Eater cooperates with Multiplexin to drive the formation of hematopoietic compartments
Figures
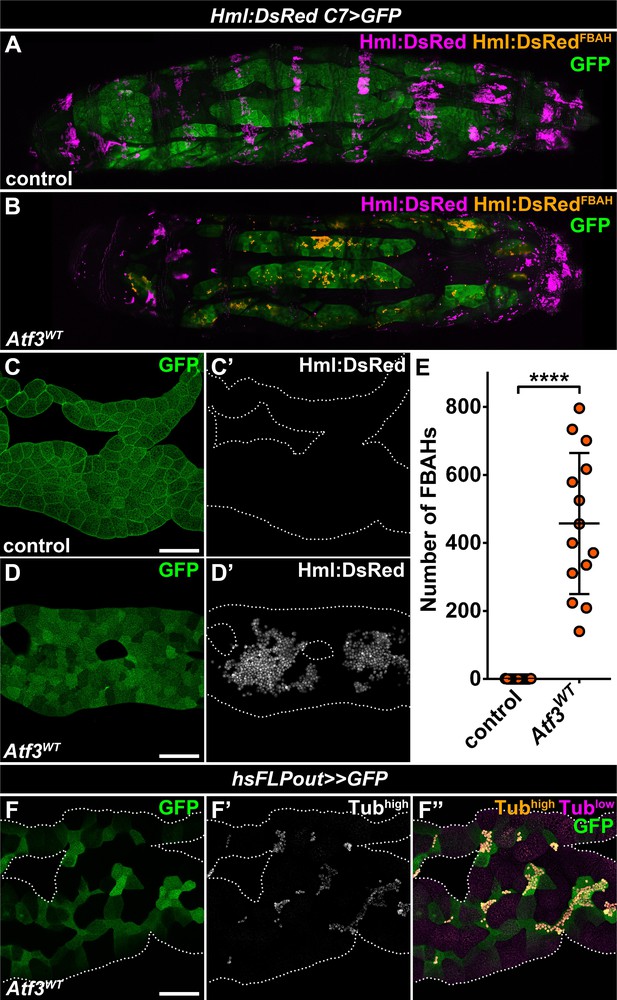
Adipose tissue-specific Atf3 overexpression redirects hemocytes to the fat body surface.
(A–B) Sessile hemocytes present in control larvae as segmental stripes (A) are redirected to the surface of the fat body overexpressing Atf3 (B). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. The images are stitched from multiple Z-projections, where FBAHs are colored amber, all other hemocytes magenta and the fat body green. (C–D) In control larvae, no hemocytes are present on the fat body (C). Atf3 overexpression induces the attachment of hemocytes, which form large clusters on the fat body surface (D). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. (E) Quantification of fat body-attached hemocyte numbers upon Atf3 overexpression with the C7-GAL4 driver. Data points represent individual fat bodies. Unpaired nonparametric two-tailed Mann-Whitney test was used to calculate p-values. Error bars indicate SD, n = 15, ****p < 0.0001. (F) Clonal overexpression of Atf3 induced with the hsFLPout system causes selective hemocyte attachment (F', white, F'', amber) to the clonal adipocytes (marked with GFP). Hemocytes are identified based on strong Tubulin staining (F'), while the weak Tubulin staining in the fat body was pseudocolored magenta (F''). Fat bodies are outlined with dotted lines (C, D, F). The images are projections of multiple confocal sections. Scale bars: 100 μm (C, D, F). See also Figure 1—figure supplement 1 and Figure 1—source data 1.
-
Figure 1—source data 1
Quantification of FBAH number in control and C7>Atf3WT larvae.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Circulating hemocyte counts from control and Atf3 overexpressing larvae.
For graphical representation the data was normalized to the average of the control.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig1-data2-v1.xlsx
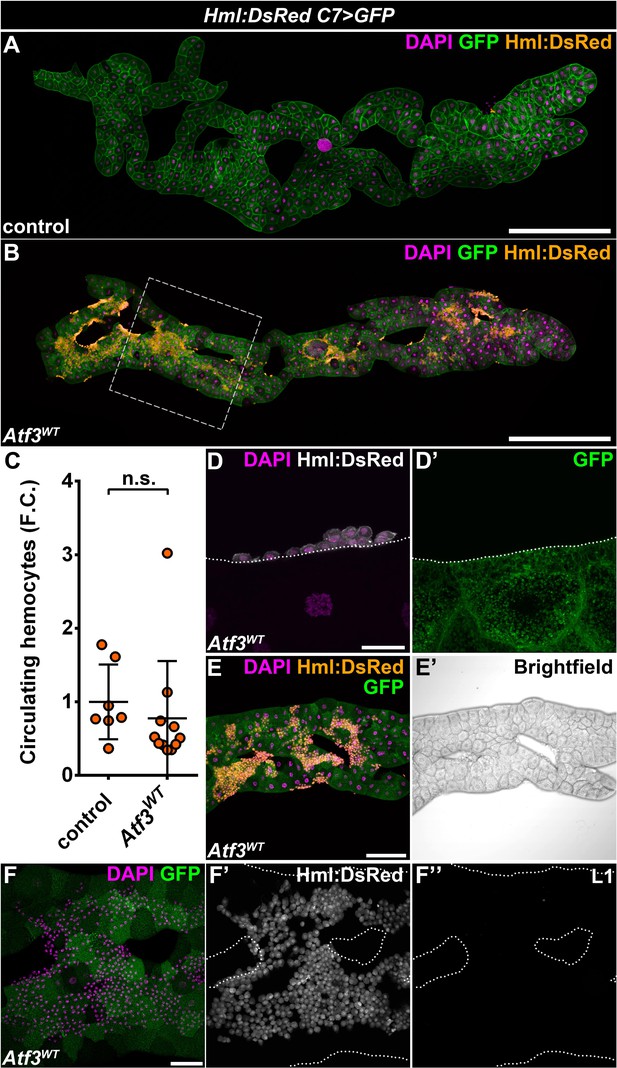
Hemocytes redirected to the fat body are not engaged in anti-tissue response.
(A–B) While under homeostatic conditions the fat body is devoid of hemocytes (A), fat body-specific Atf3 overexpression induces hemocyte adhesion along the entire length of the adipose tissue (B). Images are stitches from multiple Z-stacks. The area used for imaging and quantifications is outlined with dashed lines. Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. (C) Circulating hemocyte numbers are not affected by Atf3 overexpression in the fat body. Data points represent individual replicates. Hemocyte numbers were normalized to the control mean (shown as 1). Unpaired nonparametric two-tailed Mann-Whitney test was used to calculate p-values. Error bars indicate SD, n ≥ 7, n.s. = non significant. (D) A closeup image shows that FBAHs do not contain GFP-positive material originating from the adipose tissue. Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. (E) Brightfield image of an Atf3 overexpressing fat body (E') shows that the adipose tissue does not undergo melanization due to hemocyte attachment (E). Transgene and GFP expression was driven by the fat-body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. (F) No lamellocytes adhere to the Atf3 overexpressing fat body (F, F''), as indicated by the lack of the lamellocyte-specific L1 antibody staining (F''). Transgene and GFP expression was driven by the fat-body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. Nuclei were counterstained with DAPI (A, B, D, E, F). The images are projections of multiple confocal sections (A, B, E, F) or represent single sections (D), fat bodies are outlined with dotted lines (D, F). Scale bars: 500 μm (A, B), 20 μm (D), 100 μm (E), 50 μm (F). See also Figure 1—source data 2.
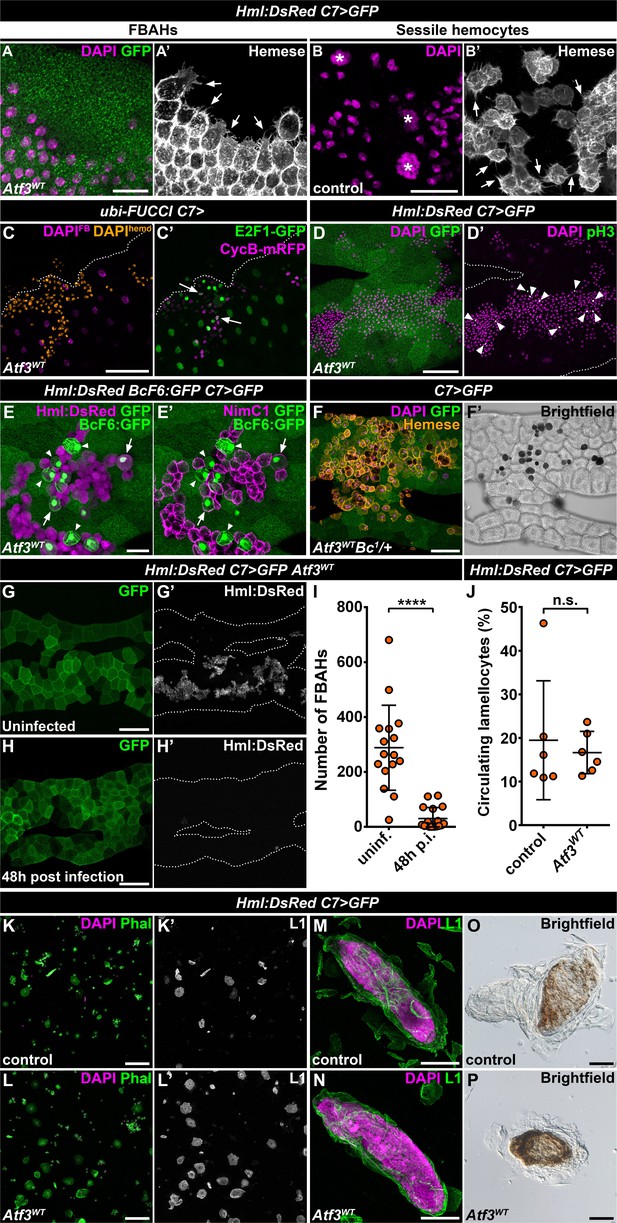
Fat body-associated hemocytes share features with sessile hemocytes.
(A–B) Similar to sessile hemocytes in control larvae (B), FBAHs tightly cluster and form filopodia on the fat body surface (A). Images are depicting the fat body (A) and the epidermis (B). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver (A). Hemocytes were stained with pan-hemocyte anti-Hemese antibody to reveal membrane morphology. Arrows indicate filopodia emanating from hemocytes, asterisks indicate nuclei of epidermal cells (B). (C–D) The ubiquitously expressed FUCCI cell cycle reporter (C) shows FBAHs in G1 (green), S (magenta) and G2/M (white, arrows) phases of the cell cycle. Mitotic FBAHs are highlighted by pH3 staining (D, arrowheads). Nuclei were pseudocolored (see Materials and methods) to indicate hemocytes (amber) or adipocytes (magenta) (C). (E–F) Crystal cells (arrowheads), and plasmatocyte-crystal cell intermediary hemocytes (arrows) are interspersed among plasmatocytes in FBAH clusters (E). Melanized crystal cells are attached to Atf3 overexpressing fat bodies (F, black cells). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver. Crystal cells were identified by expression of the BcF6:GFP transgene (E) or melanization due to the presence of the Bc1 mutation (F). Plasmatocytes were revealed with staining against NimC1 (E'), while Hml:DsRed (E) or anti-Hemese immunostaining (F) was used to show all FBAHs. (G–I) FBAH numbers decline 48 hr after L. boulardi infection (H, I) compared to uninfected controls (G). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. Data points represent individual replicates. Unpaired nonparametric two-tailed Mann-Whitney test was used to calculate p-values. Error bars indicate SD, n ≥ 16, ****p < 0.0001 (I). (J–L) The lamellocyte differentiation 24 hr after parasitic infection (J, L) is not impaired in larvae expressing Atf3 in the fat body when compared to infected controls (J, K). Data points represent individual replicates, showing the percentage of lamellocytes (L1-positive cells) in all hemocytes (total DAPI count). Statistical significance was determined with two-tailed Student's t-test, error bars indicate SD, n = 6, n.s. = non significant (J). Images depict circulating immune cells bled from L. boulardi infected larvae. Phalloidin staining (K, L, green) labels all hemocytes, L1 staining (K', L', white) shows the lamellocytes. Transgene expression was driven by the fat body-specific C7-GAL4 driver. (M–P) Encapsulation (M) and melanization (O) of parasitic eggs are not hindered by Atf3 overexpression in the fat body (N, P). Lamellocytes surrounding the eggs 24 hr after infection were visualized with L1 staining (M, N). Brown coloration of the encapsulated eggs 48 hr following infestation indicates melanization (O, P). Nuclei were counterstained with DAPI (A–D, F, K–N). The images are projections of multiple confocal sections, fat bodies are outlined with dotted lines (C, D, G, H). Scale bars: 20 μm (A, B, E), 50 μm (C, D, F, M, N), 100 μm (G, H, K, L, O, P). See also Figure 2—source datas 1 and 2.
-
Figure 2—source data 1
Quantification of FBAH number in C7>Atf3WT larvae without parasitoid wasp infection and 48 hr post-infection.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig2-data1-v1.xlsx
-
Figure 2—source data 2
The percentage of lamellocytes 24 hr after L. boulardi infestation in control and C7>Atf3WT larvae.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig2-data2-v1.xlsx
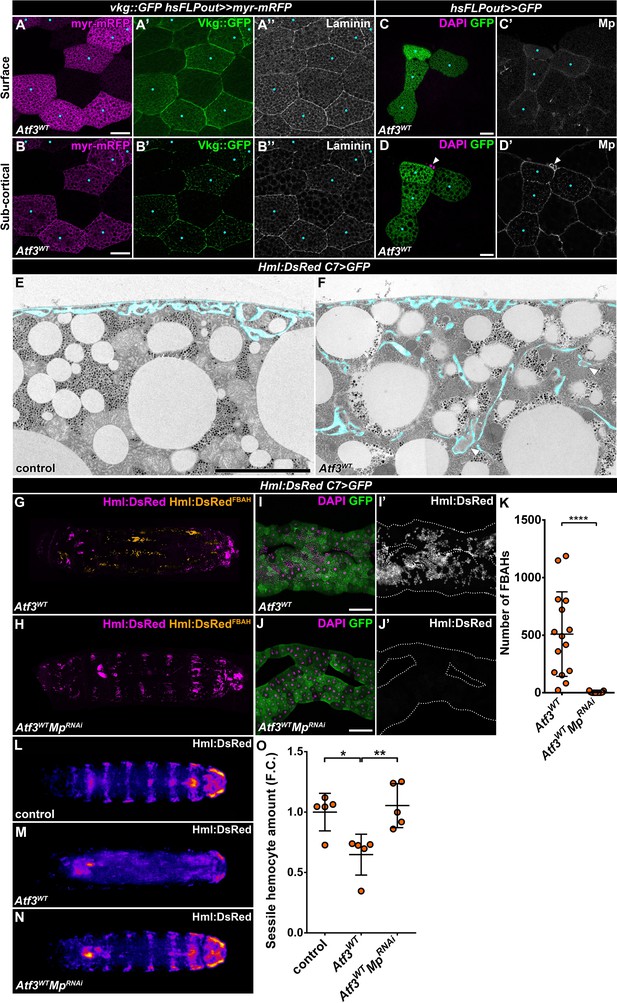
Pericellular accumulation of basement membrane components underlies hemocytes attachment to Atf3 overexpressing fat bodies.
(A–D) Clonal Atf3 overexpression in the fat body leads to the enrichment of Collagen IV, Laminin and Mp on the surface of the adipocytes (A, C), and their accumulation in foci below the cell membrane (B, D). Clonal cells overexpressing Atf3 were induced with the hsFLPout technique, and are marked by the expression of myr-mRFP (A, B, magenta) or GFP (C, D, green). The expression of Collagen IV was visualized with the Vkg::GFP reporter, while Laminin and Mp expression was determined by immunostaining. The clonal adipocytes are indicated with cyan dots. The arrowhead points to hemocytes attached to the clonal cells (D). Images are single confocal planes taken at the adipose tissue surface (A, C) or ~5 μm below the tissue surface (B, D). (E–F) Transmission electron micrographs show that compared to controls (E), fat body-specific Atf3 overexpression causes the formation of cell membrane folds (F) and the entrapment of ECM material (arrowheads). The transgene expression was driven by the fat body-specific C7-GAL4 driver. Pericellular spaces between the cell membrane and the basement membrane are colored cyan. (G–K) Knockdown of Mp in Atf3 overexpressing fat bodies abolishes the attachment of hemocytes to the adipose tissue (H, J, K) when compared to Atf3 overexpression alone (G, I, K). Whole larval images are stitches of multiple Z-projections, where FBAHs are colored amber, all other hemocytes magenta (G, H). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. For whole larvae images, the localization of the DsRed signal was determined on every confocal section, and the hemocytes situated on the fat body surface were colored amber, the rest magenta (G, H). Data points represent individual replicates. Unpaired nonparametric two-tailed Mann-Whitney test was used to determine p-values, error bars indicate SD, n = 15, ****p < 0.0001 (K). (L–O) Knockdown of Mp in Atf3 overexpressing fat bodies restores the structure of the sessile hematopoietic pockets. Compared to controls (L, O), the overexpression of Atf3 in the adipose tissue disrupts the striped sessile hemocyte pattern (M, O). The pattern is restored following the simultaneous knockdown of Mp in the fat body (N, O). Transgene expression in the fat body was driven with the C7-GAL4 driver, while the Hml:DsRed reporter was used to determine hemocyte location. Images represent the stereotypical DsRed pattern generated from the alignment of five individual larvae (L–N) and were used for quantification (O). Data points represent the total fluorescence intensity of four regions encompassing the four posterior-most sessile bands, which were normalized to the mean of controls (represented as 1) and shown as fold change. One-way ANOVA multiple comparison with Tukey's correction was used to determine significance, error bars indicate SD, n = 5. **p = 0.0067, *p = 0.0165. Nuclei were counterstained with DAPI (C, D, I, J). The images are single confocal sections (A–D), or represent projections of multiple confocal sections (G–J), fat bodies are outlined with dotted lines (I, J). Scale bars: 20 μm (A–D), 5 μm (E, F), 100 μm (I, J). See also Figure 3—figure supplements 1–2 and Figure 3—source data 1 – 2.
-
Figure 3—source data 1
Quantification of FBAH number in C7>Atf3WT and C7>Atf3WTMpRNAi larvae.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Quantification of FBAH number in C7>Atf3WT, C7>Atf3WTCol4a1RNAi, C7>Atf3 WTtrolRNAi, C7>Atf3WTvkgRNAi, C7>Atf3WTLanARNAi, C7>Atf3WTLanB1RNAi and C7>Atf3WTLanB2RNAi larvae.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig3-data2-v1.xlsx
-
Figure 3—source data 3
Quantification of FBAH number in control, C7>Atf3WT, C7>Atf3WTMpRNAi, C7>Atf3WTSPARCRNAi and C7>Atf3WTSPARCRNAiMpRNAi larvae.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Quantification of sessile hemocyte intensity in control, C7>Atf3WT and C7>Atf3WTMpRNAi larvae.
For graphical representation, the data was normalized to the average of the control.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig3-data4-v1.xlsx
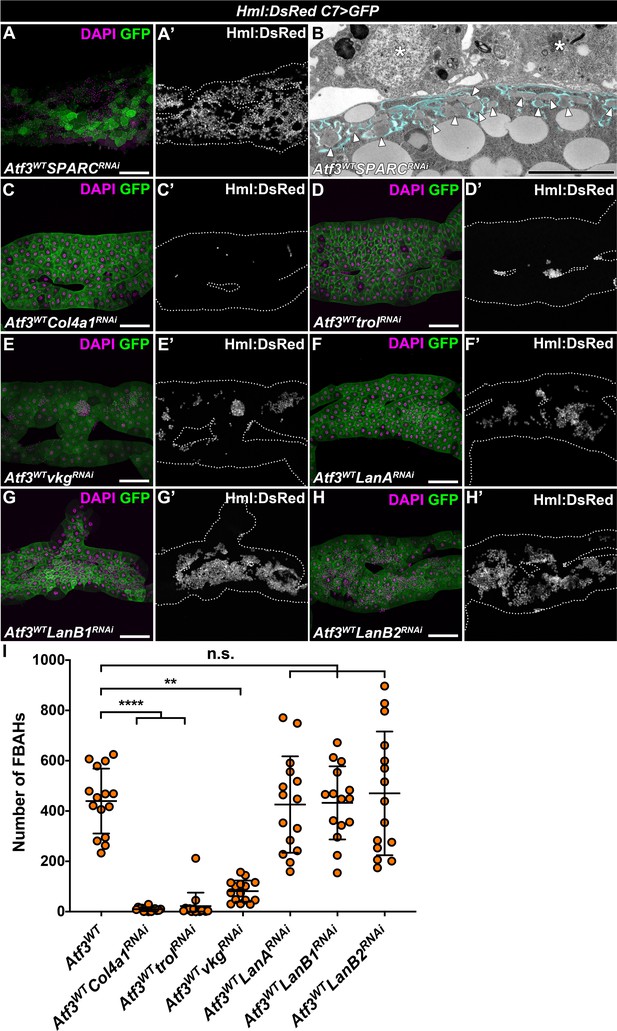
Knockdown of Collagen IV and Trol suppresses hemocyte adhesion to C7>Atf3WT fat bodies.
(A–B) Loss of SPARC in the Atf3 overexpressing fat body enhances hemocyte adhesion (A, A'), but also causes a severe expansion of pericellularly retained ECM aggregates (B). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. Pericellular space between the cell membrane and the basement membrane is highlighted in cyan, and ECM accumulations are indicated with arrowheads, while asterisks indicate attached hemocytes (B). (C–I) Knockdown of Col4a1 (C) and Trol (D) suppresses FBAH adhesion to Atf3 overexpressing fat bodies, while Vkg knockdown (E) leads to a partial suppression of the phenotype. Loss of LanA (F), LanB1 (G) and LanB2 (H) does not impact FBAH presence (I). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. Data points represent individuals replicates. Nonparametric one-way Kruskal-Wallis test with Dunn’s multiple comparison was used to determine significance. Error bars indicate SD, n = 15. **adjusted p < 0.0078, ****adjusted p < 0.0001, n.s. = non significant (I). Tissues were counterstained with DAPI (A, C–H). Images are maximum projections of multiple confocal sections (A, C–H). Fat bodies are outlined with dotted lines (A, C–H). Scale bars: 100 μm (A, C–H), 5 μm (B). See also Figure 3—source data 3.
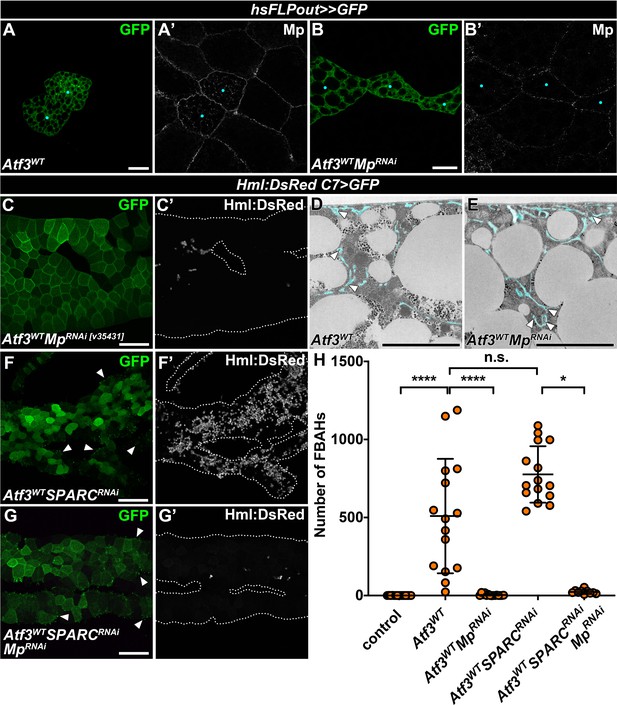
Mp is an ECM component crucial for FBAH adhesion.
(A–B) While in Atf3 overexpressing adipocyte clones Mp accumulates below the basement membrane (A), simultaneous Mp knockdown with the RNAi line used throughout the study strongly reduces pericellular accumulation (B). Clones were generated with the hsFLPout system, which drives the expression of transgenes and GFP (green). The expression of Mp was determined by antibody staining. Images are single confocal planes taken at ~5 μm below the tissue surface, where clonal cells are indicated with cyan dots. (C) Knockdown of Mp in the Atf3 overexpressing fat body with an independent RNAi line impairs FBAH formation. Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. (D–E) Transmission electron micrographs indicate no change in the formation of pericellular lacunae around the adipocytes after Mp knockdown (E) compared to Atf3 overexpression alone (D). Transgene expression was driven by the fat body-specific C7-GAL4 driver. The lacunae are highlighted in cyan, and arrowheads indicate ECM accumulations. (F–H) Loss of SPARC in the Atf3 overexpressing fat body (F', H) causes a formation of GFP-positive membrane blebs (F, arrowheads). The exacerbated FBAH numbers on C7>Atf3WTSPARCRNAi fat bodies (F, H) are suppressed to control levels following Mp knockdown (G, H), however GFP positive blebbing is still present (small green foci, compare G and F). Transgene and GFP expression was driven by the fat body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. Arrows indicate GFP positive membrane blebs (F, G). Nonparametric one-way Kruskal-Wallis test with Dunn’s multiple comparison was used to determine significance. Error bars represent SD, n ≥ 10, ****adjusted p < 0.0001, *adjusted p = 0.0303, n.s. = non significant (H). Images are single confocal sections (A, B) or represent maximum projections of multiple confocal sections (C, F, G). Fat bodies are outlined with dotted lines (C, F, G). Scale bars: 20 μm (A, B), 100 μm (C, F, G), 5 μm (D, E). See also Figure 3—source data 4.
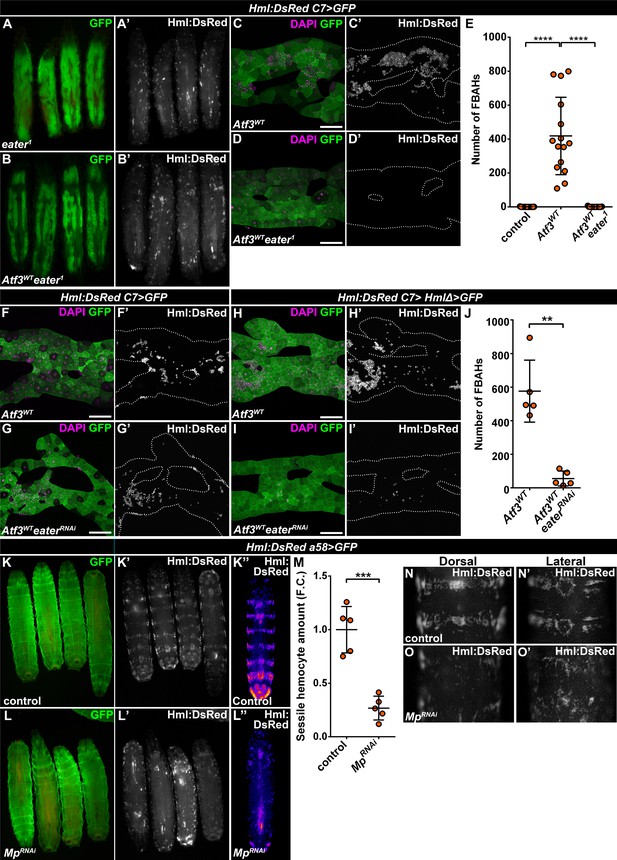
Mp and Eater control hemocyte attachment to the sessile hematopoietic pockets and on the surface of Atf3 overexpressing fat bodies.
(A–E) Eater loss abrogates the association of hemocytes both to the sessile pockets (A), and to Atf3 overexpressing fat bodies (B, D, E) compared to Atf3 overexpression alone (C, E). Transgene and GFP expression was driven by the fat-body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. Data points represent individual replicates. Nonparametric one-way Kruskal-Wallis test with Dunn’s multiple comparison was used to determine significance. Error bars indicate SD, n = 15. ****adjusted p < 0.0001 (E). (F–J) The knockdown of Eater in the Atf3 overexpressing fat body does not noticeably influence the association of hemocytes (G) compared to Atf3 overexpression alone (F). The combined expression of Atf3 in the fat body and the hemocytes does not markedly alter FBAH cluster formation (H), while simultaneous Eater knockdown suppresses hemocyte attachment (I, J), indicating hemocyte-specific requirement for Eater function. Transgene and GFP expression was driven either by the combination of the fat-body-specific C7-GAL4 driver and the hemocyte-specific HmlΔ-GAL4 (H–J), or with the C7-GAL4 alone (F, G), while Hml:DsRed marks the hemocytes. Data points represent individual replicates. Significance was determined by unpaired nonparametric two-tailed Mann-Whitney test, error bars represent SD, n = 5, **p = 0.0097 (J). (K–L) Knockdown of Mp in epidermal cells (L) disrupts the stereotypical banded sessile hemocyte pattern (K). Transgene and GFP expression was driven by the epidermis-specific a58-GAL4 driver, while Hml:DsRed marks the hemocytes. Images represent individual larvae (K, K', L, L'), or the stereotypical DsRed pattern generated from the alignment of five individual larvae (K'', L''). (M) Sessile hemocyte amounts significantly decrease following epidermal knockdown of Mp. Transgene expression was driven by the a58-GAL4 driver, while Hml:DsRed marks the hemocytes. Data points represent the total fluorescence intensity of four regions encompassing the four posterior-most sessile bands, which were normalized to the mean of controls (represented as 1) and shown as fold change. Statistical significance was determined with two-tailed student's t-test, error bars indicate SD, n = 5. ****p < 0.0001. (N–O) Compared to controls (N), the structure of both the dorsal stripe and lateral patches of the sessile hematopoietic tissue is disrupted upon knockdown of Mp in the epidermis (O). Note that hemocyte accumulation on the lateral side of larvae with epidermal-specific Mp knockdown is likely the consequence of decreased hemolymph flow due to the immobilization process (O’). Transgene expression was driven by the a58-GAL4 driver, while Hml:DsRed marks the hemocytes. Images depict the dorsal (N, O) and lateral (N’, O’) views of the A5-A6 larval segments from the same larvae. Tissues were counterstained with DAPI (C, D, F–I). Images are maximum projections of multiple confocal sections (C, D, F–I). Fat bodies are outlined with dotted lines (C, D, F–I). Scale bars: 100 μm (C, D, F–I). See also Figure 4—figure supplement 1 and Figure 4—source datas 1–3.
-
Figure 4—source data 1
Quantification of FBAH number in control, C7>Atf3WT and C7>Atf3WTeater1 larvae.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Quantification of FBAH number in C7>Hml>Atf3WT and C7>Hml>Atf3WTeaterRNAi larvae.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Quantification of sessile hemocyte intensity in control and a58 >MpRNAi larvae.
For graphical representation, the data was normalized to the average of the control.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig4-data3-v1.xlsx
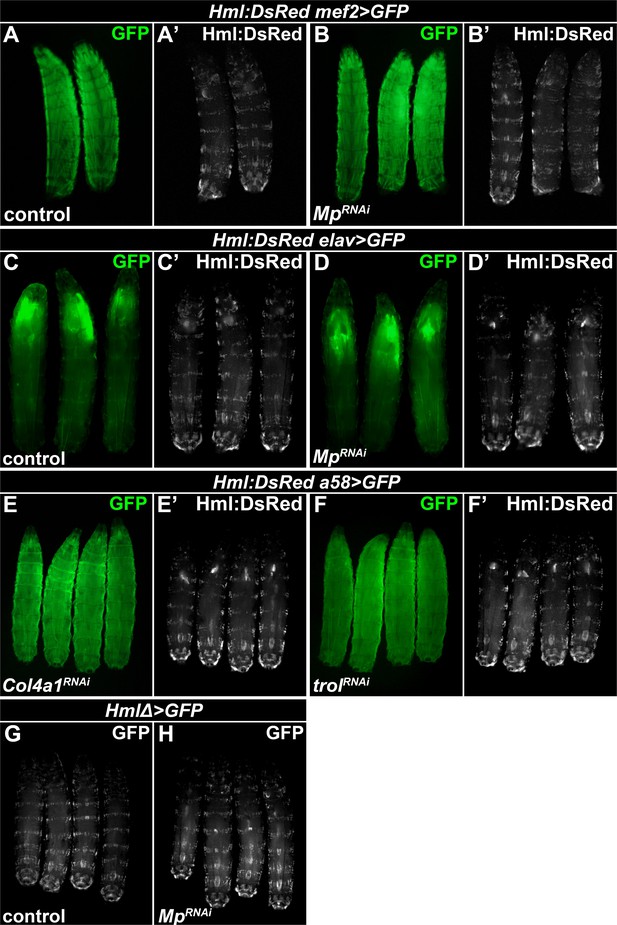
Col4a1 and Trol knockdown in the epidermis or Mp knockdown in the hemocytes, the muscles and the neurons do not affect the epidermal sessile hematopoietic compartment.
(A–D) Compared to control larvae (A, C), sessile hematopoietic pocket integrity is not compromised markedly upon muscle- (B) or neuronal-specific (D) silencing of Mp. Transgene and GFP expression was driven by the muscle-specific mef2-GAL4 driver (A, B), or with the neuronal elav-GAL4 driver (C, D), while Hml:DsRed marks the hemocytes. (E–F) The integrity of sessile hematopoietic pockets was not noticeably affected by epidermal knockdown of Col4a1 (E) or Trol (F). Transgene and GFP expression was driven with the epidermal a58-GAL4 driver, and hemocytes were visualized with the Hml:DsRed reporter. (G–H) Hemocyte-specific knockdown of Mp (H) did not cause apparent disruption of the sessile tissue pattern compared to controls (G). The hemocyte-specific HmlΔ-GAL4 driver was used to control the expression of the UAS-MpRNAi transgene and UAS-GFP.
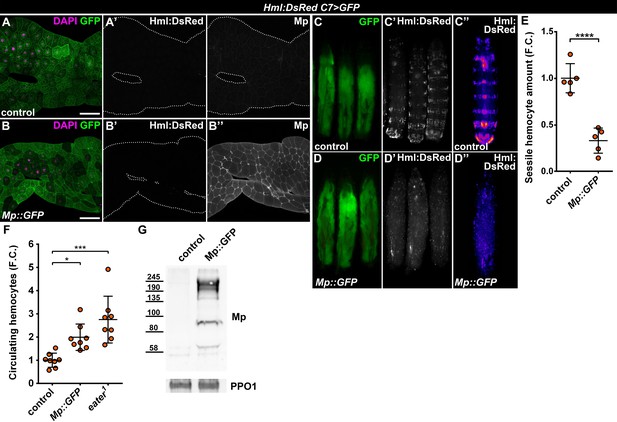
Fat body-wide Mp overexpression causes detachment of the sessile hemocytes.
(A–B) Similar to controls (A) fat bodies overexpressing Mp::GFP do not attract hemocytes (B), even though Mp integrates into the basement membrane of the adipose tissue (A'', B''). Transgene and GFP expression was driven by the fat-body-specific C7-GAL4 driver, while Hml:DsRed marks the hemocytes. The expression of Mp was determined with immunostaining. Fat bodies are outlined with dotted lines. Nuclei were counterstained with DAPI. Scale bars: 100 μm. (C–E) Fat body-specific Mp::GFP expression disrupts the segmentally organized sessile hematopoietic compartment (D) as observed in controls (C). Images represent individual larvae (C, C', D, D') or the stereotypical Hml:DsRed pattern generated from the alignment of five individual larvae (C'', D''). Data points represent the total fluorescence intensity of four regions encompassing the four posterior-most sessile bands, which were normalized to the mean of controls (represented as 1) and shown as fold change. Statistical significance was determined with two-tailed Student's t-test, error bars indicate SD, n = 5, ****p < 0.0001 (E). (F) Sessile hemocyte detachment following fat body-wide overexpression of Mp::GFP coincides with the elevation of circulating hemocyte numbers similar to eater deficiency. Data points represent individual replicates, which were normalized to control mean (represented as 1). Nonparametric one-way Kruskal-Wallis test with Dunn’s multiple comparison was used to determine significance, error bars indicate SD, n = 8, ***adjusted p = 0.0003, *adjusted p = 0.0175. (G) Mp levels increase in the circulation upon fat body-specific overexpression. Immunoblot against Mp shows multiple bands in cell-free hemolymph extracts, indicating extensive post-translational processing. Molecular weights (in kDa) are shown. Prophenoloxidase 1 (PPO1) served as a loading control. The expression of transgenes and GFP was driven with the fat body-specific C7-GAL4 driver, while hemocytes were recognized based on the expression of the Hml:DsRed reporter (A–G). See also Figure 5—figure supplement 1 and Figure 1—source datas 1–2.
-
Figure 5—source data 1
Quantification of sessile hemocyte intensity in control and C7>Mp::GFP larvae.
For graphical representation, the data was normalized to the average of the control.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Circulating hemocyte counts from control, Mp::GFP overexpressing and eater1 mutant larvae.
For graphical representation, the data was normalized to the average of the control.
- https://cdn.elifesciences.org/articles/57297/elife-57297-fig5-data2-v1.xlsx
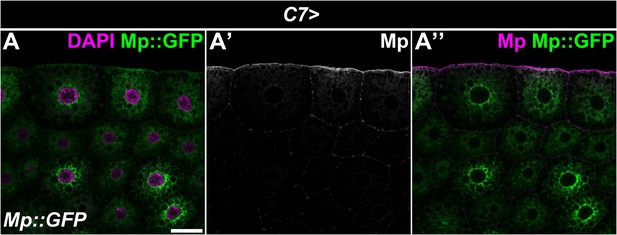
The Mp::GFP transgenic protein is incorporated into the ECM without the GFP tag.
(A) Upon fat body-specific overexpression of the Mp::GFP transgene, the GFP signal is cytoplasmic (A), while the Mp staining is increased in the basement membrane and the intercellular ECM accumulations (A'). The fat body-specific C7-GAL4 driver was used to drive the expression of the UAS-Mp::GFP transgene. Immunostaining against Mp was used to compare the localization of the GFP tag (green) with the Mp protein (A’, white, A’’, magenta). Images show single confocal planes. Nuclei were counterstained with DAPI. Scale bar: 20 μm.
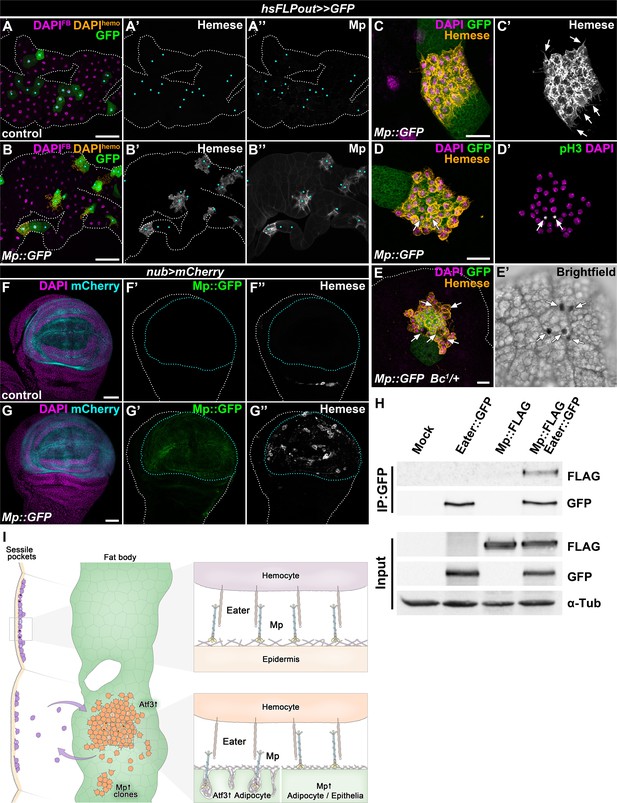
The interaction of Mp and Eater underlies hemocyte attachment to tissue surfaces.
(A–B) In contrast to expression in the whole adipose tissue, clonal overexpression of Mp::GFP in the fat body attracts hemocytes and causes local incorporation of Mp into adipocyte basement membrane, (B) compared to controls (A). Heat-shock induced FLPout clones were distinguished based on their expression of GFP. Clonal adipocytes are indicated with cyan dots. Nuclei were pseudocolored amber to indicate FBAH nuclei, and magenta to show adipocyte nuclei. (C–D) Hemocytes attached to the surface of Mp::GFP overexpressing adipocyte clones (C, D) cluster tightly together and extend filopodia (C', arrows), and some undergo cell division (D’). Immunostaining against Hemese visualizes hemocytes (C, D, amber, C', white), phospho-histone H3 staining shows mitotic nuclei (D', white, indicated with arrows). (E) Crystal cells (indicated with arrows) are present on the surface of Mp::GFP overexpressing adipocyte clones (E). Immunostaining against Hemese was used to visualize hemocytes, and melanized crystal cells can be identified due to the presence of the Bc1 mutation (E', black cells). (F–G) While in control wing discs no hemocytes can be observed on the basal side of the wing pouch (F), overexpression of Mp::GFP in this domain using the nub-GAL4, UAS-mCherry driver (F, G, cyan, outlined with cyan dotted lines) is sufficient to cause hemocyte attachment (G). The hemocytes were visualized with immunostaining against Hemese. Images represent projections of multiple confocal sections from the basal side of the wing disc. (H) Mp::FLAG co-precipitates with Eater::GFP from Drosophila S2 cells lysates. The GFP-tagged Eater served as the bait (IP:GFP). Eater and Mp proteins were detected with the anti-GFP and anti-FLAG tag-specific antibodies. The lower panel shows input extracts with α-Tubulin serving as a loading control. (I) Adipocyte-specific overexpression of Atf3 redirects hemocytes (orange) to the fat body surface (green) from the sessile hematopoietic pockets (purple), where they proliferate (cells with two nuclei) and trans-differentiate into crystal cells (cyan) and can detach from upon immune challenge, similar to their natural hematopoietic environment. The presence of Mp in the basement membrane promotes hemocyte attachment both in the sessile compartment and on the fat body surface upon Atf3 or clonal Mp expression through its interaction with the phagocytosis receptor Eater (right panels). Tissues were counterstained with DAPI (A–G). Images are projections of multiple confocal sections (A–G). Fat bodies (A, B, E) or wing discs (F) are outlined with white dotted lines. Scale bars: 100 μm (A, B), 20 μm (C–E), 50 μm (F, G). See also Figure 6—figure supplement 1.
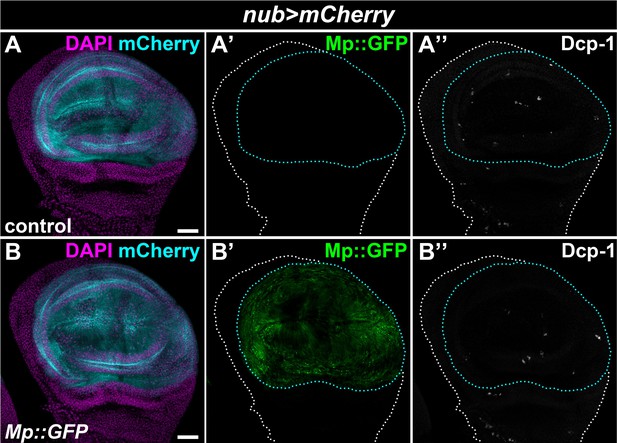
The overexpression of Mp:GFP in the wing pouch region does not induce apoptosis.
(A–B) Similar to controls (A), Mp::GFP overexpression in the wing pouch region did not result in increased apoptosis (B). Wing pouch-specific Mp::GFP and mCherry expression was driven by the nub-GAL4 driver. Apoptotic cells were visualized with immunostaining against cleaved Death caspase-1 (Dcp-1). Wing discs and the pouch regions are outlined with white and cyan dotted lines, respectively. Images are projections of multiple confocal sections from the basal side of the wing disc. Tissues were counterstained with DAPI. Scale bars: 50 μm.
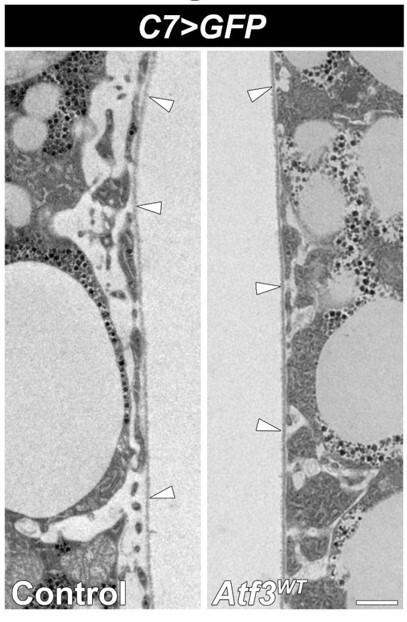
Transmission electron micrographs of control (left) and Atf3 overexpressing (right) adipocytes.
Transgene and GFP expression was driven by the fat bodyspecific C7-GAL4 driver. The average thickness of the BM is 65-85 nms in both genotypes. Basement membranes are indicated with arrowheads Scale bars: 2 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Drosophila melanogaster) | w1118 | BDSC | RRID:BDSC_3605 | |
| Strain, strain background (Drosophila melanogaster) | w; C7-GAL4 | Rynes et al., 2012 | ||
| Strain, strain background (Drosophila melanogaster) | w; UAS-Atf3WT | Sekyrova et al., 2010 | ||
| Strain, strain background (Drosophila melanogaster) | w; Hml:DsRed | Makhijani et al., 2011 | ||
| Strain, strain background (Drosophila melanogaster) | w; hsFLP, act>y+>GAL4, UAS-GFP | Sekyrova et al., 2010 | ||
| Strain, strain background (Drosophila melanogaster) | w; hsFLP, act>y+>GAL4, UAS-myr.mRFP | Sekyrova et al., 2010 | ||
| Strain, strain background (Drosophila melanogaster) | w;; Ubi-GFP.E2f11-230, Ubi-mRFP1.NLS.CycB1-266 | Zielke et al., 2014 | RRID:BDSC_55124 | |
| Strain, strain background (Drosophila melanogaster) | w;; BcF6:GFP | Tokusumi et al., 2009 | ||
| Strain, strain background (Drosophila melanogaster) | w; Bc1 | Rizki et al., 1980 | ||
| Strain, strain background (Drosophila melanogaster) | w; P[PTT-un1]vkgG454 | Morin et al., 2001 | DGRC 11069 | |
| Strain, strain background (Drosophila melanogaster) | w; MpRNAi | VDRC | v38189 | |
| Strain, strain background (Drosophila melanogaster) | w; MpRNAi II | VDRC | v35431 | |
| Strain, strain background (Drosophila melanogaster) | w; Cg25CRNAi | VDRC | v28369 | |
| Strain, strain background (Drosophila melanogaster) | w, trolRNAi | VDRC | v22642 | |
| Strain, strain background (Drosophila melanogaster) | w; vkgRNAi | VDRC | v16986 | |
| Strain, strain background (Drosophila melanogaster) | w; LanARNAi | VDRC | v18873 | |
| Strain, strain background (Drosophila melanogaster) | w; LanB1RNAi | VDRC | v23119 | |
| Strain, strain background (Drosophila melanogaster) | w; LanB2RNAi | VDRC | v42559 | |
| Strain, strain background (Drosophila melanogaster) | w; SPARCRNAi | VDRC | v16677 | |
| Strain, strain background (Drosophila melanogaster) | w;; a58-GAL4 | Galko and Krasnow, 2004 | ||
| Strain, strain background (Drosophila melanogaster) | w;; mef2-GAL4 | Ranganayakulu et al., 1996 | RRID:BDSC_27390 | |
| Strain, strain background (Drosophila melanogaster) | w, elav-GAL4 | Lin and Goodman, 1994 | RRID:BDRSC_458 | |
| Strain, strain background (Drosophila melanogaster) | eater1 | Bretscher et al., 2015 | RRID:BDSC_68388 | |
| Strain, strain background (Drosophila melanogaster) | w; HmlΔ-GAL4 | Sinenko and Mathey-Prevot, 2004 | RRID:BDSC_30139 | |
| Strain, strain background (Drosophila melanogaster) | w; nub-GAL4, UAS-mCherry | BDSC | RRID:BDSC_63148 | |
| Strain, strain background (Drosophila melanogaster) | w;; UAS-eaterRNAi | VDRC | v4301 | |
| Strain, strain background (Drosophila melanogaster) | w;; UAS-Mp::GFP | This study | ||
| Cell line (Drosophila melanogaster) | Schneider 2 (S2) cells | Drosophila Genomic Resource Center | RRID:CVCL_Z992 | |
| Recombinant DNA reagent | pENTR4-dual | Thermo Fisher | A10465 | |
| Recombinant DNA reagent | pTWG | DGRC | 1076 | |
| Recombinant DNA reagent | pTWF | DGRC | 1116 | |
| Recombinant DNA reagent | pTWG-Mp | This study | Used to generate UAS-Mp::GFP Drosophila line | |
| Transfected construct (Drosophila melanogaster) | pTWF-Mp | This study | Used to transfect S2 cells | |
| Transfected construct (Drosophila melanogaster) | pTWG-Eater | This study | Used to transfect S2 cells | |
| Transfected construct (Drosophila melanogaster) | pWA-GAL4 | Oda and Tsukita, 1999 | Used to transfect S2 cells | |
| Antibody | Anti-Hemese (mouse monoclonal) | Kurucz et al., 2003 (I. Ando) | H2 | IF (1:100) |
| Antibody | Anti-L1 (mouse monoclonal) | Kurucz et al., 2007b (I. Ando) | H10 | IF (1:100) |
| Antibody | Anti-NimrodC1 (mouse monoclonal) | Kurucz et al., 2007a (I. Ando) | N1+N47 | IF (1:100) |
| Antibody | Anti-Laminin (rabbit polyclonal) | Abcam | ab47651, RRID:AB_880659 | IF (1:500) |
| Antibody | Anti-alpha-Tubulin (mouse monoclonal) | DSHB | AA4.3, RRID:AB_579593 | IF (1:200), WB (1:1000) |
| Antibody | Anti-phospho-histone H3 (rabbit polyclonal) | Cell Signaling | Cat# 9701, RRID:AB_331535 | IF (1:500) |
| Antibody | Anti-cleaved-Dcp-1 (rabbit polyclonal) | Cell Signaling | Cat# 9578, RRID:AB_2721060 | IF (1:500) |
| Antibody | Anti-Endostatin (rat polyclonal) | Harpaz et al., 2013 (T. Volk) | IF (1:200), WB (1:1000) | |
| Antibody | Anti-FLAG M2 (mouse monoclonal) | Sigma-Aldrich | F1804, RRID:AB_439685 | WB (1:1000) |
| Antibody | Anti-GFP (rabbit polyclonal) | Acris | TP401, RRID:AB_2313770 | WB (1:5000) |
| Antibody | Anti-PPO1 (rabbit polyclonal) | Jiang et al., 1997 (M. Kanost) | WB (1:750) | |
| Antibody | Anti-mouse IgG Cy3 (donkey polyclonal) | Jackson Immuno Research | 715-165-1511 | IF (1:500) |
| Antibody | Anti-mouse IgG Cy5 (donkey polyclonal) | Jackson Immuno Research | 715-175-1510 | IF (1:500) |
| Antibody | Anti-rabbit IgG Cy3 (donkey polyclonal) | Jackson Immuno Research | 711-165-152 | IF (1:500) |
| Antibody | Anti-rabbit IgG Cy5 (donkey polyclonal) | Jackson Immuno Research | 711-175-152 | IF (1:500) |
| Antibody | Anti-rat IgG Cy5 (donkey polyclonal) | Jackson Immuno Research | 712-175-153 | IF (1:500) |
| Antibody | Anti-mouse IgG HRP (donkey polyclonal) | Jackson Immuno Research | 715-035-150 | WB (1:5000) |
| Antibody | Anti-rabbit IgG HRP (donkey polyclonal) | Jackson Immuno Research | 711-035-152 | WB (1:5000) |
| Antibody | Anti-rat IgG HRP (donkey polyclonal) | Jackson Immuno Research | 712-035-153 | WB (1:5000) |
| Chemical compound, drug | 4′,6-Diamidine-2′-phenylindole (DAPI) | Carl Roth GmBH. | 6335,1 | 1 mg/ml |
| Chemical compound, drug | N-Phenylthiourea | Sigma-Aldrich | P7629 | 0.01% w/V |
| Software, algorithm | FIJI | Schindelin et al., 2012 | http://fiji.sc, RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism 6 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Photoshop CS5.5 | Adobe Systems, Inc | RRID:SCR_014199 | |
| Software, algorithm | FluoView FV-10ASW | Olympus | RRID:SCR_014215 | |
| Software, algorithm | cellSens standard v1.11 | Olympus | RRID:SCR_014551 | |
| Software, algorithm | CellProfiler | Kamentsky et al., 2011 | RRID:SCR_007358 | |
| Other reagent | Phalloidin-Alexa 488 | Molecular Probes | A12379 | Used for F-actin staining |
| Other reagent | Dabco-Mowiol | Sigma-Aldrich | D27802,81381 | Mounting medium |
| Other reagent | TransIT-Insect Reagent | Mirus | MIR6100 | Tranfection reagent |
| Other reagent | GFP-trap beads | Chromotek | gtma-20, RRID:AB_2631406 | GFP trap beads for co-immunoprecipitation |
| Commercial assay or kit | Gateway LR Clonase II | Thermo Fisher | 11791–020 | Gateway clonase for entry-destination (LR) recombination |





