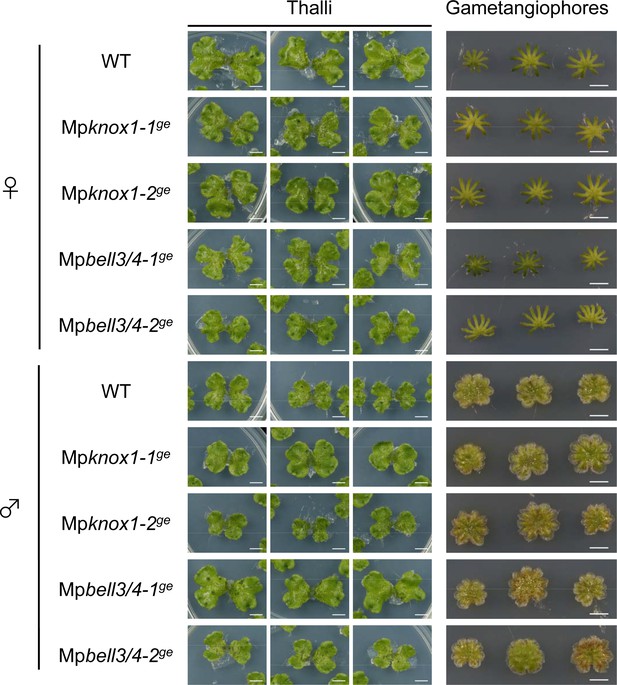
MpKNOX1 and MpBELL3/4 are dispensable for gametophyte development.
Three samples of vegetative thalli and gametangiophores from wild-type (WT), Mpknox1, and Mpbell3/4 mutant lines are shown. Bars, 5 mm.
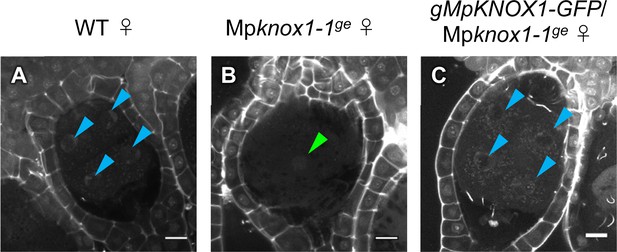
Expression of MpKNOX1-GFP by the MpKNOX1 promoter complements the karyogamy defects of Mpknox1 mutants.
(A–C) 5 days after fertilization (DAF) zygotes obtained by crossing the indicated female lines with wild-type males. Green and blue arrowheads indicate a male pronucleus and embryo nuclei, respectively. Bars, 10 µm.
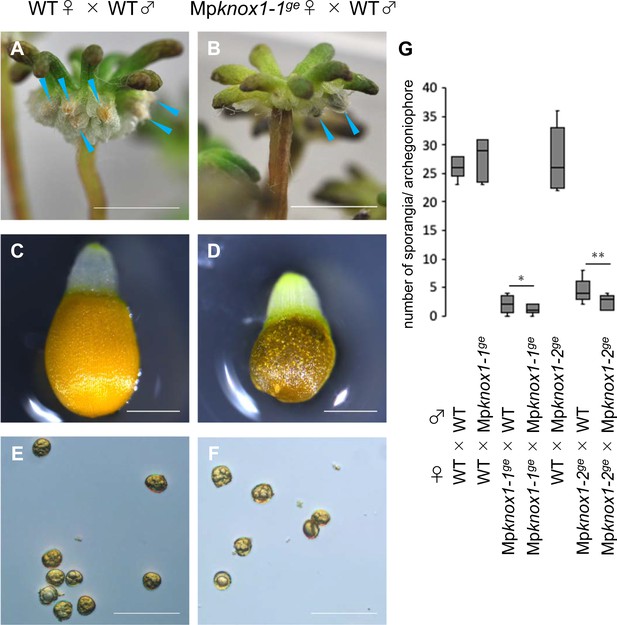
Sporangium and spore formation in wild-type and Mpknox1 plants.
(A, B) Archegoniophores of wild-type (A) and Mpknox1-1ge (B) female plants 4 weeks after crossing with wild-type males. Blue arrowheads indicate sporangia harboring expanded capsules. (C–F) Capsule (C, D) and spores (E, F) in wild-type (C, E) and Mpknox1-1ge (D, F) female plants crossed with wild-type males. (G) A box plot showing the number of sporangia per archegoniophore from indicated crosses. Five gametangiophores were analyzed for each genotype. *p=0.355* and **0.128, Student’s t-test. Bars, 5 mm (A, B), 0.5 mm (C, D), and 50 µm (E, F).
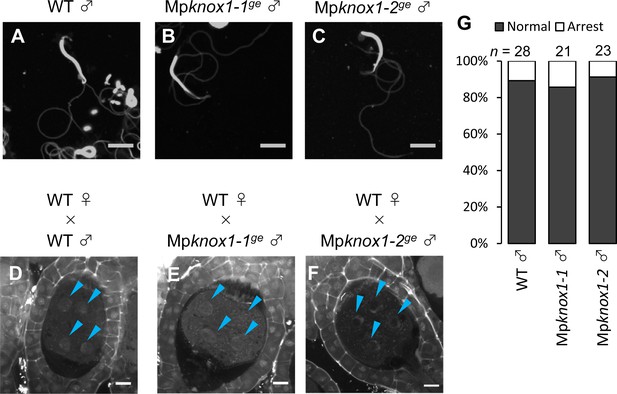
MpKNOX1 is dispensable for sperm differentiation and embryogenesis.
(A–C) DAPI-stained sperm cells from wild-type (WT; A) and Mpknox1 male plants (B, C) show indistinguishable morphology. (D–F) 5 days after fertilization (DAF) sporophytes in WT female plants crossed with WT (D) or Mpknox1males of distinct alleles (E, F). Blue arrowheads indicate nuclei of embryo cells. (G) Proportion of developed and arrested zygotes in WT female plants crossed with WT or Mpknox1 males of distinct alleles. Numbers of observed zygotes are indicated above each bar. Bars, 5 µm (A–C) and 10 µm (D–F).
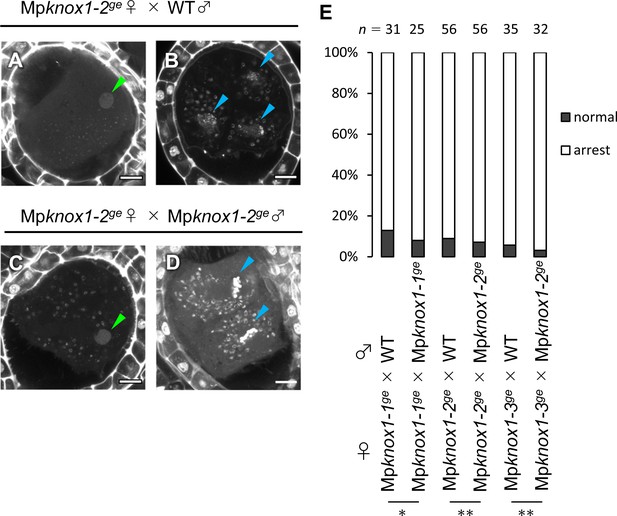
Paternally inherited Mpknox1 does not enhance the zygote arrest phenotype caused by maternally inherited Mpknox1.
(A–D) 7 days after fertilization (DAF) sporophytes in Mpknox1-2ge female plants crossed with wild-type (A, B) or Mpknox1-2ge male plants (C, D). Shown are representative images of zygotes arrested at karyogamy (A, C), developing sporophytes (B), and developing sporophytes under mitosis (D). Green and blue arrowheads indicate a male pronucleus and embryo nuclei, respectively. Bars, 10 µm. (E) A bar graph showing the ratios of developed vs. arrested zygotes derived from the crosses of Mpknox1 eggs with either wild-type or Mpknox1 sperm. Numbers of observed zygotes are indicated above each bar. *p=0.681 and **1, Fisher’s exact test.
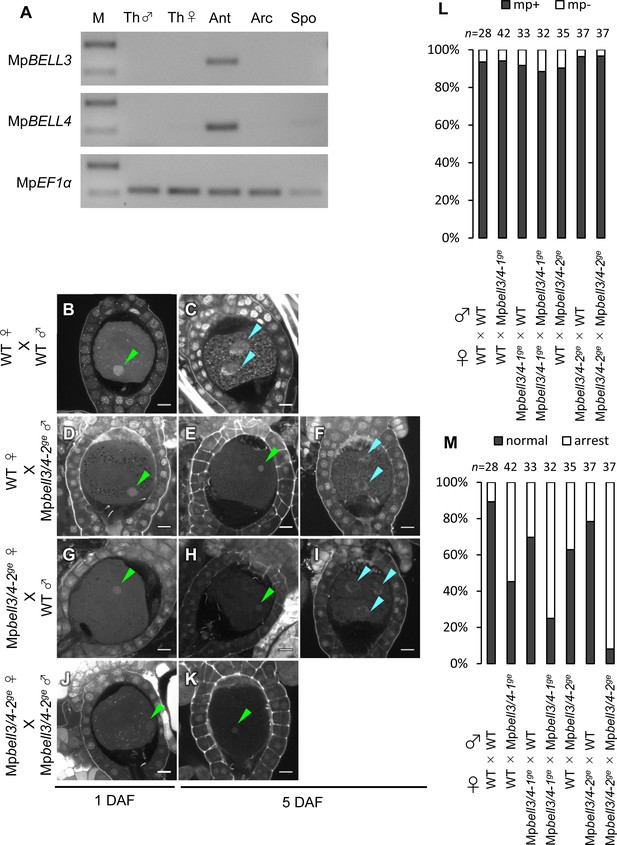
Both paternally and maternally inherited MpBELL genes are required for karyogamy.
(A) RT-PCR analysis indicating that MpBELL3 and MpBELL4 are specifically expressed in antheridiophores. The lanes are labeled as in Figure 2A. Shown is a representative result from the experiments using three independently collected samples each with two technical replicates. See Figure 4—figure supplement 1A for the primer position. (B–K) Zygotes at 1 day after fertilization (DAF; B, D, G, J) and 5 DAF (C, E, F, H, I, K) from crosses between a wild-type female and wild-type male (B, C) or Mpbell3/4-2ge male (D–F) and a Mpbell3/4-2ge female and a wild-type male (G–I) or Mpbell3/4-2ge male (J, K). The presence of male pronuclei (green arrowheads) in zygotes of all genotypes at 1 DAF (B, D, G, J) indicates that MpBELL3 and MpBELL4 are dispensable for plasmogamy. Note that zygotes produced from both or one Mpbell3/4 parent exhibit a variable degree of karyogamy arrest as visualized by the retention of male pronuclei (green arrowheads) among those starting embryonic division (nuclei labeled with blue arrowheads). Bars, 10 µm. (L) A bar graph showing the ratios of zygotes containing male pronuclei (mp+) vs. those not containing male pronuclei (mp-) from indicated crosses at 1 DAF. Numbers of observed zygotes are shown above each bar. (M) A bar graph showing the ratios of developed vs. arrested zygotes at 5 DAF from the indicated crosses. Numbers of observed zygotes are shown above each bar.
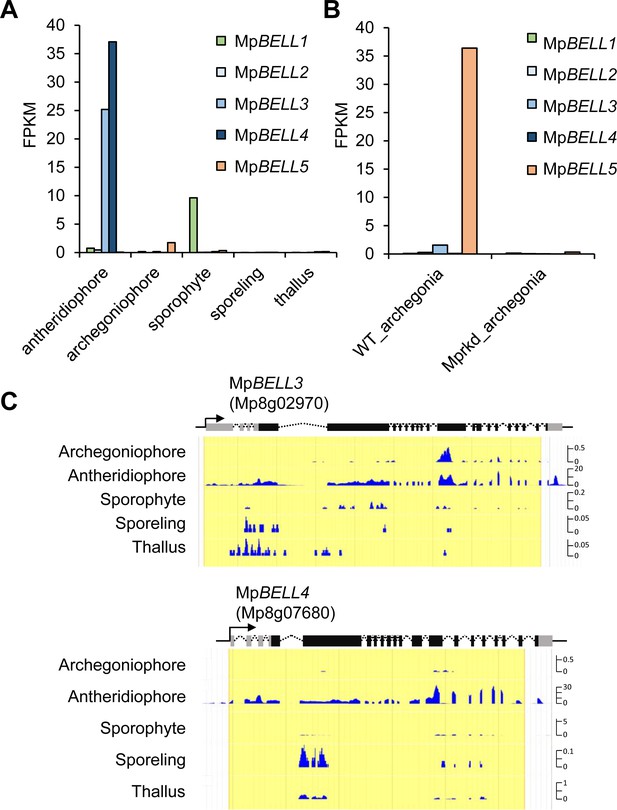
MpBELL3 and MpBELL4 are preferentially expressed in antheridiophores.
(A, B) Bar graphs showing the expression levels of MpBELL genes in the indicated organs, constructed from publicly available transcriptome data (Bowman et al., 2017) (A) and the RNA-seq data obtained in this study (B). (C) Snapshots of Genome Browser views of the MpBELL3 and MpBELL4 loci displaying their preferential expression in antheridiophores. The unit for Y axis is a read count normalized against 1× genome coverage.
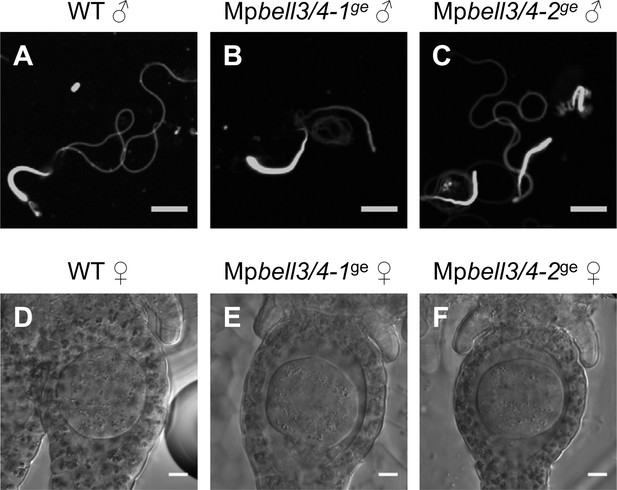
MpBELL3 and MpBELL4 are dispensable for gamete differentiation.
( A–C) DAPI-stained sperm cells from wild-type (A), Mpbell3/4-1ge (B), and Mpbell3/4-2ge (C) male plants. (D–F) Differential Interference contrast (DIC) images of archegonia in wild-type (D), Mpbell3/4-1ge (E), and Mpbell3/4-2ge (F) female plants. Bars, 5 µm (A–C) and 10 µm (D–F).
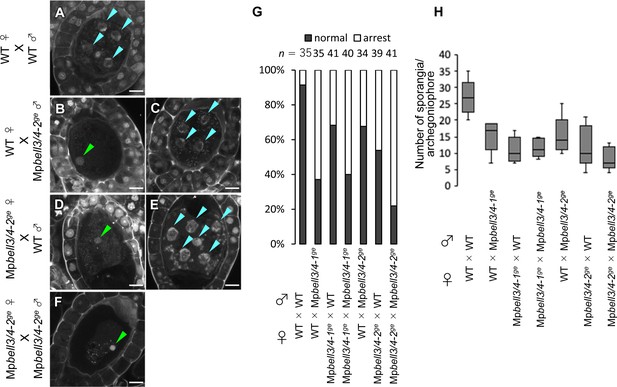
MpBELL3 and MpBELL4 are dispensable for gamete differentiation.
(A–F) 7 days after fertilization (DAF) sporophytes from the indicated crosses. Note that zygotes inheriting Mpbell3/4 from either or both parent(s) were arrested at karyogamy with variable ratios. Arrested zygotes were recognized with retention of male pronuclei (green arrowheads), whereas developing zygotes with divided nuclei (blue arrowheads). Bars, 10 µm. (G) A bar graph showing the ratios of developed vs. arrested zygotes at 7 DAF from the indicated crosses. Numbers of observed zygotes are shown above each bar. (H) A box plot showing the number of sporangia per archegoniophore from indicated crosses. Five gametangiophores were analyzed for each genotype.
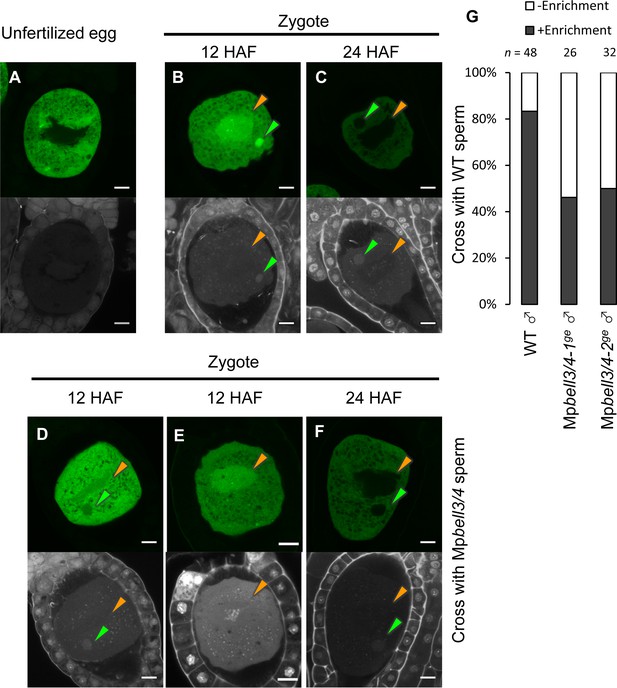
MpKNOX1 transiently localizes to female and male pronuclei prior to karyogamy.
(A–F) GFP (upper panels) and DAPI (lower panels) signals from gMpKNOX1-GFP/Mpknox1-1ge eggs (A) and zygotes obtained by crossing a gMpKNOX1-GFP/Mpknox1-1ge female with a wild-type (B, C) or Mpbell3/4-2ge (D–F) male. Note that before fertilization MpKNOX1-GFP was exclusively localized to the cytosol (A). At 12 HAF, MpKNOX1-GFP signals were enriched in female (orange arrowhead) and male (green arrowhead) pronuclei in the wild-type background (B). In the absence of paternally inherited MpBELL3 and MpBELL4, about a half of zygotes did not exhibit nuclear-enriched MpKNOX1-GFP signals at 12 HAF (D, E). At 24 HAF, weak and exclusively cytosolic GFP signals were detected in both genotypes (C, F). Bars, 10 µm. (G) A bar graph showing the ratios of zygotes with and without pronuclei-enriched MpKNOX1-GFP signals from the indicated crosses. Numbers of observed zygotes are shown above each bar.
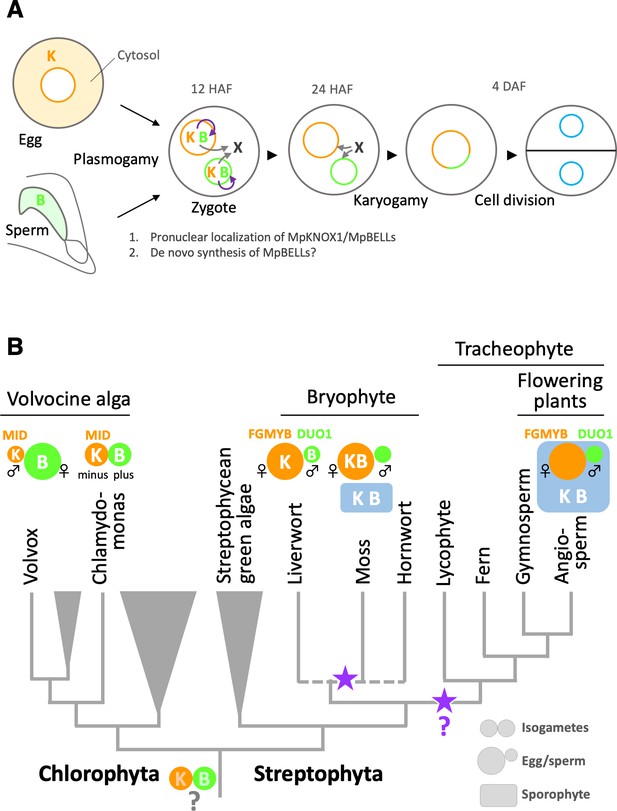
Functions and expression patterns of KNOX/BELL transcription factors in green plants.
(A) Expression patterns of KNOX and BELL proteins and their predicted role in zygote activation in M. polymorpha. ‘K’ and ‘B’ represent MpKNOX1 and MpBELL protein subunits, respectively. Orange, green, and blue circles represent female pronuclei, male pronuclei, and nuclei of embryo cells, respectively. Purple curved arrows indicate auto-amplification of BELL levels by KNOX/BELL-mediated transcriptional control. X indicates unknown karyogamy-promoting factor(s) whose expression and/or functions are activated by KNOX/BELL-mediated transcription. (B) Predicted evolutionary trajectory of KNOX/BELL expression patterns along the green plant lineages. Orange/green circles and blue rectangles represent gametes and sporophyte bodies, respectively. ‘K‘ and ‘B’ indicate the expression of KNOX and BELL proteins, respectively. Purple stars indicate the predicted positions at which the functional transition of KNOX/BELL from zygote activation to sporophyte morphogenesis occurred. Also indicated are the expression patterns of evolutionarily conserved regulators of sexual differentiation; a male-determinant factor MID of volvocine algae, and female- and male-differentiation factors, FGMYB and DUO1, respectively, of land plants. Note that the KNOX/BELL expression patterns in ancestral plants at the bottom of the tree are an inference.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Marchantia polymorpha) | MpKNOX1 | MalpolBase | Mp5g01600 | |
| Gene (Marchantia polymorpha) | MpBELL1 | MalpolBase | Mp8g18310 | |
| Gene (Marchantia polymorpha) | MpBELL2 | MalpolBase | Mp4g09650 | |
| Gene (Marchantia polymorpha) | MpBELL3 | MalpolBase | Mp8g02970 | |
| Gene (Marchantia polymorpha) | MpBELL4 | MalpolBase | Mp8g07680 | |
| Gene (Marchantia polymorpha) | MpBELL5 | MalpolBase | Mp5g11060 | |
| Gene (Marchantia polymorpha) | MpSUN | MalpolBase | Mp5g02400 | |
| Gene (Marchantia polymorpha) | MpECpro | MalpolBase | Mp5g18000 | |
| Strain, strain background (Marchantia polymorpha, male) | Tak-1 | DOI:10.1016/j.cell.2017.09.030 | Male wild-type strain of Marchantia | |
| Strain, strain background (Marchantia polymorpha, female) | Tak-2 | DOI:10.1016/j.cell.2017.09.030 | Female wild-type strain of Marchantia | |
| Recombinant DNA reagent | pMpGE_En03 | Addgene | RRID:Addgene_71535 | Entry plasmid for gRNA |
| Recombinant DNA reagent | pMpGE010 | Addgene | RRID:Addgene_71536 | Destination plasmid for CRISPR/Cas9 |
| Recombinant DNA reagent | pMpGE_En04 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pBC-GE12 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pBC-GE23 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pBC-GE34 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pMpGE017 | DOI:10.15252/embj.2018100240 | Destination plasmid for Cas9 nickase | |
| Recombinant DNA reagent | pMpGE010_MpKNOX1ge | This paper | Plasmid to create Mpknox1 mutants | |
| Recombinant DNA reagent | pMpGE017_MpBELL4ge-MpBELL3ge | This paper | Plasmid to create Mpbell3/4 mutants | |
| Recombinant DNA reagent | pMpSL30 | DOI:10.15252/embj.2018100240 | Destination plasmid | |
| Recombinant DNA reagent | pMpSL30_MpKNOX1pro-H2B-GFP-3’MpKNOX1 | This paper | MpKNOX1 promoter reporter | |
| Recombinant DNA reagent | pMpSL30_gMpKNOX1-GFP | This paper | MpKNOX1 complementation | |
| Recombinant DNA reagent | pMpSL30_ ECpro-MpSUN-GFP | This paper | Egg cell-specific nuclear envelop marker | |
| Genetic reagent (Marchantia polymorpha) | Mpknox1-1ge | This paper | Mpknox1 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpknox1-2ge | This paper | Mpknox1 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpknox1-3ge | This paper | Mpknox1 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpbell3/4-1ge | This paper | Mpbell3/4 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpbell3/4-2ge | This paper | Mpbell3/4 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | MpKNOX1pro:H2B-GFP/WT ♀ | This paper | pMpSL30_MpKNOX1pro-H2B-GFP-3′MpKNOX1 was transformed into Tak-2 | |
| Genetic reagent (Marchantia polymorpha) | gMpKNOX1-GFP/Mpknox1-1ge♀ | This paper | pMpSL30_gMpKNOX1-GFP was transformed into Mpknox1-1ge | |
| Genetic reagent (Marchantia polymorpha) | ECpro:MpSUN-GFP | This paper | pMpSL30_ ECpro-MpSUN-GFP was transformed into Tak-2 or Mpknox1-2ge | |
| Genetic reagent (Marchantia polymorpha) | Mprkd-1 | DOI:10.1016/j.cub.2016.05.013 | ||
| Genetic reagent (Marchantia polymorpha) | Mprkd-3 | DOI:10.1016/j.cub.2016.05.013 | ||
| Commercial assay or kit | RNeasy Plant Mini Kit | Qiagen | 74904 | |
| Commercial assay or kit | TruSeq RNA Sample Prep Kit v2 | Illumina | RS-122-2001 | |
| Commercial assay or kit | Gateway LR Clonase II Enzyme mix | Thermo Fisher Scientific | 11791020 | |
| Software, algorithm | TopHat ver. 2.0.14 | doi:10.1093/bioinformatics/btp120 | RRID:SCR_013035 | |
| Software, algorithm | DESeq2 | doi:10.1186/s13059-014-0550-8 | RRID:SCR_015687 | |
| Software, algorithm | TCC | DOI:10.1186/1471-2105-14-219 | RRID:SCR_001779 |
Additional files
-
Supplementary file 1
List of genes with changed mRNA levels in Mprkd archegonia.
Genes with reduced and increased expression levels in Mprkd archegonia are listed in sheet 1 and sheet 2, respectively.
- https://cdn.elifesciences.org/articles/57090/elife-57090-supp1-v2.xlsx
-
Supplementary file 2
Primers used in this study.
Lowercase and uppercase letters indicate synthetic adaptor and target DNA sequences, respectively.
- https://cdn.elifesciences.org/articles/57090/elife-57090-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57090/elife-57090-transrepform1-v2.docx





