Echinoderms provide missing link in the evolution of PrRP/sNPF-type neuropeptide signalling
Figures
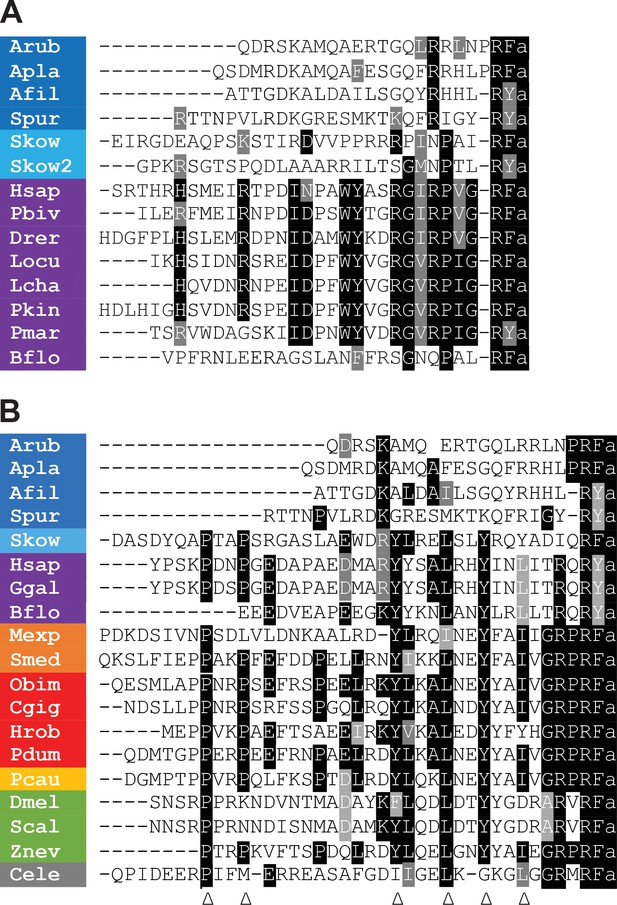
Comparison of the sequences of echinoderm NPY/NPF/PrRP-like peptides with related peptides in other taxa.
(A) Comparison with PrRP-type neuropeptides. Conserved residues are highlighted in black (identical) or grey (conservative substitutions) (B) Comparison with NPY/NPF-type neuropeptides. Conserved residues are highlighted in black (identical) or grey (conservative substitutions). The arrowheads indicate residues that have been shown to be important for the three-dimensional structure of the NPY/NPF-type peptides but which are not present in the echinoderm peptides. The colour coding of phyla is as follows: dark blue (Echinodermata), light blue (Hemichordata), purple (Chordata), orange (Platyhelminthes), red (Lophotrochozoa), yellow (Priapulida), green (Arthropoda), grey (Nematoda). The full names of the species and the accession numbers of the sequences are listed in Figure 1—source data 1.
-
Figure 1—source data 1
Accession numbers of the precursor sequences used for the peptide alignments in Figure 1.
Accession numbers of the precursor sequences used for the peptide alignments in Figure 1.
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig1-data1-v1.docx
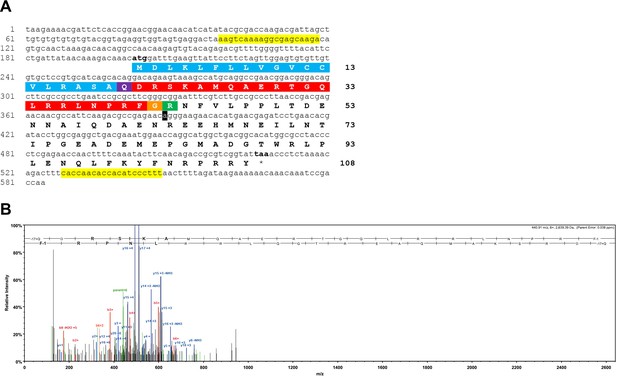
Sequencing of the A. rubens NPY/NPF/PrRP-like precursor cDNA and determination of the structure of the neuropeptide derived from the precursor.
(A) Sequence of a cDNA encoding the precursor of a NPY/NPF/PrRP-like peptide in A. rubens. The cDNA sequence (lowercase) comprises an open reading frame of 324 bases that encode a 108-residue protein (uppercase). The predicted signal peptide is shown in blue, the predicted cleavage site is shown in green and the predicted neuropeptide is shown in red, with an N-terminal glutamine that is a potential substrate for pyroglutamation shown in purple and with a C-terminal glycine that is a potential substrate for amidation shown in orange. The cDNA was amplified by PCR from A. rubens radial nerve cord cDNA using primers corresponding to the sequences highlighted in yellow. The cDNA was cloned in the vector pBluescript II SK (+) and the T3 and T7 primers were used for the sequencing. A single nucleotide that differs from a contig sequence (1060225) identified from A. rubens radial nerve cord transcriptome data is highlighted in black, but this is a synonymous substitution. This sequence has been deposited in GenBank under the accession number MK033631.1 (B) Annotated mass spectrum showing the structure of a NPY/NPF/PrRP-like peptide isolated from an A. rubens radial nerve cord extract. The peptide QDRSKAMQAERTGQLRRLNPRF, with Q1-Pyroglutamate (−17.02655 Da) and F22-amidated (−0.98402 Da), was observed at charge state +6, monoisotopic m/z 440.90631 Da with a precursor mass error of 0.12 ppm [MH+ 2640.40148 Da] and with a retention time (RT) of 66.3484 min. The b series of peptide fragment ions are shown in red, the y series in blue and additional identified peptide fragment ions in green. The peptide was identified with: Sequest HT (v1.17); XCorr: 4.27, Percolator q-Value: 0.0e0, Percolator PEP:1.6e-2. The fragment match tolerance used for the search was 0.8 Da. The fragmentation table for this mass spectrum can be found in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Fragmentation table of the mass spectrum for the A. rubens neuropeptide ArPrRP (QDRSKAMQAERTGQLRRLNPRF) shown in Figure 1—figure supplement 1B.
F22-amidated (−0.98402 Da), Q1-Pyroglutamate (−17.02655 Da), observed at charge state +6, monoisotopic m/z 440.90631 Da with a precursor mass error of 0.12 ppm).
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig1-figsupp1-data1-v1.xls
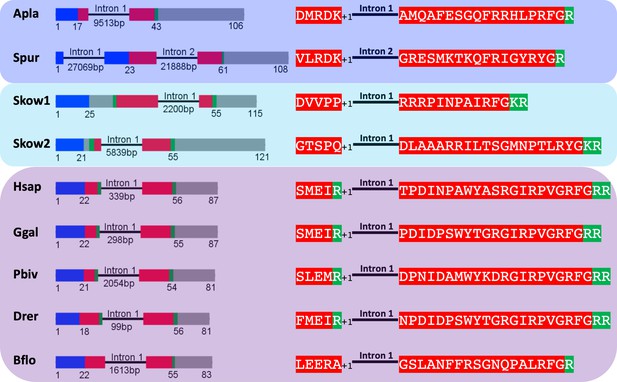
Comparison of exon/intron structure of genes encoding precursors of PrRP-like peptides in echinoderms and hemichordates with precursors of PrRP-type peptides in chordates.
Schematic representations of the gene structures are shown, with protein-coding exons shown as rectangles and introns shown as lines (with intron length stated underneath). The protein-coding exons are colour-coded to show regions that encode the N-terminal signal peptide (blue), the neuropeptide (red), monobasic or dibasic cleavage sites (green) and other regions of the precursor protein (grey). Note that a common characteristic is that an intron interrupts the coding sequence in the N-terminal or central region of the neuropeptide, with the intron consistently located between the first and second nucleotides (phase one intron represented by +1) of the codon for the amino acid shown after intron. Taxa are highlighted in phylum-specific colours: dark blue (Echinodermata), light blue (Hemichordata), purple (Chordata). The full names of the species and the accession numbers of the sequences are listed in Figure 2—source data 1.
-
Figure 2—source data 1
Accession numbers of the sequences used for the gene structure analysis in Figure 2 and Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig2-data1-v1.docx
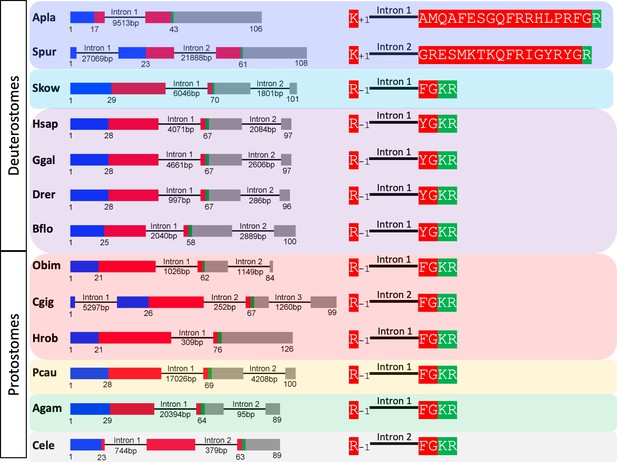
Comparison of the exon/intron structure of genes encoding echinoderm precursors of PrRP-like peptides and genes encoding NPY/NPF-type precursors in other taxa.
The left side of the figure shows schematic representations of the gene structures, with protein-coding exons shown as rectangles and introns shown as lines (with intron length stated underneath). The protein-coding exons are colour-coded to show regions that encode the N-terminal signal peptide (blue), the neuropeptide (red), monobasic or dibasic cleavage sites (green) and other regions of the precursor protein (grey). The right side of the figure shows how an intron interrupts the coding sequence of the neuropeptides, but the position of the intron is different for echinoderm PrRP-like peptides and bilaterian NPY/NPF-type neuropeptides. In genes encoding echinoderm PrRP-like peptides the intron interrupts the coding sequence in the N-terminal or central region of the PrRP-like peptide, with the intron located between the first and second nucleotides of the codon (phase one intron) for an alanine (A. planci) or glycine (S. purpuratus) residue and the position of the intron in the reading frame is therefore represented as +1. In contrast, in bilaterian NPY/NPF genes the intron interrupts the coding sequence for NPY/NPF-type peptides between the second and third nucleotide of the codon for the arginine residue of the C-terminal RF or RY dipeptide and the position of this phase two intron in the reading frame is represented as −1. Taxa are highlighted in phylum-specific colours: dark blue (Echinodermata), light blue (Hemichordata), purple (Chordata), red (Lophotrochozoa), yellow (Priapulida), green (Arthropoda), grey (Nematoda). Species names are as follows: Apla (Acanthaster planci), Spur (Strongylocentrotus purpuratus), Skow (Saccoglossus kowalevskii), Hsap (Homo sapiens), Ggal (Gallus gallus), Drer (Danio rerio), Bflo (Branchiostoma floridae), Obim (Octopus bimaculoides), Cgig (Crassostrea gigas), Hrob (Helobdella robusta), Pcau (Priapulus caudatus), Agam (Anopheles gambiae), Cele Caenorhabditis elegans.
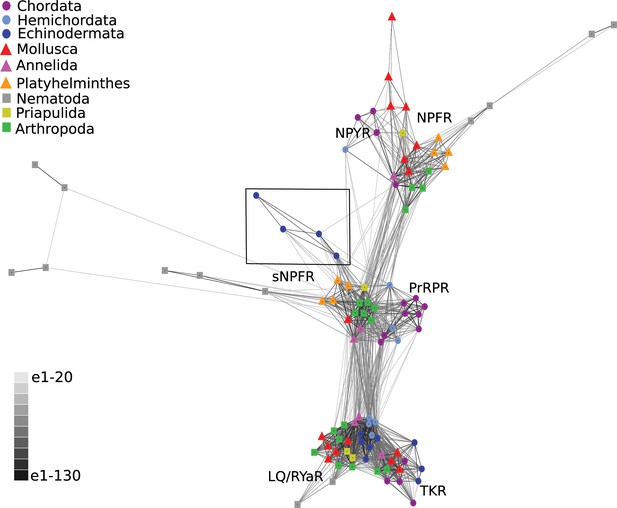
BLOSUM62 cluster map of NPY/NPF/PrPR/sNPF-type receptors and closely related tachykinin-type receptors (TKR) and luqin/RYamide-type receptors (LQ/RYaR).
Nodes are labelled with phylum-specific colours, as shown in the key, and connections represent BLAST relationships with a P value > 1e-65. Note that the echinoderm receptors (boxed) have more connections with PrRP/sNPF-type receptors than with NPY/NPF-type receptors. The sequences of the receptors included in this figure are listed in Figure 3—source data 1.
-
Figure 3—source data 1
Accession numbers of the receptor sequences used for the CLANS analysis in Figure 3.
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig3-data1-v1.docx

Asterias rubens sNPF/PrRP-type receptor (Ar-sNPF/PrRPR) transcript.
Sequence of a transcript (contig 1120879 from A. rubens radial nerve cord transcriptome) that encodes a sNPF/PrRP-type receptor. The transcript sequence (lowercase; numbering on the left) and the deduced amino acid sequence (uppercase and highlighted in grey; numbering on the right) are shown. A cDNA encoding the open reading frame was cloned by PCR using primers corresponding to the sequences highlighted in yellow, ligated into the vector pCDNA 3.1 (+) and sequenced. The sequence of the cloned cDNA was identical to the open reading frame of contig 1120879, with the exception of a single nucleotide substitution (t to c; highlighted in black), which results in replacement of an isoleucine residue (codon: ata in contig 1120879) with a threonine residue (codon: aca in the cloned cDNA). This sequence has been deposited in GenBank under the accession number MH807444.1.
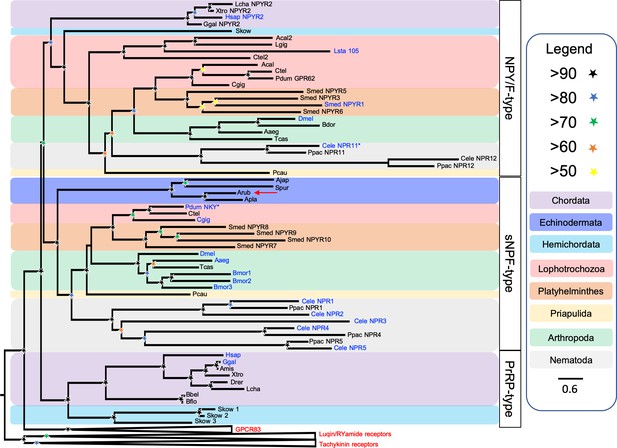
Phylogenetic tree showing that a candidate receptor for the A. rubens neuropeptide ArPrRP is an ortholog of protostome sNPF-type receptors.
The tree includes NPY/NPF-type receptors, chordate PrRP-type receptors and protostome sNPF-type receptors, with GPR83-type, luqin-type, and tachykinin-type receptors as outgroups to root the tree. Interestingly, the candidate receptor for the A. rubens neuropeptide ArPrRP (red arrow) and orthologs from other echinoderms are positioned in a clade comprising protostome sNPF-type receptors, whereas candidate receptors for PrRP-type peptides in the hemichordate S. kowalevskii are positioned in a clade containing chordate PrRP-type receptors. Note that NPY/NPF-type receptors form a distinct clade that includes an NPY/NPF-type receptor from the hemichordate S. kowalevskii, but no echinoderm receptors are present in this clade. The tree was generated in W-IQ-tree 1.0 using the Maximum likelihood method. The stars represent bootstrap support (1000 replicates, see legend) and the coloured backgrounds represent different taxonomic groups, as shown in the key. The names with text in blue represent the receptors for which ligands have been experimentally confirmed. The asterisks highlight receptors where the reported ligand is atypical when compared with ligands for receptors in the same clade. Species names are as follows: Aaeg (Aedes aegypti), Acal (Aplysia californica), Ajap (Apostichopus japonicus), Amis (Alligator mississippiensis), Apla (Acanthaster planci), Arub (Asterias rubens), Bbel (Branchiostoma belcheri), Bdor (Bactrocera dorsalis), Bflo (Branchiostoma floridae), Bmor (Bombyx mori), Cele (Caenorhabditis elegans), Cgig (Crassostrea gigas), Ctel (Capitella teleta), Dmel (Drosophila melanogaster), Drer (Danio rerio), Ggal (Gallus gallus), Hsap (Homo sapiens), Lcha (Latimeria chalumnae), Lgig (Lottia gigantea), Lsta (Lymnaea stagnalis), Pcau (Priapulus caudatus), Pdum (Platynereis dumerilii), Ppac (Pristionchus pacificus), Skow (Saccoglossus kowalevskii), Smed (Schmidtea mediterranea), Spur (Strongylocentrotus purpuratus), Tcas (Tribolium castaneum), Xtro (Xenopus tropicalis). The accession numbers of the sequences used for this phylogenetic tree are listed in Figure 4—source data 1.
-
Figure 4—source data 1
Accession numbers of the receptor sequences used for the phylogenetic analysis shown in Figure 4.
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig4-data1-v1.docx

Comparison of the sequences of ArPrRP and orthologs from other echinoderms with protostome sNPF-type peptides.
Conserved residues are highlighted in black or grey. Species names are highlighted in phylum-specific or superphylum-specific colours: blue (Echinodermata), red (Lophotrochozoa), orange-red (Platyhelminthes), green (Arthropoda) and grey (Nematoda). Species names are as follows: Aaeg (Aedes aegypti), Afil (Amphiura filiformis), Apla (Acanthaster planci), Arub (Asterias rubens), Bmor (Bombyx mori), Cele (Caenorhabditis elegans), Cgig (Crassostrea gigas), Dmel (Drosophila melanogaster), Lsta (Lymnaea stagnalis), Oara (Ophiopsila aranea), Pdum (Platynereis dumerilii), Smed (Schmidtea mediterranea), Spur (Strongylocentrotus purpuratus), Tcas (Tribolium castaneum). The accession numbers of the sequences included in this alignment are listed in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Accession numbers of the precursor sequences used for the peptide alignments in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig4-figsupp1-data1-v1.docx
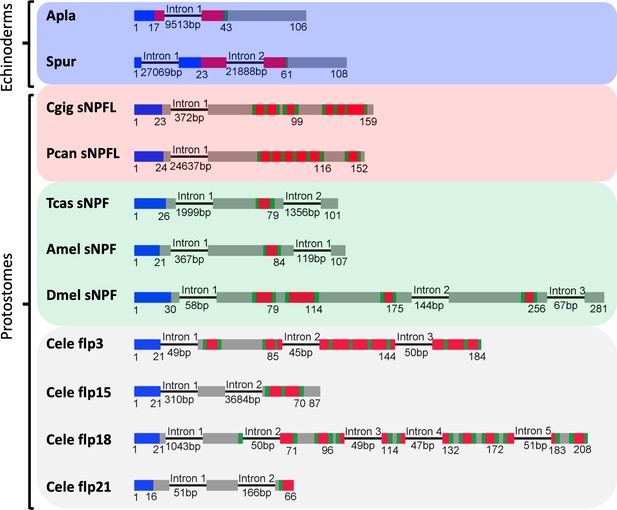
Comparison of the exon/intron structure of genes encoding echinoderm orthologs of the ArPrRP precursor and genes encoding protostome sNPF-type precursors.
Schematic representations of the gene structures are shown, with protein-coding exons shown as rectangles and introns shown as lines (with intron length stated underneath). The protein-coding exons are colour-coded to show regions that encode the N-terminal signal peptide (blue), the neuropeptide(s) (red), monobasic or dibasic cleavage sites (green) and other regions of the precursor protein (grey). The coloured backgrounds label the following the taxonomic groups: echinoderms (blue), lophotrochozoa (red), arthropods (green) and nematodes (grey). Species abbreviations: Apla (Acanthaster planci), Spur (Strongylocentrotus purpuratus), Cgig (Crassostrea gigas), Pcan (Pomacea canaliculata), Tcas (Tribolium castaneum), Amel (Apis mellifera), Dmel (Drosophila melanogaster), Cele (Caenorhabditis elegans). The accession numbers for the sequences of the precursors shown in this figure are listed in Figure 4—figure supplement 2—source data 1.
-
Figure 4—figure supplement 2—source data 1
Accession numbers of the precursor sequences used for the gene structure analysis in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig4-figsupp2-data1-v1.docx
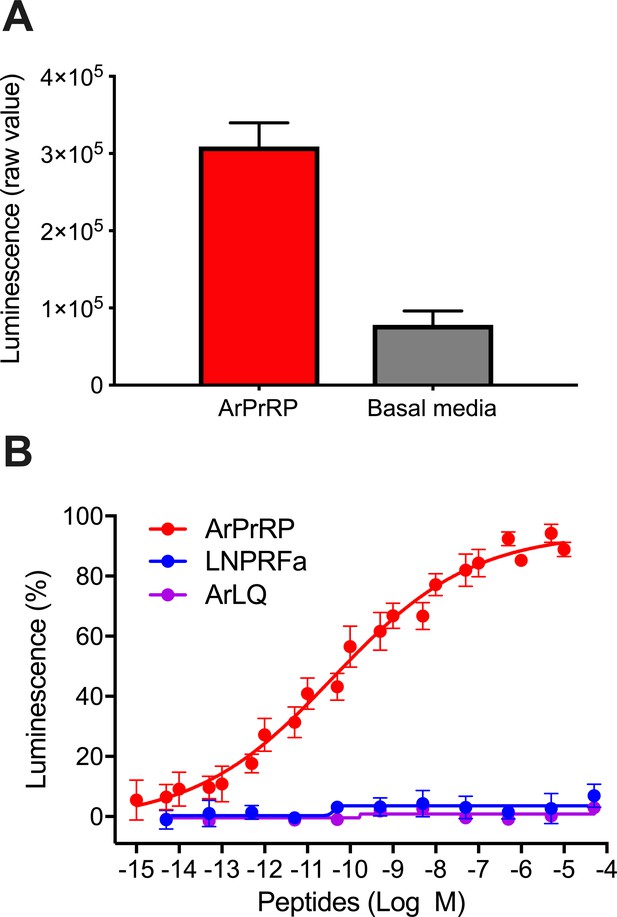
A. rubens PrRP-like peptide ArPrRP acts as a ligand for the A. rubens sNPF/PrRP-type receptor Ar-sNPF/PrRPR.
(A) The A. rubens PrRP-like peptide ArPrRP (10−5 M; red bar) triggers luminescence in CHO-K1 cells expressing the A. rubens PrRP/sNPF-type receptor Ar-sNPF/PrRPR, the promiscuous G-protein Gα16 and the calcium-sensitive luminescent GFP-apoaequorin fusion protein G5A. For comparison, the background luminescence of cells that were not exposed to ArPrRP is shown (basal media; grey bar). Mean values (± S.E.M) were determined from three independent experiments performed in triplicate (B). Graph showing the selectivity of ArPrRP as a ligand for Ar-sNPF/PrRPR. ArPrRP causes dose-dependent luminescence in CHO-K1 cells expressing Ar-sNPF/PrRPR, with an EC50 of 0.15 nM. Ar-sNPF/PrRPR is not activated by a C-terminal pentapeptide fragment of ArPrRP (LNPRFamide) or by the A. rubens luqin-type peptide ArLQ. Each point represents mean values (± S.E.M) from at least three independent experiments done in triplicate. The raw data for the experiments shown in Figure 5 and in Figure 5—figure supplement 2 can be found in Figure 5—source data 1.
-
Figure 5—source data 1
Data for the graphs shown in Figure 5 and Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/57640/elife-57640-fig5-data1-v1.xlsx
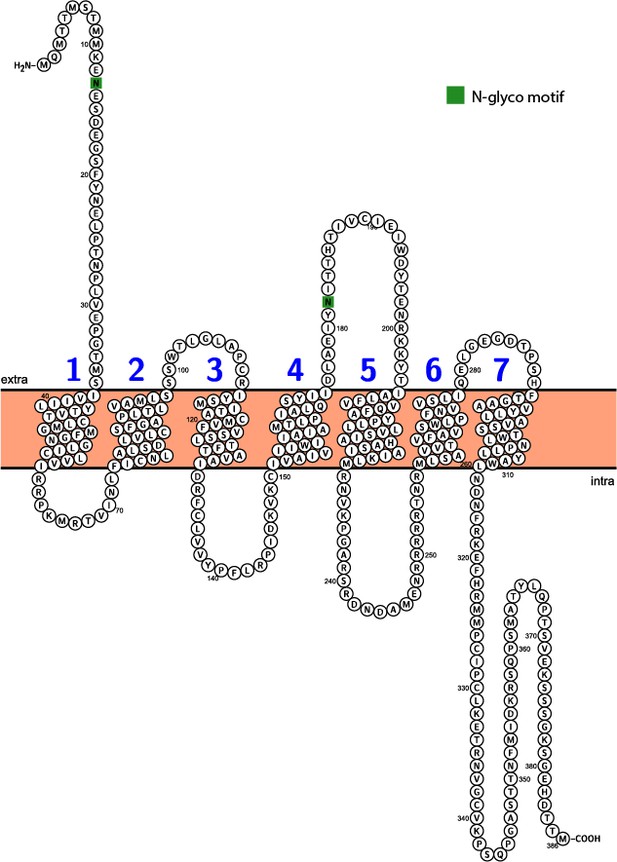
Predicted topology of Ar-sNPF/PrRPR.
Seven predicted transmembrane domains are numbered successively in blue and two predicted N-glycosylation sites are shown with green boxes. This figure was generated using Protter (http://wlab.ethz.ch/protter/start/).
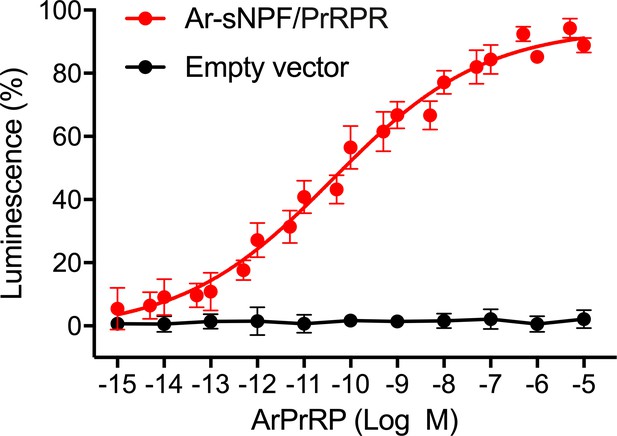
ArPrRP does not trigger luminescence in CHO-K1 cells transfected with an empty pcDNA 3.1(+) vector.
ArPrRP triggers dose-dependent luminescence in CHO-K1 cells transfected with pcDNA 3.1 (+) vector containing the coding sequence for the A. rubens sNPY/PrRP-type receptor Ar-sNPF/PrRPR (red circles; as also shown in Figure 5) but ArPrRP does not trigger luminescence in CHO-K1 cells transfected with an empty pcDNA 3.1(+) vector (black circles). Each point represents mean values (± S.E.M.) from at least three independent experiments, with each experiment performed in triplicate.
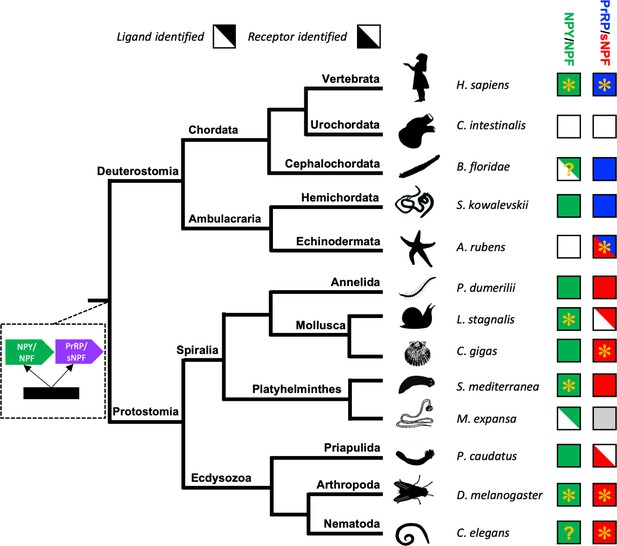
Phylogenetic diagram showing the occurrence of NPY/NPF-type, sNPF-type and PrRP-type neuropeptide signalling in the Bilateria.
The tree shows the phylogenetic relationships of selected bilaterian phyla. A gene duplication event giving rise to the paralogous NPY/NPF-type (green) and PrRP/sNPF (purple) signalling systems is shown at a position in the tree corresponding to the common ancestor of the Bilateria. Phyla in which NPY/NPF-type peptides/precursors and NPY/NPF-type receptors have been identified are labelled with green-filled squares. Phyla in which PrRP-type peptides/precursors and PrRP-type receptors have been identified are labelled with blue-filled squares. Phyla in which sNPF-type peptides/precursors and sNPF-type receptors have been identified are labelled with red-filled squares. The inclusion of an asterisk in filled squares indicates that activation of a receptor by a peptide ligand has been demonstrated experimentally. Note that in the starfish Asterias rubens (this study) a PrRP-type peptide (blue triangle) is the ligand for receptor that has been found to be an ortholog sNPF/PrRP-type receptors (Figure 3) or an ortholog of sNPF-type receptors (Figure 4); hence this receptor is represented here as a red triangle. Note also the mutually exclusive patterns in the phylogenetic distribution of sNPF-type signalling and PrRP-type signalling, with the former found in protostomes and the latter found in vertebrates, cephalochordates and hemichordates, which is supportive of the hypothesis that these signalling systems are orthologous. Our discovery of a PrRP/sNPF-type signalling system in echinoderms provides a missing link in the evolution of this neuropeptide signalling system. NPY/NPF-type signalling occurs in most phyla, but it has been lost in echinoderms and urochordates. The inclusion of a question mark for the putative NPY/NPF-type peptide identified in the cephalochordate B. floridae (Mirabeau and Joly, 2013; Elphick and Mirabeau, 2014) signifies that it is atypical of NPY/NPF-type peptides, which may explain why NPY/NPF-type receptors have yet to be identified in cephalochordates. The inclusion of a question mark in the C. elegans green square indicates that the peptide identified as a ligand for the C. elegans NPY/NPF-type receptor (Chalasani et al., 2010) does not have the typical features of an NPY/NPF-type peptide. The grey square for sNPF in M. expansa, for which only transcriptome sequence data are available, indicates that sNPF-type peptides and sNPF-type receptor(s) are likely to be present in this species because sNPF-type peptides and sNPF-type receptors have been identified in another platyhelminth species, S. mediterranea, for which a genome sequence is available. Species names are as follows: H. sapiens (Homo sapiens), C. intestinalis (Ciona intestinalis), B. floridae (Branchiostoma floridae), S. kowalevskii (Saccoglossus kowalevskii), A. rubens (Asterias rubens), P. dumerilii (Platynereis dumerilii), L. stagnalis (Lymnaea stagnalis), M. expansa (Moniezia expansa), S. mediterranea (Schmidtea mediterranea), C. gigas (Crassostrea gigas), D. melanogaster (Drosophila melanogaster), C. elegans (Caenorhabditis elegans). Silhouettes of representative animals from each phylum are from www.openclipart.com and they are free from copyright.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | pBluescript II KS (+) plasmid (cloning vector) | Invitrogen | Cat# K280002 | |
| Recombinant DNA reagent | pcDNA3.1(+) with neomycin selectable marker (mammalian expression vector) | Invitrogen | Cat# V790-20 | |
| Commercial assay, kit | Lipofectamine 3000 | Invitrogen | Cat# L3000015 | |
| Transfected construct (Asterias rubens) | Asterias rubens sNPF/PrPR receptor cDNA cloned into an expression vector | This paper | Genbank: MH807444 | Cloned in the plasmid pcDNA3.1+ from Invitrogen |
| Transfected construct (Aequorea victoria) | Chimeric green fluorescent protein-aequorin fusion protein (G5A) | Baubet et al., 2000 | N/A | Cloned into the pEGFP-C1 vector (CLONTECH) |
| Transfected construct (Homo sapiens) | Human guanine nucleotide binding protein, alpha 15 (16) (Gq class) | cDNA resource center | Cat# GNA1500000 | HGNC ID:4383 Human GNA15 cloned into the plasmid pcDNA3.1+. |
| Cell line (Cricetulus griseus) | Chinese hamster ovary cells (CHO-K1) | Sigma-Aldrich | RRID:CVCL_0214 | Cat. No. 85051005 |
| Software, algorithm | Prism | GraphPad | Version 7.0 | |
| Software, algorithm | Sequest Proteome Discoverer | Thermo Fisher Scientific | Version 2.2 | |
| Software, algorithm | Scaffold | Proteome Software | Version 4.8.4 |





