Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens
Figures
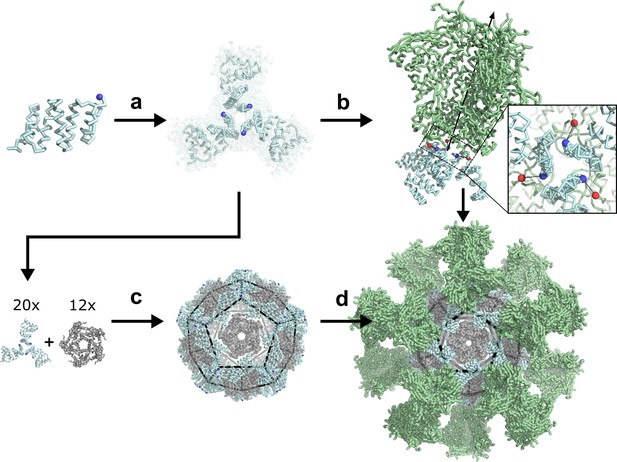
De novo design of protein nanoparticles tailored for multivalent antigen presentation.
(a) Computational docking of monomeric repeat proteins into C3-symmetric trimers using the RPX method. (b) Selection of trimers for design based on close geometric match between their N termini (blue spheres) and C termini (red spheres) of a viral antigen (green, BG505 SOSIP shown for illustration). (c) Design of two-component nanoparticles incorporating a fusion component (cyan) and assembly component (gray). (d) Nanoparticle assembled with antigen-fused trimeric component yields multivalent antigen-displaying nanoparticle.
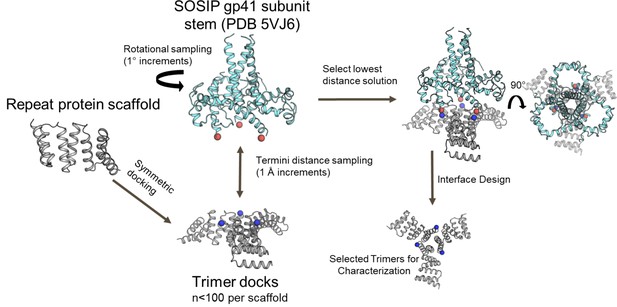
Computational docking and design of trimers for fusion to a specific viral glycoprotein.
Design models for C3-symmetric trimers (gray) with labeled N termini (blue spheres) screened against the BG505 SOSIP trimeric glycoprotein subunit gp41 (cyan) with C-terminal residues labeled (red spheres).
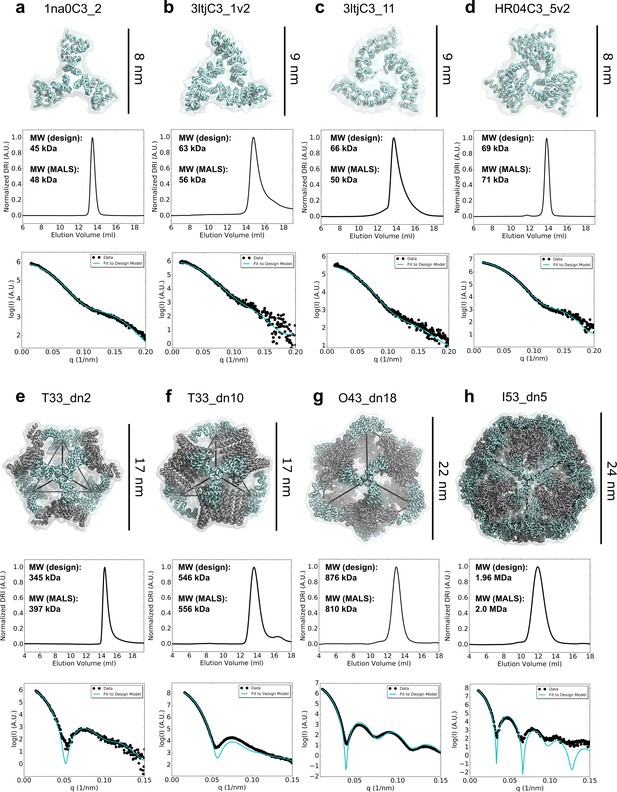
Biophysical characterization of antigen-tailored trimers and nanoparticles.
Top rows, design models. Middle rows, SEC chromatograms and calculated molecular weights from SEC-MALS. Bottom rows, comparisons between experimental SAXS data and scattering profiles calculated from design models. (a) 1na0C3_2. (b) 3ltjC3_1v2. (c) 3ltjC3_11. (d) HR04C3_5v2. (e) T33_dn2. (f) T33_dn10. (g) O43_dn18. (h) I53_dn5.
-
Figure 2—source data 1
Biophysical properties of designed trimers and two-component nanoparticles.
Experimentally-measured data (exp) is compared to predicted design data (model). Molecular weights (MW) were obtained using the ASTRA software. Rg and Dmax calculations performed in Scatter3 SAXS analysis software with the determined qmax values. X values computed from the FoXS online SAXS web server between the designed model and the experimental scattering data.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data1-v1.docx
-
Figure 2—source data 2
1na0C3_2 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data2-v1.txt
-
Figure 2—source data 3
3ltjC3_1v2 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data3-v1.txt
-
Figure 2—source data 4
3ltjC3_11 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data4-v1.txt
-
Figure 2—source data 5
HR04C3_5v2 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data5-v1.txt
-
Figure 2—source data 6
1na0C3_2 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data6-v1.txt
-
Figure 2—source data 7
3ltjC3_1v2 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data7-v1.txt
-
Figure 2—source data 8
3ltjC3_11 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data8-v1.txt
-
Figure 2—source data 9
HR04_5v2 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data9-v1.txt
-
Figure 2—source data 10
T33_dn2 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data10-v1.txt
-
Figure 2—source data 11
T33_dn10 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data11-v1.txt
-
Figure 2—source data 12
O43_dn18 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data12-v1.txt
-
Figure 2—source data 13
I53_dn5 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data13-v1.txt
-
Figure 2—source data 14
T33_dn2 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data14-v1.txt
-
Figure 2—source data 15
T33_dn10 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data15-v1.txt
-
Figure 2—source data 16
O43_dn18 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data16-v1.txt
-
Figure 2—source data 17
I53_dn5 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-data17-v1.txt
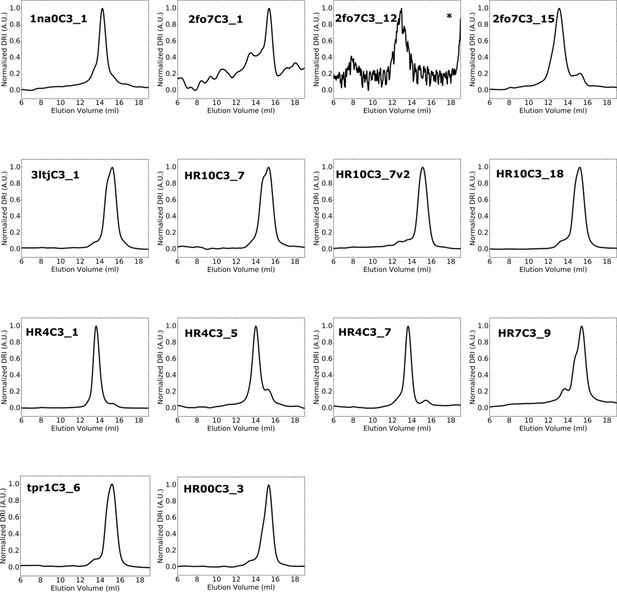
SEC-MALS chromatograms for designed trimers occupying an off-target oligomeric state.
Predominant oligomeric species for each design were collected by fractionation from initial SEC purification, and fourteen chromatograms are presented here from a subsequent round of high-performance SEC-MALS using a Superdex 200 column. *Chromatogram obtained using a Superdex 75 10/300 GL column.
-
Figure 2—figure supplement 1—source data 1
SEC-MALS data for off-target designed trimers.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-figsupp1-data1-v1.docx
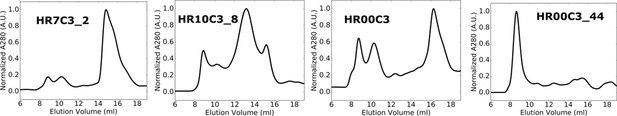
SEC chromatograms for designed trimers with off-target retention volumes after Ni2+ IMAC.
Primary SEC chromatograms obtained from a Superdex 200 column for soluble proteins after purification by Ni2+ IMAC. Designs presented here formed off-target or polydisperse assemblies based on retention volume.
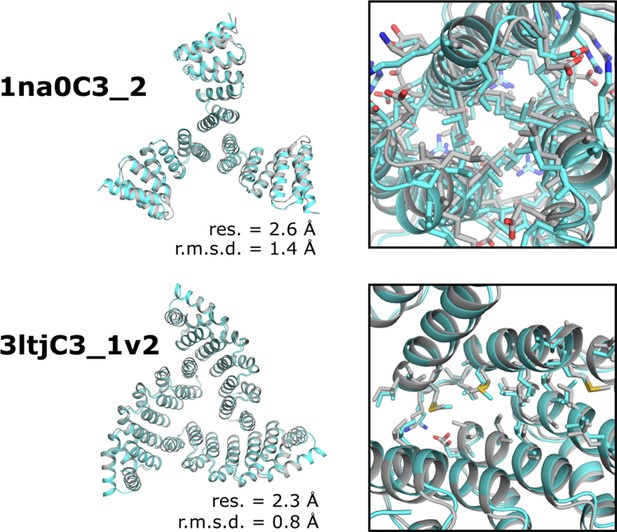
Comparison between the experimentally determined crystal structures and corresponding models of two designed trimers.
Left, design models (gray) and crystal structures (gray) superposed indicating resolution of structure (res.) and backbone r.m.s.d. between structure and design model. Right, magnified view of the de novo designed interface and side chain packing.
-
Figure 2—figure supplement 3—source data 1
Crystallography data collection and refinement statistics for designed trimers 1na0C3_2 and 3ltjC3_1v2.
Statistics for the highest-resolution shell are shown in parentheses.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-figsupp3-data1-v1.docx
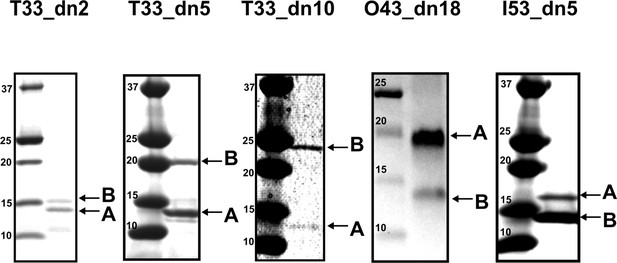
SDS-PAGE of bicistronically-expressed designed nanoparticles eluted from Ni2+ IMAC.
For each designed nanoparticle: Left - protein standard (Precision Plus Dual Xtra, Bio-Rad). Right - labeled bands corresponding to the expected size of each component.
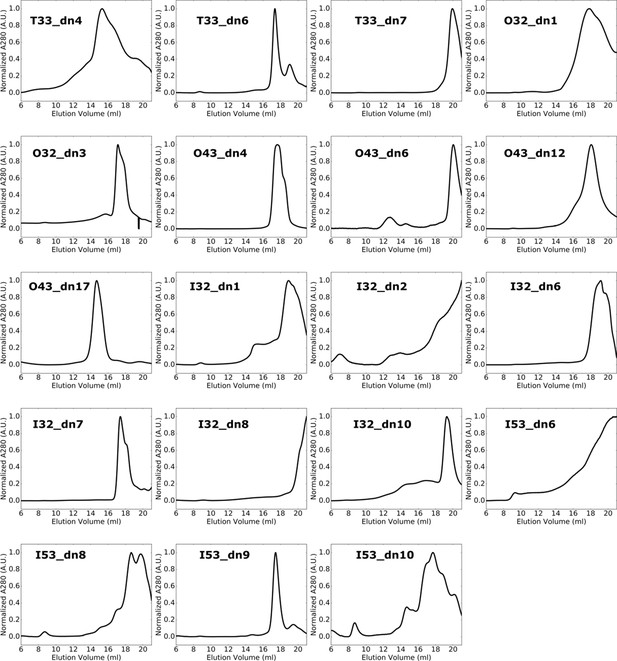
SEC profiles for two-component nanoparticles with off-target retention volumes after Ni2+ IMAC.
Primary SEC chromatograms obtained from a Superose six column for soluble proteins directly after purification by Ni2+ IMAC. Designs presented here formed off-target or polydisperse assemblies based on retention volume.
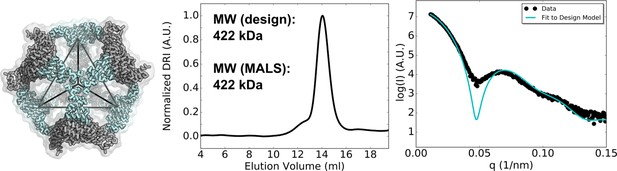
Biophysical characterization of T33_dn5.
Left - designed model. Middle - SEC chromatograms and calculated molecular weights from SEC-MALS. Right - comparisons between experimental SAXS data and scattering profile computed from the design model.
-
Figure 2—figure supplement 6—source data 1
T33_dn5 SEC-MALS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-figsupp6-data1-v1.txt
-
Figure 2—figure supplement 6—source data 2
T33_dn5 SAXS.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig2-figsupp6-data2-v1.txt
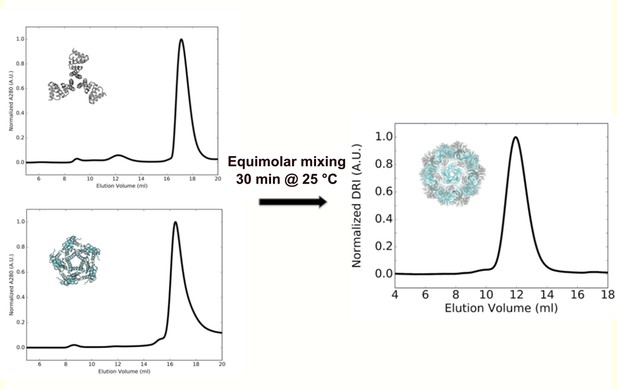
In vitro assembly of I53_dn5.
SEC chromatograms of individual components I53_dn5A (pentameric assembly component, cyan) and I53_dn5B (trimeric fusion component, gray), and nanoparticle purified from equimolar assembly run on a Superose 6 10/300 GL column.
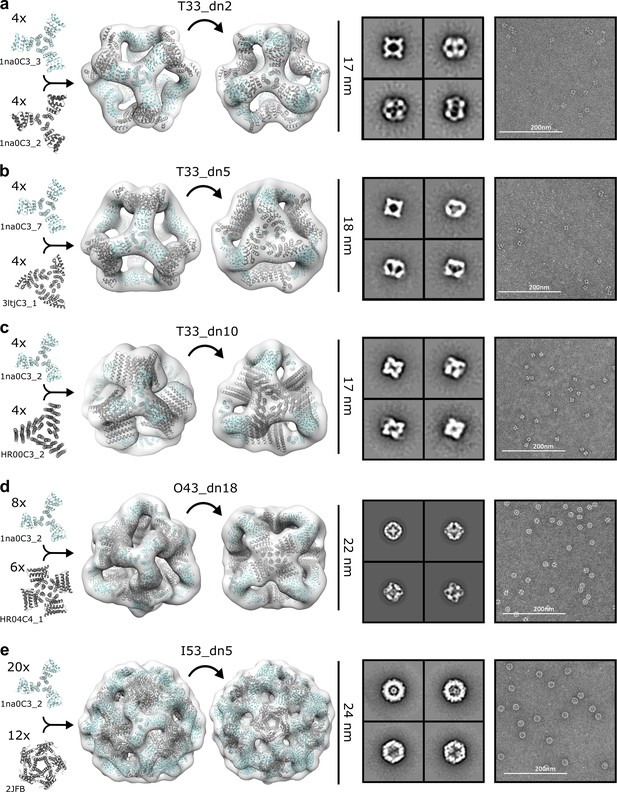
NS-EM analysis of antigen-tailored nanoparticles.
From left to right: designed trimers incorporated in each designed nanoparticle, nanoparticle design models fit into NS-EM density (views shown down each component axis of symmetry), designed nanoparticle 2D class-averages, raw electron micrographs of designed nanoparticles. (a) T33_dn2. (b) T33_dn5. (c) T33_dn10. (d) O43_dn18. (e) I53_dn5.
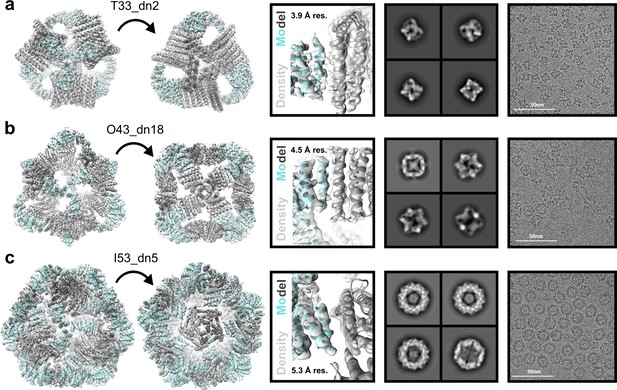
Cryo-EM analysis of antigen-tailored nanoparticles.
From left to right: cryo-EM maps with refined nanoparticle design models fit into electron density, view of designed nanoparticle interface region fit into cryo-EM density with indicated resolution (res.), designed nanoparticle 2D class-averages, raw cryo-EM micrographs of designed nanoparticles. (a) T33_dn10. (b) O43_dn18. (c) I53_dn5.
-
Figure 4—source data 1
Cryo-EM data acquisition metrics for designed nanoparticles T33_dn10, O43_dn18, and I53_dn5.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig4-data1-v1.docx
-
Figure 4—source data 2
Cryo-EM model building and refinement statistics for designed nanoparticles T33_dn10, O43_dn18, and I53_dn5.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig4-data2-v1.docx
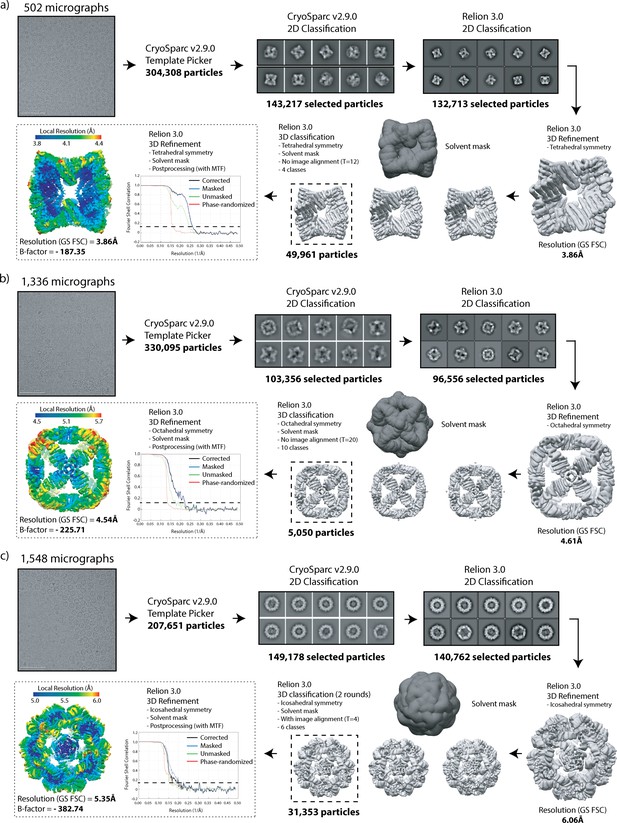
Cryo-EM data processing workflow.
Statistics are presented for (a) T33_dn10, (b) O43_dn18, and (c) I53_dn5.
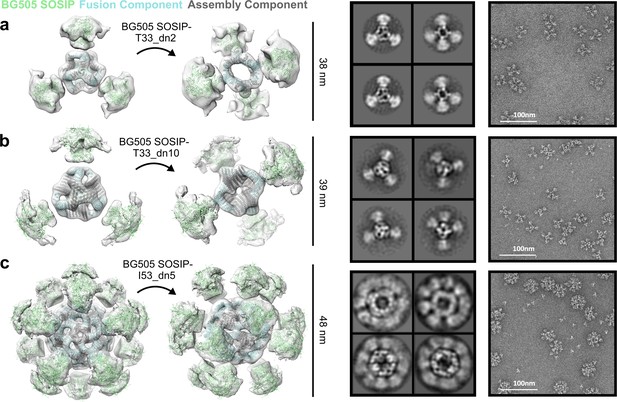
NS-EM analysis of BG505 SOSIP-displaying nanoparticles.
From left to right: BG505 SOSIP-displaying nanoparticle models fit into NS-EM density, 2D class-averages, raw NS-EM micrographs of assembled BG505 SOSIP-displaying nanoparticles. (a) BG505 SOSIP–T33_dn2. (b) BG505 SOSIP–T33_dn10. (c) BG505 SOSIP–I53_dn5.
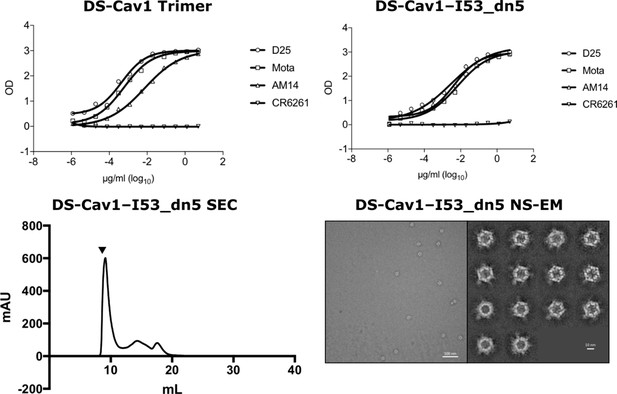
Structural and antigenic characterization of DS-Cav1–I53_dn5.
Top panel: ELISA using anti-DS-Cav1 antibodies D25, Motavizumab (Mota), AM14, or negative control CR6261 added to DS-Cav1 trimer with foldon or antigen-displaying nanoparticle DS-Cav1–I53_dn5. Bottom panel: SEC chromatogram of DS-Cav1–I53_dn5 on a Superose six column and NS-EM field image and 2D class averages for DS-Cav1–I53_dn5.
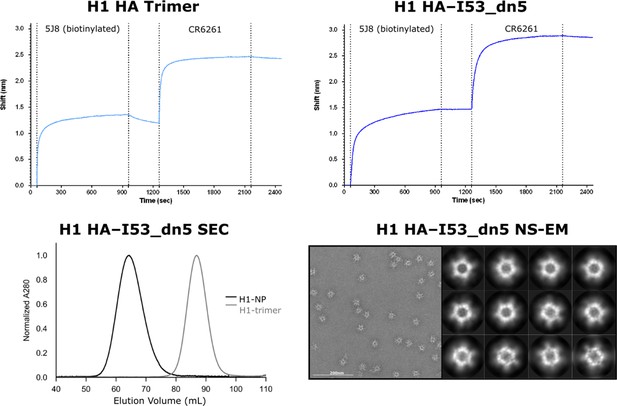
Structural and antigenic characterization of HA–I53_dn5.
Top panel: Octet bio-layer interferometry using plate-coated head-directed mAb 5J8 for antigen capture, and subsequent stem-directed mAb CR6261 addition to both antigen-fused trimeric component HA–I53_dn5B and antigen-displaying nanoparticle HA–I53_dn5. Bottom panel: SEC chromatogram for nanoparticle HA–I53_dn5 compared to trimer HA–I53_dn5B on a Sephacryl S-500 column, and NS-EM field image and 2D class averages for HA–I53_dn5.
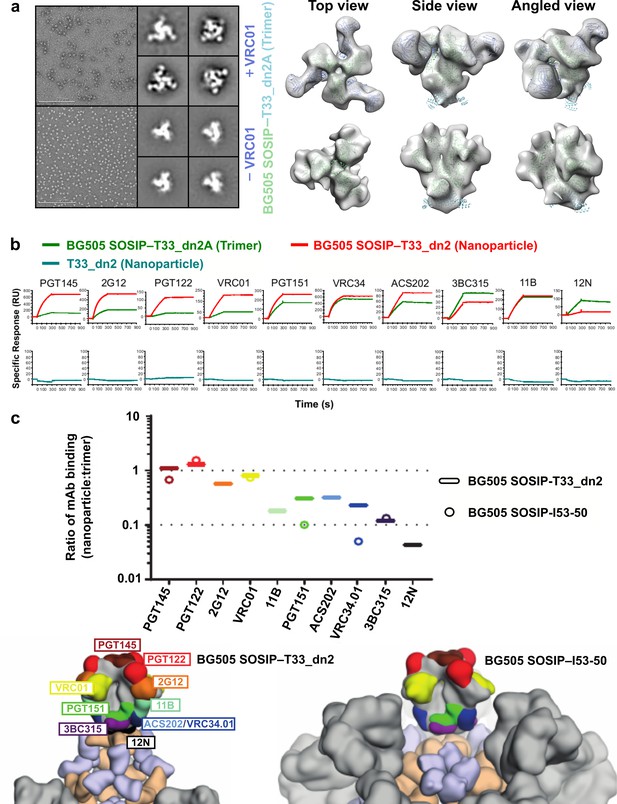
BG505 SOSIP epitope accessibility compared between tetrahedral and icosahedral presentation geometries.
(a) NS-EM micrographs of BG505 SOSIP–T33_dn2A with and without VRC01 Fab bound, 2D class averages, and models fit into NS-EM density. (b) Representative sensorgrams of indicated proteins binding to anti-Env mAbs. (c) Relative accessibility of epitopes on BG505 SOSIP–T33_dn2 nanoparticles and BG505 SOSIP–I53-50 nanoparticles as determined by mAb binding (top). Ratio of moles of macromolecules are means of 2–4 experimental replicates. Epitopes mapped onto BG505 SOSIP are presented on models of T33_dn2 and I53-50 (bottom). Wheat, antigen-fused trimeric component; purple, assembly component; gray, neighboring BG505 SOSIP trimers on the nanoparticle surface.
-
Figure 6—source data 1
BG505 SOSIP-T33_dn2 SPR Data.
- https://cdn.elifesciences.org/articles/57659/elife-57659-fig6-data1-v1.xlsx
Tables
Summary of the experimental characterization for designed trimers and two-component nanoparticles.
1na0C3_2 and 3ltjC3_1v2 structures determined by X-ray crystallography and T33_dn10, O43_dn18, and I53_dn5 structures determined by cryo-EM.
| Design | Targeted Antigens | Experimental Molecular Weight (kDa) | Target Molecular Weight (kDa) | SAXS X value | Resolution, backbone r.m.s.d. structure (Å, Å) |
|---|---|---|---|---|---|
| 1na0C3_2 | HA, SOSIP, DS-Cav1 | 48 | 45 | 1.4 | 2.6, 1.4 |
| 3ltjC3_1v2 | SOSIP, DS-Cav1 | 56 | 63 | 1.1 | 2.3, 0.8 |
| 3ltjC3_11 | SOSIP, DS-Cav1 | 50 | 66 | 1.6 | -- |
| HR04C3_5v2 | SOSIP | 71 | 69 | 1.5 | -- |
| T33_dn2 | HA, SOSIP, DS-Cav1 | 397 | 345 | 4.8 | -- |
| T33_dn5 | HA, SOSIP, DS-Cav1 | 422 | 422 | 1.7 | -- |
| T33_dn10 | HA, SOSIP, DS-Cav1 | 546 | 556 | 2.3 | 3.9, 0.65 |
| O43_dn18 | HA,SOSIP, DS-Cav1 | 810 | 876 | 2.9 | 4.5, 0.98 |
| I53_dn5 | HA, SOSIP, DS-Cav1 | 2000 | 1960 | 1.2 | 5.3, 1.30 |
-
Table 1—source data 1
Summary of the experimental characterization for designed trimers and two-component nanoparticles.
- https://cdn.elifesciences.org/articles/57659/elife-57659-table1-data1-v1.docx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | RPX Method | PMID:28338692 | Symmetric docking and scoring protocol | |
| Software, algorithm | Sic_axle | PMID:30849373 | Protein structure alignment protocol | |
| Software, algorithm | Rosetta Macromolecular Modeling Suite | PMID:28430426 | RRID:SCR_015701 | Version 3 |
| Software, algorithm | Relion | PMID:23000701 | RRID:SCR_016274 | Cryo-EM structure determination software |
| Strain, strain background (E. coli) | BL21 | New England Biolabs | Cat. #:C2527H | Competent T7 expression strain |
| Strain, strain background (E. coli) | Lemo21 | New England Biolabs | Cat. #:C2527H | Competent T7 expression strain |
| Strain, strain background (E. coli) | HEK293F | PMID:26779721 | RRID:CVCL_6642 | Suspension-based cells for high yield expression of recombinant proteins |
| Chemical compound, drug | IPTG | Sigma | Cat. #:I6758 | Induces protein expression through T7 promoter |
| Chemical compound, drug | Kanamycin | Sigma | Cat. #:K1377 | Antibiotic |
| Chemical compound, drug | Carbenicillin | Sigma | Cat. #:C1389 | Antibiotic |
| Chemical compound, drug | Expifectamine | ThermoFisher | Cat. #:A38915 | Transfection reagent |
| Chemical compound, drug | Polyethylenimine | Polysciences Inc | Cat. #:23966 | Transfection reagent |
| Recombinant DNA reagent | pET21b(+) | Genscript | Addgene Cat. #:69741–3 | Bacterial expression vector |
| Recombinant DNA reagent | pET28b(+) | Gen9 | Addgene Cat. #:69865–3 | Bacterial expression vector |
| Recombinant DNA reagent | pPPI4 | Progenics Pharmaceuticals Inc PMID:10623724 | Mammalian secretion vector, containing codon-optimized stabilized gp140 | |
| Recombinant DNA reagent | CMV/R | PMID:15994776 | Mammalian secretion vector, containing CMV enhancer/promoter with HTLV-1 R region | |
| Antibody | PGT145 human monoclonal | PMID:21849977 | RRID:AB_2491054 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) |
| Antibody | PGT122 human monoclonal | PMID:21849977 | RRID:AB_2491042 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) |
| Antibody | 2G12 human monoclonal | PMID:8551569 | RRID:AB_2819235 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) |
| Antibody | VRC01 human monoclonal | PMID:20616233 | RRID:AB_2491019 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) |
| Antibody | ACS202 human monoclonal | PMID:27841852 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) | |
| Antibody | VRC34 human monoclonal | PMID:27174988 | RRID:AB_2819228 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) |
| Antibody | PGT151 human monoclonal | PMID:24768347 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) | |
| Antibody | 3BC315 human monoclonal | PMID:22826297 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) | |
| Antibody | 11B rabbit monoclonal | PMID:27545891 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) | |
| Antibody | 12N rabbit monoclonal | PMID:27545891 | anti-HIV-1 Env (anti-Fc immobilization level of 320 ± 1.5 RU) | |
| Antibody | 5J8 human monoclonal | PMID:21849447 | anti-HA (20 μg/mL) | |
| Antibody | CR6261 human monoclonal | PMID:19079604 | anti-HA (20 μg/mL) | |
| Antibody | D25 human monoclonal | PMID:24179220 | anti-RSV F (1 pg/mL - 10 μg/mL) | |
| Antibody | Motavizumab mouse-human chimeric monoclonal | PMID:20065632 | anti-RSV F (1 pg/mL - 10 μg/mL) | |
| Antibody | AM14 human monoclonal | PMID:26161532 | anti-RSV F (1 pg/mL - 10 μg/mL) |
Additional files
-
Supplementary file 1
Sequences for all designed trimers, homo-oligomers, two-component nanoparticles, and antigen-fused components.
(A) Amino acid sequences for all designed trimers and de novo homo-oligomers used for two-component nanoparticle design. Sequences include initiating methionines and His6-tags. Designed trimers that expressed solubly are denoted in bold, and experimental methods used for characterization are included in parentheses. *Components from previously described designed homo-oligomers in Fallas et al., 2017 or the Protein Data Bank (PDB ID). (B) Amino acid sequences for all designed two-component nanoparticles. Sequences include initiating methionines and His6-tags. Designs that expressed solubly and co-eluted from IMAC are denoted in bold. Input oligomers from (A) are included in parentheses. (C) Amino acid sequences for all antigen-fused trimeric nanoparticle components. Sequences include initiating methionines and signal peptides.
- https://cdn.elifesciences.org/articles/57659/elife-57659-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57659/elife-57659-transrepform-v1.docx





