Repurposing of KLF5 activates a cell cycle signature during the progression from a precursor state to oesophageal adenocarcinoma
Figures
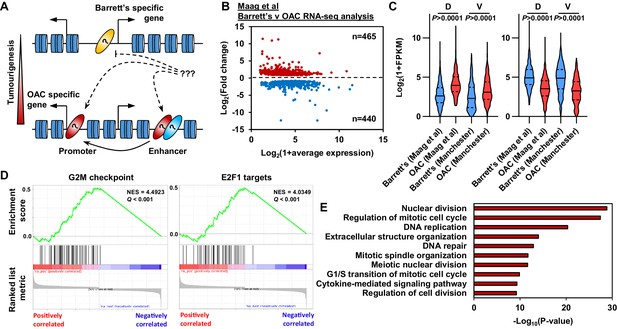
Oesophageal adenocarcinoma (OAC) tumourigenesis is associated with enhanced cell cycle gene activity.
(A) Schematic of possible transcription factor network induction during OAC development. Coloured ovals represent different transcription factors. (B) Scatter plot of significant differentially (±1.5 x, Q-value <0.05) expressed genes between human Barrett’s oesophagus n = 13 and human OAC n = 12 samples (Maag et al., 2017). (C) Violin plots of expression of differentially expressed genes between Barrett’s oesophagus (n = 13) and oesophageal adenocarcinoma (n = 12) from discovery dataset (D; Maag et al., 2017) and validation dataset (V; BO = 3; OAC n = 4). Genesets are shown for upregulated (left) and downregulated (right) in OAC. (D) Gene set enrichment analysis of differentially expressed genes. The top two upregulated gene sets are shown with normalised enrichment score (NES) and Q-value. (E) Biological pathway GO term analysis of upregulated genes. The top 10 terms are shown. See also Figure 1—figure supplement 1.
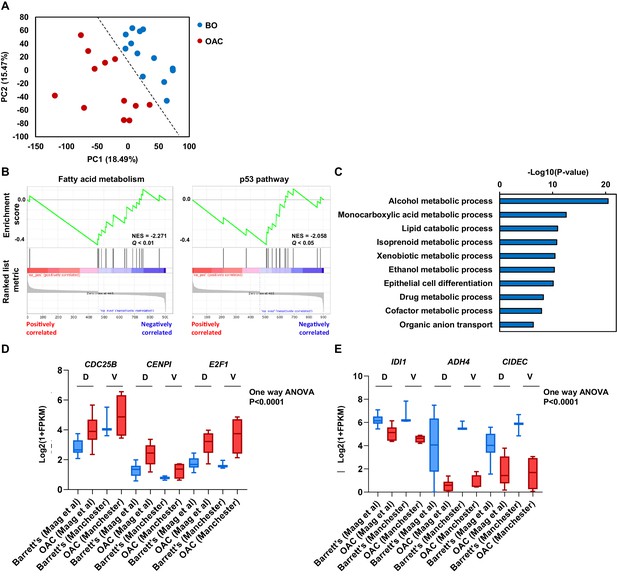
Differential gene expression analysis between Barrett’s and OAC patient samples.
(A) PCA plot of 13 Barrett’s oesophagus (blue) and 12 OAC (red) RNA-seq samples (Maag et al., 2017). (B) GSEA plots of significantly downregulated gene sets in OAC compared to Barrett’s oesophagus. Normalised enrichment score (NES) and Q-value are shown. (C) Biological pathway GO term of significantly downregulated genes in OAC compared to Barrett’s oesophagus. (D) Box plot of Log2(1+FPKM) expression from BO (blue) and OAC (red) samples in discovery (D; Maag et al., 2017) and validation (V; this study) datasets for upregulated example genes (D) and downregulated example genes (E). Whiskers represent minimum and maximum values and one-way ANOVA p-value is shown.
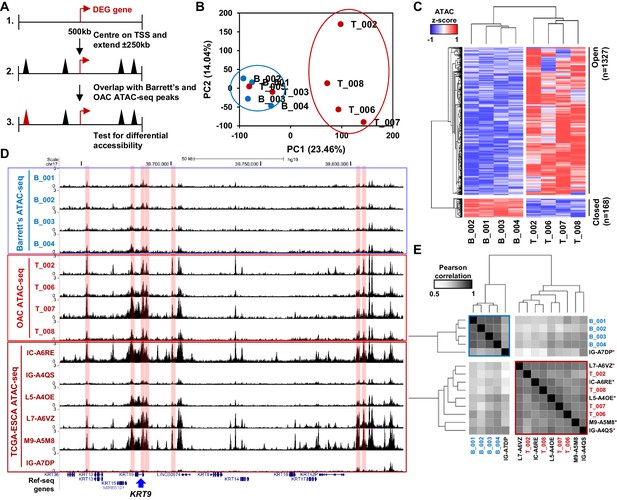
Altered chromatin accessibility landscape during OAC carcinogenesis.
(A) Schematic of ATAC-seq analysis. All peaks within ±250 kb of the TSS of a differentially expressed genes were assessed for differential accessibility between Barrett’s oesophagus and OAC. (B) Principal Component Analysis plot of log2(1+FPKM) ATAC-seq signal from all accessible regions within ±250 kb of a differentially expressed gene TSS from all human Barrett’s oesophagus (B; n = 4) and oesophageal adenocarcinoma samples (T; n = 6). (C) Heatmap of z-score ATAC-seq signal from human Barrett’s oesophagus (B; n = 4) and OAC (T; n = 4) samples at differentially accessible regions (±2 x; Q < 0.1). Hierarchical clustering of samples and regions performed using 1-Pearson correlation. (D) Example UCSC browser view of BO, OAC and TCGA ESCA ATAC-seq data surrounding the KRT19 locus with differentially accessible regions highlighted in red. (E) Correlation plot of Pearson correlation of log2(1+FPKM) ATAC-seq signal at differentially accessible regions. Hierarchical clustering performed using 1-Pearson correlation and the two main clusters are highlighted blue (BO) and red (OAC). TCGA samples are indicated by asterisks. See also Figure 2—figure supplement 1.
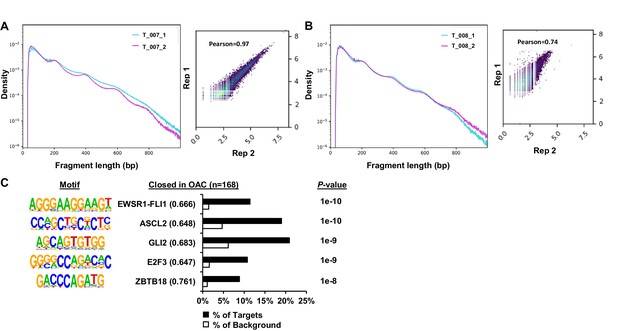
ATAC-seq analysis of patient OAC samples.
(A) Fragment length plot of log10 transformed ATAC-seq signal from technical replicates 1 and 2 of the T_007 OAC sample (left). Correlation plot of ATAC-seq signal from technical replicates 1 and 2 of the T_007 OAC sample at ATAC-seq peaks called from replicate 1 (right). Pearson correlation coefficient is shown. (B) Fragment length plot of log10 transformed ATAC-seq signal from technical replicates 1 and 2 of the T_008 OAC sample (left). Correlation plot of ATAC-seq signal from technical replicates 1 and 2 of the T_008 OAC sample at ATAC-seq peaks called from replicate 1 (right). Pearson correlation coefficient is shown. (C) Bar chart of percentage targets and percentage background of de novo discovered motifs at regions with significant decreased chromatin accessibility in OAC. De novo motifs, called transcription factor with motif match score (in brackets) and P-value are shown.
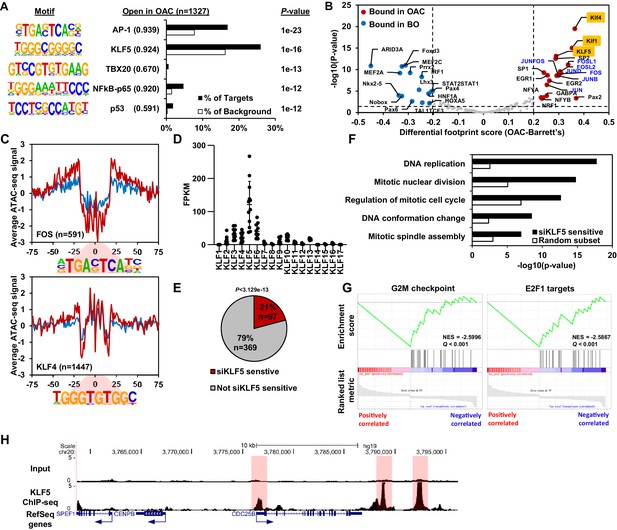
KLF5 control a cell cycle gene expression programme in OAC.
(A) Bar chart of percentage targets and percentage background of de novo discovered motifs at increased accessible regions in OAC compared to Barrett’s oesophagus. De novo motif, called transcription factor with motif match score (brackets) and P-value are shown. (B) Scatter plot of differential footprinting depth around human transcription factor motifs in differential accessible regions in BO and OAC tissue. Significant motifs with more footprint depth in BO are labelled blue and in OAC labelled red. KLF TF-binding motifs are highlighted in orange and AP1 motifs are in blue font. (C) BO (blue) and OAC (red) ATAC-seq signal at FOS (AP1) and KLF4 (KLF) motifs in differentially accessible regions. (D) Expression (FPKM) of KLF family transcription factors in OAC RNA samples (n = 12; Maag et al., 2017). (E) Pie chart of percentage of upregulated genes in patient OAC samples that are also downregulated with siKLF5 treatment. p-Value shown. (F) Biological pathway GO term analysis of OAC upregulated and siKLF5 downregulated genes and a random gene selection. (G) Gene set enrichment analysis of genes that are upregulated in OAC and downregulated by siKLF5 treatment. Top two downregulated gene sets are shown. Normalised enrichment scores (NES) and Q-values are shown. (H) Example UCSC Genome Browser view of KLF5 ChIP-seq binding at the CDC25B locus. KLF5 peaks highlighted in red. See also Figure 3—figure supplements 1 and 2.
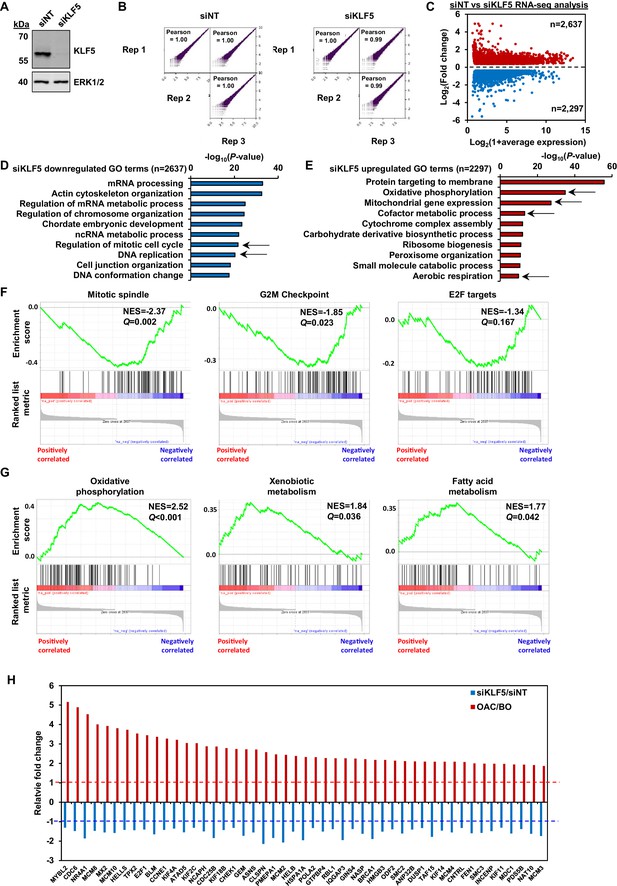
Identification of the KLF5-regulated cistrome in OE19 cells.
(A) Immunoblot of protein lysate from OE19 cells treated with either siNT or siKLF5 and probed with antibodies against KLF5 and ERK1/2. (B) Correlation plots of RNA-seq data from three biological replicates. Pearson correlations are shown. (C) Scatter plot of significantly differentially expressed genes (red are up- and blue are down-regulated genes;±1.3 x, Q-value <0.05) in OE19 cells with siKLF5 treatment. (D) Biological processes GO term analysis of downregulated genes with siKLF5 treatment. Cell cycle related GO terms are indicated with arrows. (E) Biological processes of GO term analysis of upregulated genes with siKLF5. Metabolism related GO terms are indicated with arrows. (F) Gene set enrichment analysis of downregulated genes with siKLF5 treatment. Cell cycle related gene sets, normalised enrichment scores (NES) and Q-values are shown. (G) Gene set enrichment analysis of upregulated genes with siKLF5 treatment. Metabolism-related gene sets, normalised enrichment scores (NES) and Q-values are shown. (H) Bar chart of fold change in expression of cell-cycle-related genes in siKLF5-treated OE19 cells (blue; n = 3) or patient OAC samples (red).
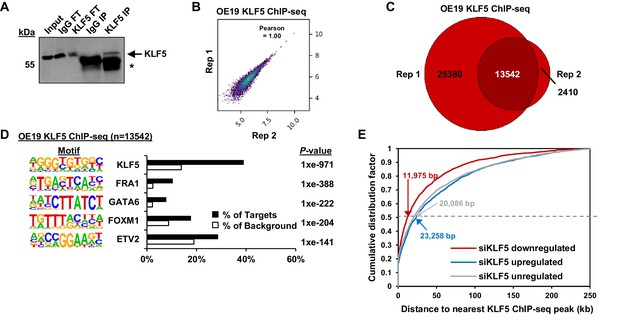
KLF5 ChIP-seq analysis in OE19 cells.
(A) Immunoblot of KLF5 from OE19 cells following ChIP in different fractions; IgG FT (flow through from non-specific IgG precipitation), KLF5 FT (flow through from KLF5 precipitation), IgG elute (material eluted following IgG precipitation) KLF5 IP (material eluted following KLF5 immunoprecipitation). Arrows indicating immunoprecipitated protein (KLF5) and asterisks are IgG heavy chain. (B) Correlation plot of KLF5 ChIP-seq data from OE19 cells from two biological replicates. Pearson correlation coefficient is shown. (C) Venn diagrams showing overlap of peaks called by each replicate (Rep) for KLF5 ChIP-seq from OE19 cells. The numbers of peaks in each sector are given. (D) Bar chart of percentage targets and percentage background of de novo discovered motifs in OE19 KLF5 ChIP-seq peaks. De novo motif, called transcription factor with motif match score (in brackets) and p-value are shown. Note that the KLF5 motif is shown in the reverse orientation to those in other figures. (E) Cumulative distribution of the closest KLF5 ChIP-seq peaks to the TSS of genes that were significantly down or upregulated following siKLF5 treatment compared to expressed genes (FPKM >1) that were not significantly deregulated by siKLF5 treatment. Median distances to the TSS are shown.
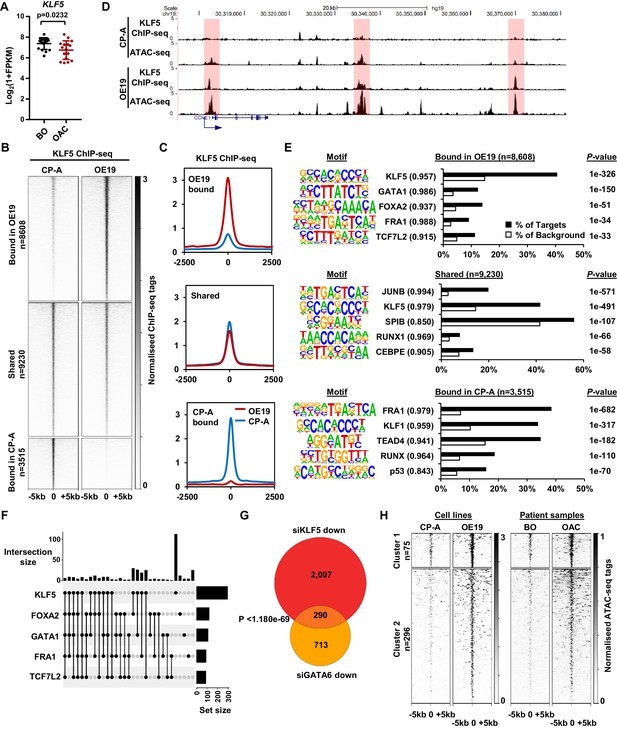
KLF5 binds to distinct regions in OE19 cells.
(A) Expression Log2(1+FPKM) of KLF5 in BO and OAC tissue. (B) Heatmap of KLF5 ChIP-seq signal at regions (peak centre ±5 kb) significantly bound in OE19 only (+2x; Q < 0.05), shared regions (no significant change) and regions bound in CP-A only (−2x; Q-value <0.05). (C) Tag density plot of KLF5 ChIP-seq signal at regions (peak centre ±2.5 kb) bound in OE19 only, shared regions and regions bound in CP-A only. (D) Genome browser tracks showing KLF5 ChIP-seq and ATAC-seq in CP-A and OE19 cells at the CCNE1 locus. Differential bound regions are highlighted in red. (E) Bar chart of percentage targets and percentage background of de novo discovered motifs at regions bound in OE19 only, shared regions and regions bound in CP-A only. De novo motifs, called transcription factor with match scores and P-values shown. (F) UPSET plot of DNA motifs found in 371 KLF5 binding regions that are specific to OE19-specific binding regions that are located within loci (+/- 250 kb) containing genes upregulated in OAC and downregulated with KLF5 depletion. The motifs identified in E (KLF5, GATA1, FOXA2, FRA1 and TCF7L2) found within each peak are shown. (G) Venn diagram showing the overlap in genes downregulated in OE19 cells following treatment with siRNAs targeting KLF5 and GATA6. (H) Heatmap of ATAC-seq signal at the KLF5 binding regions from (F) in the indicated cell lines (left) or patient derived tissue (right). Regions were subject to k-means hierarchical clustering (k = 2). See also Figure 4—figure supplements 1 and 2.
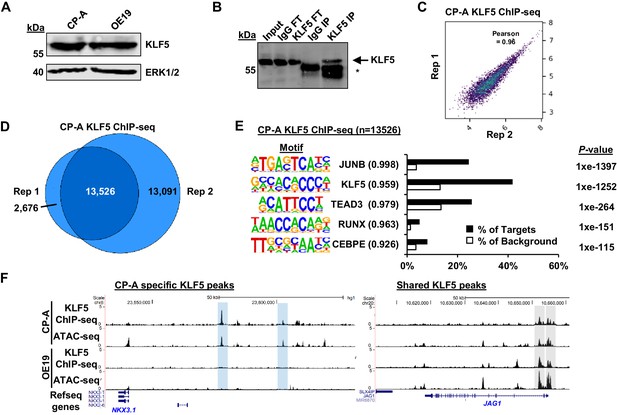
KLF5 ChIP-seq analysis in CP-A cells.
(A) Immunoblot of CP-A and OE19 protein lysate probed with antibodies against KLF5 and ERK1/2. (B) Immunoblot of KLF5 from CP-A cells following ChIP in different fractions; IgG FT (flow through from non-specific IgG precipitation), KLF5 FT (flow through from KLF5 precipitation), IgG elute (material eluted following IgG precipitation) KLF5 IP (material eluted following KLF5 immunoprecipitation). Arrows indicating immunoprecipitated protein (KLF5) and asterisks are IgG heavy chain. (C) Scatter plots of KLF5 ChIP-seq data from CP-A from two biological replicates. Pearson correlation coefficient shown. (D) Venn diagrams showing overlap of peaks called by each replicate for KLF5 ChIP-seq from CP-A cells. The numbers of peaks in each sector are given. (E) Bar chart of percentage targets and percentage background of de novo discovered motifs at CP-A KLF5 ChIP-seq peaks. De novo motif, called transcription factor with motif match score (in brackets) and p-value shown. (F) Example UCSC genome browser tracks of KLF5 ChIP-seq and ATAC-seq data from CP-A and OE19 cells at the NKX3-1 (left) and JAG1 loci (right). CP-A-specific peaks are highlighted in blue and shared peaks are highlighted in grey.
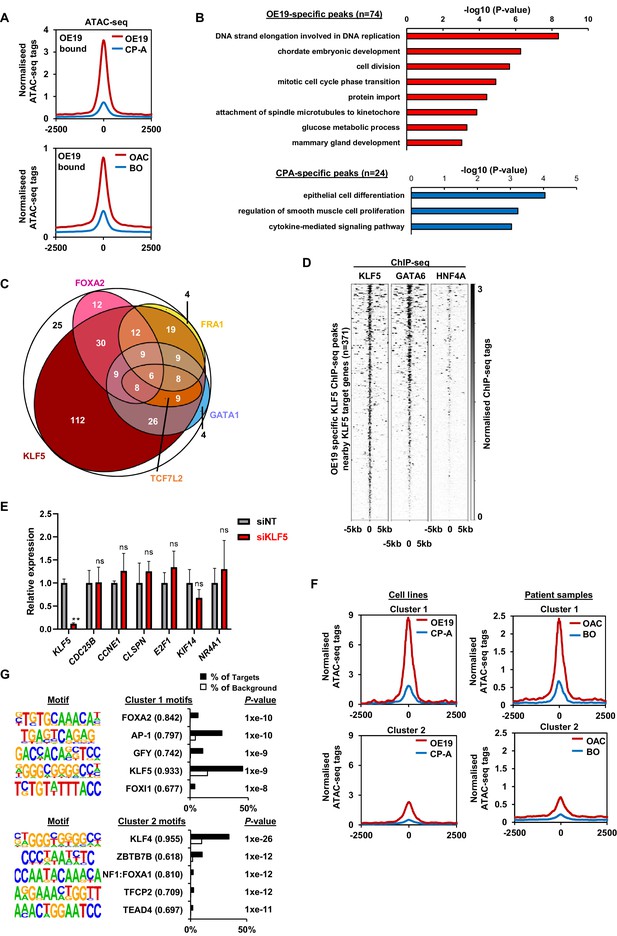
Integrative analysis of KLF5 ChIP-seq data in OE19 and CP-A cells.
(A) Tag density plots of normalised ATAC-seq signal at OE19 specific KLF5 ChIP-seq regions from CP-A (blue) and OE19 (red) cells (top) and merged BO (blue) and merged OAC (red) tissue (bottom). (B) Top GO terms of nearest genes associated with OE19-specific (top) or CP-A-specific (bottom) KLF5 peaks which show either increased (top) or decreased (bottom) expression in OAC. (C) Euler diagram of 299 KLF5-binding regions that are specific to OE19-specific binding regions that are located within loci (+/- 250 kb) containing genes upregulated in OAC and downregulated with KLF5 depletion. The motifs identified in Figure 4E (KLF5, GATA1, FOXA2, FRA1 and TCF7L2) found within each peak are shown (note that an additional 71 regions cannot be depicted due to the small numbers involved). The regions in the white circle contain none of the indicated motifs. (D) Heatmap of KLF5, GATA6 and HNF4A ChIP-seq signal from OE19 cells at OE19 specific KLF5 ChIP-seq peaks nearby KLF5-activated target genes. (E) RT-qPCR analysis of the indicated genes following treatment of CP-A cells with non-targeting (NT) or KLF5 siRNAs for 72 hr (n = 3; P-values **=<0.01; ns = not significant). (F) Tag density plots of normalised ATAC-signal at cluster 1 (left) and cluster 2 (right) regions (see Figure 4G) from CP-A (blue) and OE19 (red) cells (top) and merged BO (blue) and merged OAC (top) samples (bottom). (G) Bar chart of percentage targets and percentage background of de novo discovered motifs in cluster 1 (top) and cluster 2 (bottom) regions. De novo motif, called transcription factor with motif match score and p-value are shown.
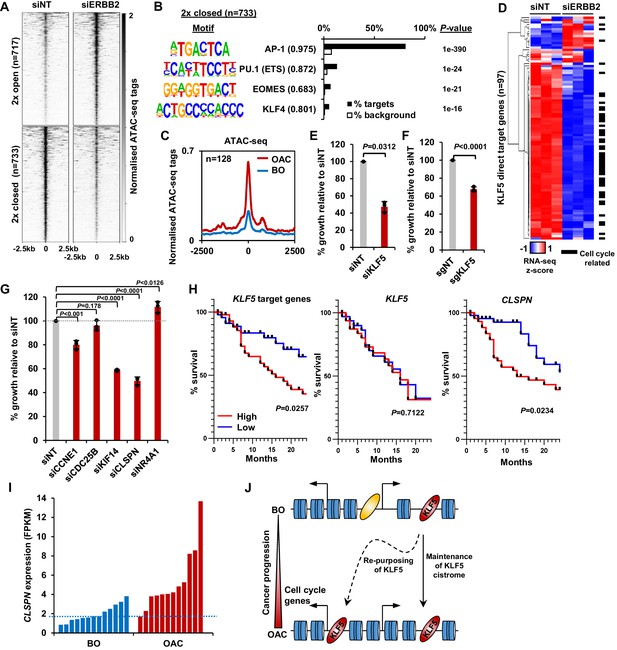
KLF5 controls cell proliferation in OAC.
(A) Heatmap of ATAC-seq signal at regions (peak centre ±2.5 kb) with significantly differential accessibility (±2 x; Q < 0.05) in OE19 cells treated with siERBB2. (B) Bar chart of percentage targets and percentage background of de novo discovered motifs at regions closed with siERBB2 treatment in OE19 cells. De novo motifs, called transcription factor with motif match scores (brackets) and p-values are shown. (C) Tag density plot of ATAC-seq signal from BO and OAC tissue at regions that demonstrate KLF5 binding in OE19 cells and reduced chromatin accessibility in OE19 cells upon siERBB2 treatment. (D) Heatmap of z-score of expression of KLF5 direct target genes in OE19 cells treated with either siNT or siERBB2. Cell cycle related genes are indicated with a black bar. (E) Bar chart showing the % relative growth of OE19 cells treated with either siNT or siKLF5. p-Value is shown (n = 3). (F) Bar chart showing the % relative growth of OE19-dCas9-KRAB cells treated with either non-targeting guides or guides targeting the KLF5 TSS. p-Value is shown (n = 3). (G) Bar chart showing the % relative growth of OE19 cells treated with either siNT or siRNA against the indicated target genes. p-Values are shown (n = 3). (H) Kaplan-Meier curves of patient survival across 24 months for high (above median; red) or low (below median; blue) expression of the 97 KLF5 target genes (left), KLF5 (middle) or CLSPN (right). p-Values are shown. (I) Expression levels (FPKM) of CLSPN expression in BO and OAC patient samples. (J) Model of KLF5 action in BO and OAC. KLF5 binds chromatin in BO and is re-purposed in OAC to bind and regulate cell-cycle-related genes. See also Figure 5—figure supplements 1, 2 and 3.
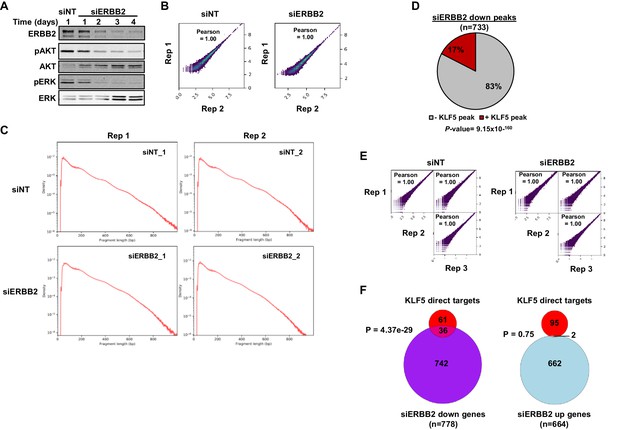
ERBB2 and KLF5 regulate an overlapping set of genes.
(A) Immunoblot of OE19 protein lysate from cells treated with siERBB2 for up to 4 days or siNT probed with antibodies against ERBB2, phosphorylated AKT (pAKT), AKT, phosphorylated ERK (pERK) and ERK. (B) Correlation plots of replicates of ATAC-seq data from OE19 cells treated with either siNT or siERBB2 for 72 hr. Pearson correlations are shown. (C) Fragment length plots of replicates of ATAC-seq data from OE19 cells treated with either siNT or siERBB2. (D) Pie chart showing the percentage of ATAC-seq peaks showing a reduction of chromatin accessibility upon siERBB2 treatment that overlap with a KLF5 ChIP-seq peak in OE19 cells. Percentages and P-value (Fisher exact) for the overlap are shown. (E) Correlation plots of replicates of RNA-seq data from OE19 cells treated with either siNT or siERBB2 for 72 hr. Pearson correlations are shown. (F) Venn diagrams showing the overlap between the 97 direct KLF5-activated genes and genes either down- (left) or up- (right) regulated following siERBB2 treatment of OE19 cells (two fold change, FDR < 0.05, FPKM > 1). p-Values are hypergeometric tests.
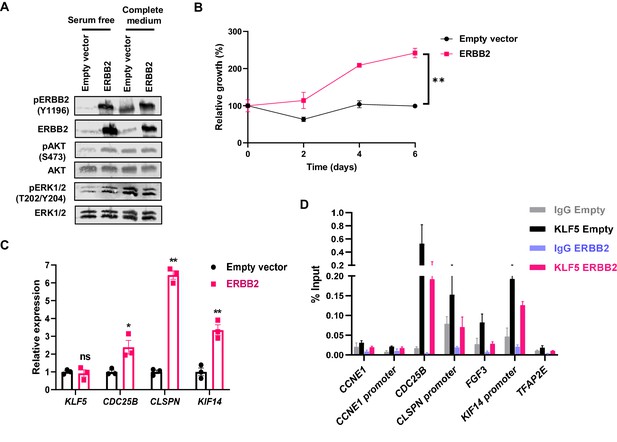
ERBB2 overexpression drives growth factor independent proliferation and gene expression in BO-derived CP-A cells.
(A) Western blots of the indicated proteins or phosphorylated (p) sites in CP-A cells stably transfected with empty or ERBB2 expressing vectors grown in serum free medium for 48 hr (lanes 1 and 2) or serum-free media and stimulated with complete media for 15 min (lanes 3 and 4). (B) Relative growth of CP-A cells stably transfected with empty or ERBB2 expressing vectors grown in serum-free media for 6 days (n = 3; ** = p value<0.01, two-way ANOVA). (C) RT-qPCR analysis of the indicated genes (normalised to GAPDH) in serum starved CP-A cells stably transfected with empty or ERBB2 expressing vectors (n = 3; p-values, *=<0.05, **=<0.01). (D) ChIP-qPCR of KLF5 binding to regulatory regions associated with the indicated genes in CP-A cells stably transfected with empty or ERBB2 expressing vectors grown in serum-free media for 48 hr. Non-specific IgG is provided as a control (n = 3; ns = non-significant increases).
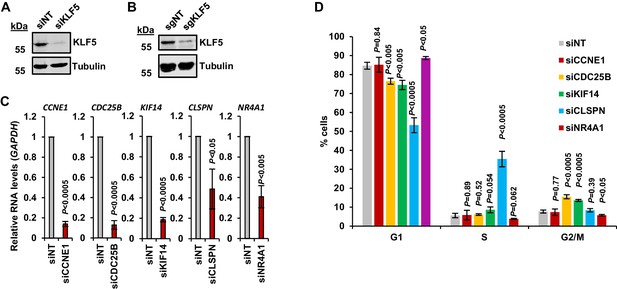
KLF5 drives cell cycle progression in OE19 cells.
(A) Immunoblot of OE19 cell lysate from cells treated with either siNT or siKLF5 for 6 days and probed with antibodies against KLF5 and Tubulin. (B) Immunoblot of OE19-dCas9-KRAB lysate of OE19-dCas9-KRAB cells treated with a non-targeting guide or a pool of three guides targeting the KLF5 TSS and probed with antibodies against KLF5 and Tubulin. (C) RT-qPCR of RNA from OE19 cells treated with either siNT, siCCNE1, siCDC25B, siKIF14, siCLSPN or siNR4A1. The gene that was knocked-down was tested relative to GAPDH and p-values are shown (n = 3). (D) Bar chart of the percentages of OE19 cells treated with either siNT, siCCNE1, siCDC25B, siKIF14, siCLSPN or siNR4A1 for 6 days in G1, S and G2/M phase of the cell cycle. P-values were calculated comparing each knockdown to siNT and p-values are shown (n = 3).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | OE19 | ACACC | 96071721 | |
| Cell line (H. sapiens) | CP-A | ATCC | KR-42421 | |
| Cell line (H. sapiens) | OE19-dCas9-KRAB | This study | OE19 transfected with vector to express dCas9-KRAB under doxycycline control | |
| Cell line (H. sapiens) | CP-A-ERBB2 | This study | CP-A stably overexpressing ERBB2 | |
| Cell line (H. sapiens) | CP-A-empty | This study | CP-A cells containing an empty vector control | |
| Biological sample (H. sapiens) | Barrett’s oesophagus biopsies | Salford NHS FT | Freshly isolated from patients undergoing endoscopy | |
| Biological sample (H. sapiens) | Oesophageal adenocarcinoma biopsies | Salford NHS FT | Freshly isolated from patients undergoing endoscopy | |
| Transfected construct (human) | SmartPool siRNA against KLF5 | Horizon discovery | L-013571-00-0005 | |
| Transfected construct (human) | SmartPool siRNA against ERBB2 | Horizon discovery | L-003126-00-0005 | |
| Transfected construct (human) | SmartPool non-targeting siRNA | Horizon discovery | D-001810-10-0020 | |
| Transfected construct (human) | Full length non-targeting guide RNA | Synthego | 5’-GUAAGGCUAUGAAGAGAUAC-3’ | |
| Transfected construct (human) | Full length guide RNAs targeting KLF5 TSS | Synthego | 5’-GUGCGCUCGCGGUUCUCUCG-3’ 5’-AGGACGUUGGCGUUUACGUG-3’ 5’-GCGUCAAGUGUCAGUAGUCG-3’ | |
| Antibody | Rabbit monoclonal KLF5 antibody | Abcam | ab137676 | (1:10000) for western blot; 5 ug for ChIP-seq |
| Antibody | Mouse monoclonal tubulin antibody | Sigma-Aldrich | T9026 | (1:2000) for western blot |
| Antibody | Spike-in antibody | Active Motif | 61686 | 1 ug for ChIP-seq |
| Antibody | Mouse monoclonal ErbB2 antibody | ThermoFisher | MA5-14057 | (1:1000) |
| Antibody | Mouse monoclonal AKT antibody | Cell signalling technology | 2920 | (1:2000) |
| Antibody | Rabbit monoclonal phosphor-Akt (S473) antibody | Cell signalling technology | 4060S | (1:2000) |
| Antibody | Rabbit monoclonal Erk1/2 antibody | Cell signalling technology | 4695S | (1:1000) |
| Antibody | Mouse monoclonal phosphor-Erk1/2 (T202,Y204) | Cell signalling technology | 9106S | (1:2000) |
| Antibody | Donkey anti-mouse secondary antibody (800CW) | Licor | 925 - 32212 | (1:10,000) |
| Antibody | Donkey anti-rabbit secondary antibody (700CW) | Licor | 925–32213 | (1:10,000) |
| Recombinant DNA reagent | pX330-U6-Chimeric_BB-CBh-hSpCas9 (plasmid) | Addgene | #42230 | AAVS guide RNA sequence 5’-GGGCCACTAGGGACAGGAT-3’ |
| Recombinant DNA reagent | pAAVS1-Puro-TRE-dCas9-KRAB-DNR (plasmid) | This study | pAS-4939 | |
| Recombinant DNA reagent | pHAGE-ERBB2 | Addgene | 116734 | |
| Recombinant DNA reagent | pHAGE-empty | This study | pAS-4940 | |
| Recombinant DNA reagent | pMD2.G | Addgene | 12259 | |
| Recombinant DNA reagent | psPAX2 | Addgene | 12260 | |
| Sequenced-based reagent | Primers | This study | See Supplementary file 11 | |
| Commercial assay or kit | Lipofectamine RNAiMAX | Thermofisher | 13778150 | |
| Commercial assay or kit | Fugene HD | Promega | E2311 | |
| Commercial assay or kit | QuantiTect SYBR Green RT-PCR Kit | Qiagen | 204243 | |
| Commercial assay or kit | RNeasy Plus Mini Kit | Qiagen | 74134 | |
| Commercial assay or kit | RNase-free DNase set | Qiagen | 79254 | |
| Commercial assay or kit | Ampure XP beads | Beckman Coulter Agencourt | A63881 | |
| Commercial assay or kit | TruSeq stranded RNA library kit v2 | Illumina | RS-122–2001 | |
| Commercial assay or kit | Nextera DNA library prep kit | Illumina | FC-121–1031 | |
| Commercial assay or kit | Nextera Index kit | Illumina | FC-121–1012 | |
| Commercial assay or kit | NEBNext high fidelity 2x PCR master mix | NEB | M0541 | |
| Commercial assay or kit | DNA Clean and Concentrator | Zymo | D4013 | |
| Commercial assay or kit | Polyfect | Qiagen | 301107 | |
| Commercial assay or kit | PEG-it | System Biosciences | LV810A-1 | |
| Commercial assay or kit | Polybrene | EMD Millipore | TR-1003 | |
| Chemical compound, drug | RS-1 | Sigma-Aldrich | R9782 | Used at final concentration 7.5 μM |
| Chemical compound, drug | SCR7 pyrazine | Sigma-Aldrich | SML1546 | Used at final concentration 1 μM |
| Chemical compound, drug | Doxycycline | Sigma-Aldrich | D3447 | Used at final concentration of 100 ng/mL |
| Chemical compound, drug | propidium iodide | Sigma | P4170 | Used at 50 μg/mL |
| Peptide, recombinant protein | RNase | Sigma | R4642 | Used at 100 μg/mL |
| Peptide, recombinant protein | EGF | ThermoFisher | 10450–013 | 5 μg/L |
| Peptide, recombinant protein | Bovine pituitary extract | ThermoFisher | 13028014 | Used at 50 mg/L |
| Software, algorithm | Trimmomatic | Bolger et al., 2014 | V0.34 | http://www.usadellab.org/cms/?page=trimmomatic |
| Software, algorithm | Bowtie2 | Langmead and Salzberg, 2012 | v2.3.0 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Software, algorithm | Star | Dobin et al., 2013 | V2.5.4 | https://github.com/alexdobin/STAR |
| Software, algorithm | Macs2 | Zhang et al., 2008 | v2.1.1 | https://github.com/taoliu/MACS |
| Software, algorithm | Cufflinks | Tarapore et al., 2013 | v2.2.1 | http://cole-trapnell-lab.github.io/cufflinks/ |
| Software, algorithm | DEseq2 | Love et al., 2014 | V1.22.2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Software, algorithm | TOBIAS | Bentsen et al., 2020 | v0.5.1 | https://github.com/loosolab/TOBIAS |
| Software, algorithm | featureCounts | Liao et al., 2014 | V1.6.2 | http://subread.sourceforge.net |
| Software, algorithm | FastQC | v0.11.4 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | |
| Software, algorithm | bedtools | Quinlan and Hall, 2010 | v2.26.0 | https://bedtools.readthedocs.io/en/latest/ |
| Software, algorithm | DeepTools | Ramírez et al., 2016 | V2.5.0 | https://deeptools.readthedocs.io/en/develop/ |
| Software, algorithm | GSEA | Subramanian et al., 2005 | V3.0 | http://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Main_Page |
| Software, algorithm | Homer | Heinz et al., 2010 | v4.9 | http://homer.ucsd.edu/homer/ |
| Software, algorithm | R | R Core Team (2018) | v3.5.1 | https://www.r-project.org/ |
| Software, algorithm | GraphPad Prism | V8.0 | www.graphpad.com | |
| Other | Crystal violet | Sigma Aldrich | HT90132 | Used at concentration of 0.1% |
| Other | Gibco RPMI 1640 | ThermoFisher | 52400 | |
| Other | Gibco fetal bovine serum | ThermoFisher | 10270 | |
| Other | Gibco penicillin/streptomycin | ThermoFisher | 15140122 | |
| Other | Keratinocyte SFM (1x) | ThermoFisher | 17005042 |
Additional files
-
Source code 1
ATAC fragment size visualisation.
- https://cdn.elifesciences.org/articles/57189/elife-57189-code1-v2.zip
-
Supplementary file 1
Differentially expressed genes in OAC.
Significantly (±1.5 x; Q-value <0.05) differentially expressed genes between BO (n = 13) and OAC (n = 12) (Maag et al., 2017).
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp1-v2.xlsx
-
Supplementary file 2
Differentially accessible regions within ±250 kb of TSS of a DEG.
(A) Total accessible regions from BO (n = 4) and OAC (n = 6) samples. (B) Significant differentially accessible open regions (+2x; Q-value <0.1). (C) Significant differentially accessible closed regions (−2x; Q-value <0.1).
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp2-v2.xlsx
-
Supplementary file 3
DNA motifs enriched in OAC-specific open chromatin regions.
Top ten motifs found by de novo motif discovery and their associated transcription factors that are enriched in ‘open in OAC’ (top) or ‘closed in OAC’ (bottom).
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp3-v2.xlsx
-
Supplementary file 4
siKLF5 RNA-seq analysis.
Significant differentially expressed genes with siKLF5 treatment (±1.3 x, Q-value <0.05)
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp4-v2.xlsx
-
Supplementary file 5
KLF5 ChIP-seq datasets.
(A) ChIP-seq peaks in OE19 cells. (B) ChIP-seq peaks in CP-A cells. (C) Differentially bound KLF5 ChIP-seq peaks (CP-A vs OE19).
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp5-v2.xlsx
-
Supplementary file 6
De novo analysis of DNA motif enrichment in KLF5 ChIP-seq peak datasets.
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp6-v2.xlsx
-
Supplementary file 7
(A) Frequency of KLF5, GATA1, FOXA2, FRA1 and TCF7L2 motifs within OE19 specific KLF5 ChIP-seq regions. one denotes present and 0 absent. (B) Overlaps of motifs and the basis of Figure 4G (A. KLF5; B. GATA1; C. FOXA2; D. FRA1; E. TCF7L2).
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp7-v2.xlsx
-
Supplementary file 8
DNA motifs enriched in Cluster one and Cluster two regions.
Top 10 motifs found by de novo motif discovery and their associated transcription factors that are enriched in cluster 1 (top) or cluster 2 (bottom).
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp8-v2.xlsx
-
Supplementary file 9
Genomic coordinates of regions on OE19 cells that show a decrease in ATAC-seq signal upon treatment of siERBB2 for 72 hr.
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp9-v2.xlsx
-
Supplementary file 10
De novo discovered motifs from regions that exhibit reduced chromatin accessibility upon treatment of siERBB2 for 72 hr.
De novo motifs, % targets and % background, called transcription factor with match score and p-value are shown.
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp10-v2.xlsx
-
Supplementary file 11
List of PCR primers used in RT-qPCR and ChIP-qPCR experiments.
- https://cdn.elifesciences.org/articles/57189/elife-57189-supp11-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57189/elife-57189-transrepform-v2.docx





