EHMT2 epigenetically suppresses Wnt signaling and is a potential target in embryonal rhabdomyosarcoma
Figures
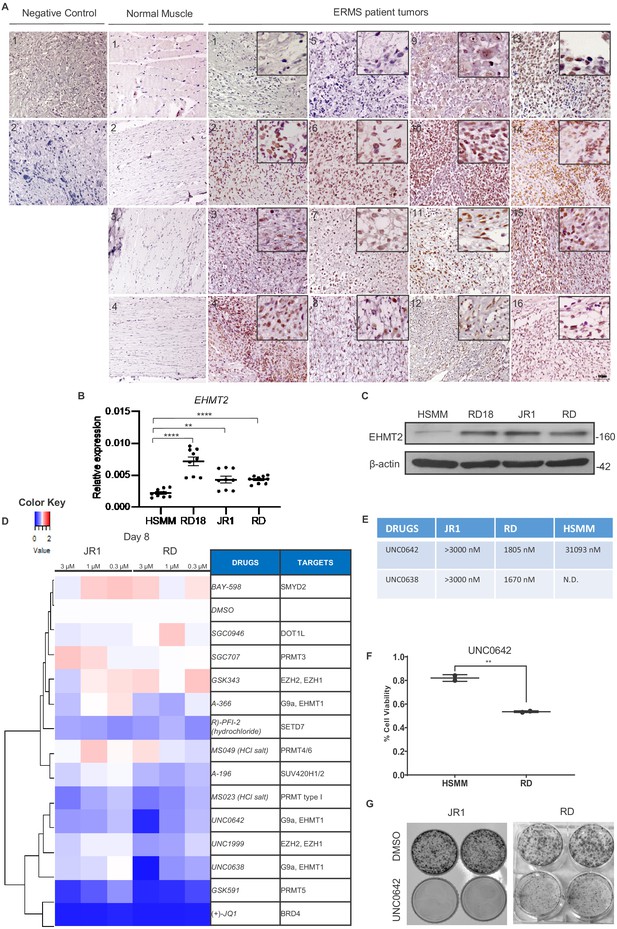
EHMT2 is overexpressed in embryonal rhabdomyosarcoma (ERMS).
(A) 16 archival ERMS patient tumor specimens and four normal muscle samples were analyzed by immunohistochemistry using anti-EHMT2 antibody. Negative control indicates staining using secondary antibody alone. Inset shows zoomed in image of nuclear EHMT2 staining. Scale bar: 100 μm. (B) EHMT2 mRNA (n = 3) were examined in three patient-derived cell lines (RD, RD18, and JR1) in comparison to primary human skeletal muscle myoblasts (HSMMs) by qPCR. Values correspond to the average ± SEM. All three ERMS cell lines examined showed an increased EHMT2 mRNA expression compared to HSMM. (C) EHMT2 protein levels were examined by western blotting in HSMM, RD18, JR1, and RD cells. A representative image of three different experiments is shown. All three ERMS cell lines examined showed an increased EHMT2 protein expression compared to HSMM. (D) ERMS cell lines JR1 and RD were treated with the indicated methyltransferase inhibitors (3, 1, and 0.3 µM). Viability on day 8 was scored by MTS assay and measured as the ratio over control cells treated with an equivalent dilution of DMSO. RED indicates viability >control; WHITE is equal to control, and BLUE is less than control. The experiment was conducted in triplicates and (+)-JQ1 was used as a positive control. GSK591, UNC0642, and UNC638 had a strong effect on viability. (E) The IC50 of EHMT2 inhibitors in JR1, RD, and HSMM is shown. (F) HSMM and RD cells were treated with DMSO or UNC0642 for 6 days. Cell viability was assessed by trypan blue staining. (G) JR1 and RD cells were treated with DMSO or UNC0642 for 9 days. Colony formation was assessed by staining with crystal violet. A representative image of three different experiments is shown. In (B) data from three independent biological replicates each with three technical replicates were plotted. Statistical significance was calculated by unpaired two-tailed t-test. **p≤0.01, ***p≤0.001. N.D. = not determined.
-
Figure 1—source data 1
qPCR data for endogenous G9a expression in ERMS cell lines.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig1-data1-v3.xlsx
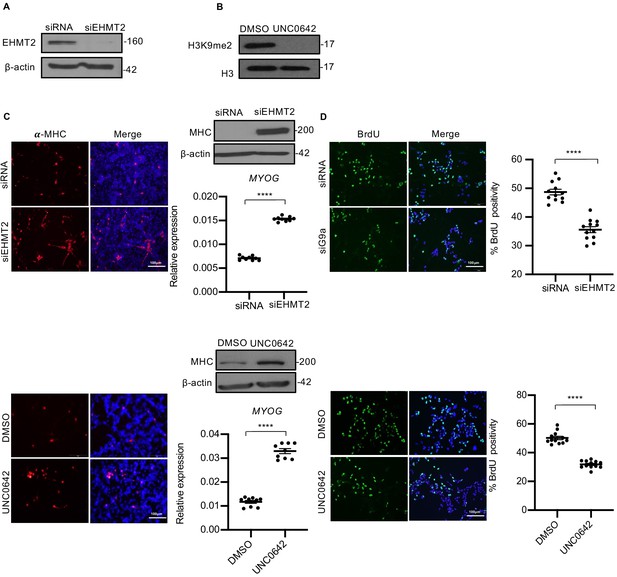
EHMT2 inhibits differentiation and promotes proliferation of myoblasts.
(A) EHMT2 was depleted in RD18 cells using siRNA. Control and siEHMT2 cells were analyzed for knockdown efficiency by western blot. β-actin was used as an internal loading control. (B) H3K9me2 levels were analyzed 48 hr after 2.5 μM of UNC0642 treatment in RD18 cells. Histone H3 was used as a loading control. (C) Differentiation was analyzed in control and siEHMT2 RD18 cells (upper panels) or DMSO and 2.5 μM of UNC0642 RD18-treated cells (lower panels) after culture for 5 days in differentiation medium (DM). Cells were analyzed by immunofluorescence and western blot using anti-MHC antibody as indicated. Nuclei were stained with DAPI. Representative images of three different experiments are shown. Expression of MYOG was analyzed by qPCR at day 2 of differentiation (n = 3). Values correspond to the average ± SEM. (D) Proliferation was analyzed in control and siEHMT2 RD18 cells (upper panels); or DMSO and 2.5 μM of UNC0642-treated RD18 cells (lower panels) by immunostaining with anti-BrdU antibody. Cells were analyzed by immunofluorescence (n = 3). The dot plots show the percentage of BrdU+ in siEHMT2 and UNC0642-treated cells relative to their respective controls. Values correspond to the average ± SEM. In (C) data from three independent biological replicates each with three technical replicates were plotted. In (D) data from three independent biological replicates each with four technical replicates were plotted. Statistical significance was calculated by unpaired two-tailed t-test. ****p≤0.
-
Figure 2—source data 1
qPCR data for day 2 myogenin expression in RD18 cells upon G9a knockdown.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig2-data1-v3.xlsx
-
Figure 2—source data 2
qPCR data for day 2 myogenin expression in RD18 cells upon G9a activity inhibition by UNC0642.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig2-data2-v3.xlsx
-
Figure 2—source data 3
BrdU quantification data in RD18 cells upon G9a knockdown.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig2-data3-v3.xlsx
-
Figure 2—source data 4
BrdU quantification data in RD18 cells upon G9a activity inhibition by UNC0642.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig2-data4-v3.xlsx
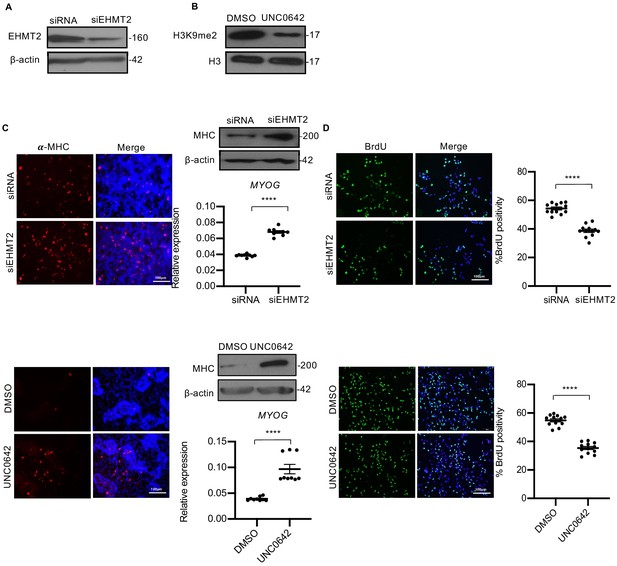
Loss of EHMT2 expression or activity in JR1 cells increases differentiation and reduces proliferation.
(A) Western blot showing EHMT2 knockdown efficiency in siEHMT2 cells. (B) Inhibition of EHMT2 activity by UNC0642 decreased H3K9me2 protein level after 48 hr of treatment as shown by western blot. (C) EHMT2 knockdown and UNC0642 treatment resulted in increased MHC+ cells as seen by immunofluorescence assay and western blot after 5 days in DM in JR1 cells. Data is representative of three independent experiments. EHMT2 knockdown and treatment with UNC0642 also increased MYOG expression as seen by qPCR analysis (n = 3). Values correspond to the average ± SEM. (D) EHMT2 knockdown and UNC0642 treatment reduced BrdU+ cells as seen by immunofluorescence assays. Data is representative of three independent experiments. The dot plots show the percentage of BrdU+ cells in siEHMT2 cells and UNC0642-treated cells relative to controls. Values correspond to the average ± SEM. In (C) data from three independent biological replicates each with three technical replicates were plotted. In (D) data from three independent biological replicates each with four technical replicates were plotted. Statistical significance in (C and D) was calculated by unpaired two-tailed t-test. ****p≤0.0001.
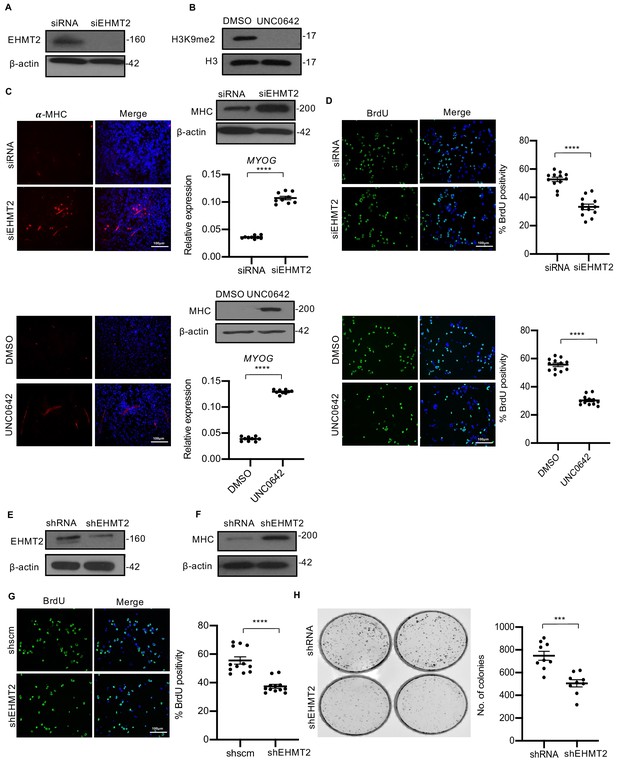
Loss of EHMT2 expression or activity in RD cells increases differentiation and reduces proliferation.
(A) Western blot showing EHMT2 knockdown efficiency. (B) Inhibition of EHMT2 activity by UNC0642 decreased H3K9me2 protein level after 48 hr of treatment as shown by western blot. (C) EHMT2 knockdown and treatment with UNC0642 increased MHC+ cells as seen by immunofluorescence assay and western blot after 5 days in DM. The image is representative of two independent experiments. EHMT2 knockdown and treatment with UNC0642 also increased MYOG expression as seen by qPCR analysis (n = 3). Values correspond to the average ± SEM. (D) EHMT2 knockdown and UNC0642 treatment reduced BrdU+ cells 48 hr in growth media as seen by immunofluorescence assays. The dot plots show the percentage of BrdU+ cells relative to controls (n = 3). Values correspond to the average ± SEM. (E) Lentiviral-mediated stable EHMT2 knockdown (shEHMT2) efficiency in RD cells is shown by western blot. (F) shEHMT2 cells showed increased MHC levels after 5 days in DM by western blot. Data is representative of two independent experiments. (G) shEHMT2 cells showed reduced BrdU+ cells compared to control. Data is representative of three independent experiments. The percentage of BrdU+ cells is shown. Values correspond to the average ± SEM. (H) Colony formation was analyzed in control and shEHMT2 cells. The number of colonies was quantified which showed reduced numbers in shEHMT2 cells. (n = 3). Values correspond to the average ± SEM. In (C and H) data from three independent biological replicates each with three technical replicates were plotted. In (D and G) data from three independent biological replicates each with four technical replicates were plotted. Statistical significance in (C, D, G, and H) was calculated by unpaired two-tailed t-test. ***p≤0.001 and ****p≤0.0001.
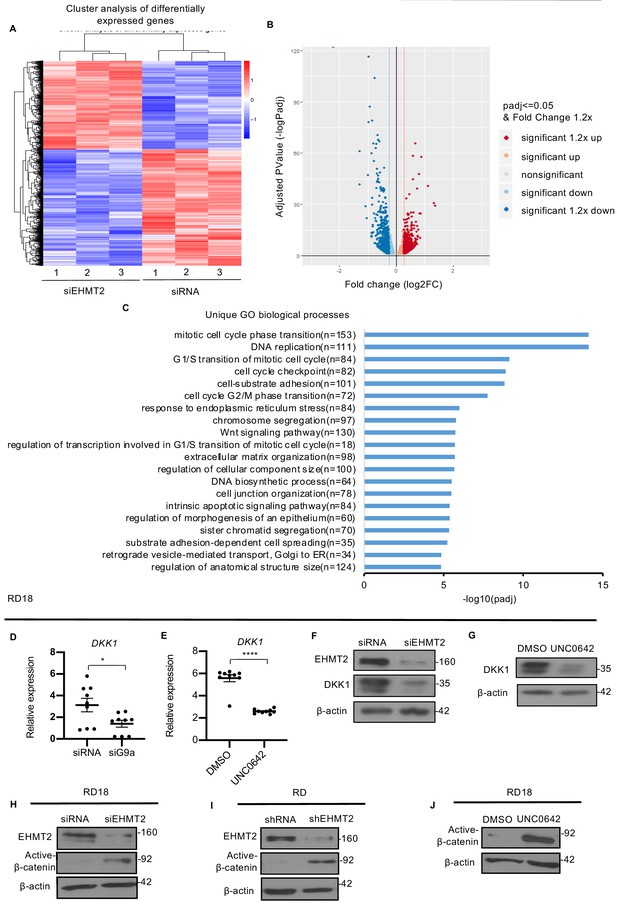
EHMT2 regulates DKK1 and Wnt signaling.
(A) RNA-seq heatmap showing hierarchical clustering of differentially expressed genes. RNA-Seq was performed with control and siEHMT2 RD cells (n = 3). Red represents high expression and blue represents low expression. (B) Volcano plot showing distribution of differentially expressed genes upon EHMT2 knockdown in RD cells. (C) GO enrichment histogram displaying top 20 unique significantly enriched biological processes upon EHMT2 knockdown in RD cells based on p-adjusted value where n signifies the number of differentially expressed genes concerning the GO term. (D and E) qPCR analysis for DKK1 mRNA in RD18 control and siEHMT2 cells and upon 2.5 μM of UNC0642 treatment (n = 3). Values correspond to the average ± SEM. (F and G) DKK1 protein was analyzed in control and siEHMT2 RD18 cells and in DMSO and 2.5 μM of UNC0642-treated RD18 cells. Representative images of three different experiments are shown. (H–J) Western blot analysis showed increased active-β-catenin in siEHMT2 RD18 cells relative to controls, in stable RD shEHMT2 cells, and upon UNC0642 treatment in RD18 cells as indicated. Representative images from three different experiments are shown. In (D and E) data from three independent biological replicates each with three technical replicates were plotted. Statistical significance was calculated by unpaired two-tailed t-test. *p≤0.05, ***p≤0.001.
-
Figure 3—source data 1
qPCR data for DKK1 expression in RD18 cells upon G9a knockdown.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig3-data1-v3.xlsx
-
Figure 3—source data 2
qPCR data for DKK1 expression in RD18 cells upon G9a activity inhibition by UNC0642.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig3-data2-v3.xlsx
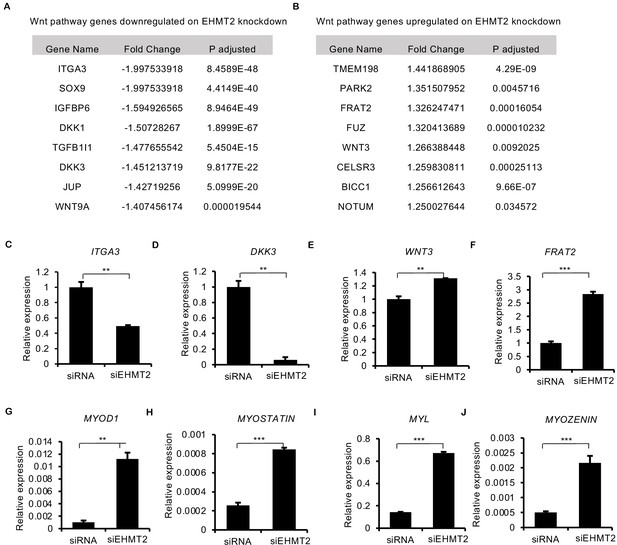
Validation of RNA-sequencing analysis.
(A and B) List of top significantly altered Wnt pathway genes identified in RNA sequencing analysis that are downregulated, or upregulated, upon EHMT2 knockdown. (C–F) qPCR validation of two downregulated genes ITGA3 and DKK3 and two upregulated genes FRAT2 and WNT3 in RD cells. Data shown is representative of two independent experiments. Error bars indicate the mean ± SD. (G–J) qPCR validation of four myogenic differentiation genes MYOD1, MYOSTATIN, MYL, and MYOZENIN that show upregulation upon EHMT2 knockdown in RNA-Seq analysis. Data shown is representative of two independent experiments. Error bars indicate the mean ± SD. In (C–J) the bar graphs are plotted for one biological representative each with three technical replicates of two biological experiments. Statistical significance in (C–J) was calculated as unpaired two-tailed t-test. **p≤0.001, ***p≤0.001 and ****p≤0.0001.
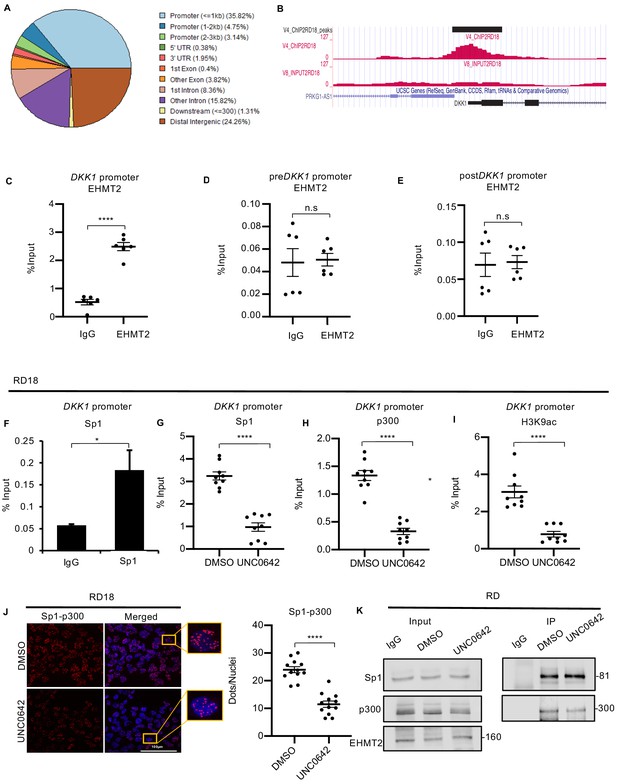
EHMT2 binds to the DKK1 promoter and regulates Sp1/p300 occupancy in a methyltransferase activity-dependent manner.
(A) ChIP-seq analysis in RD18 cells showed EHMT2 occupancy at different regions of the chromatin. (B) Snapshot of EHMT2 binding peak at the DKK1 promoter from the UCSC genome browser. (C) EHMT2 occupancy at the DKK1 promoter was validated by ChIP-PCR (n = 3) in RD18 cells. (D and E) The specificity of the EHMT2 occupancy was validated by ChIP-PCR using primers spanning the chromatin region of enrichment, before (preDKK1) and after (postDKK1) the EHMT2 peak at the DKK1 promoter (n = 3) in RD18 cells. The dot plot shows EHMT2 enrichment compared to IgG which was used as a control. Values correspond to the average ± SEM. (F) Sp1 occupancy was analyzed by ChIP-PCR at the DKK1 promoter in RD18 cells. IgG was used as a control (n = 2). Bar graph for one representative biological experiment with three technical replicates is shown. Values correspond to the average ± SD. (G–I) Sp1, p300, and H3K9ac enrichment at the DKK1 promoter was analyzed in 2.5 μM of UNC0642-treated RD18 cells compared to DMSO controls. The dot plots show reduced enrichment in UNC0642-treated cells (n = 3). Values correspond to the average ± SEM. (J) Proximity ligation assay was done to examine Sp1 and p300 teraction in control and 2.5 μM of UNC0642-treated RD18 cells. Images were captured using confocal microscopy. The dot plot shows the number of dots per nuclei in UNC0642-treated cells compared to control cells (n = 3). Each dot represents an interaction. Values correspond to the average ± SEM. (K) Immunoprecipitation with anti-Sp1 antibody was done to examine interaction with p300 in control and 2.5 μM of UNC0642-treated RD cells. 10% lysate was run as input and immunoblotted for Sp1, p300, and EHMT2 by western blotting. The numbers indicate molecular weight of proteins. In (C–E) and (G–J) data from three independent biological replicates each with three technical replicates were plotted. Statistical significance was calculated by unpaired two-tailed t-test. *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
-
Figure 4—source data 1
ChIP qPCR data for G9a occupancy on DKK1 promoter, pre DKK1 promoter region and post DKK1 promoter region in RD18 cells.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig4-data1-v3.xlsx
-
Figure 4—source data 2
ChIP qPCR data for Sp1 occupancy on DKK1 promoter in RD18 cells.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig4-data2-v3.xlsx
-
Figure 4—source data 3
ChIP qPCR data for Sp1, p300 and H3K9ac occupancy on DKK1 promoter upon G9a activity inhibition by UNC0642.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig4-data3-v3.xlsx
-
Figure 4—source data 4
PLA quantification data of Sp1-p300 interaction in RD18 cells upon G9a activity inhibition by UNC0642.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig4-data4-v3.xlsx
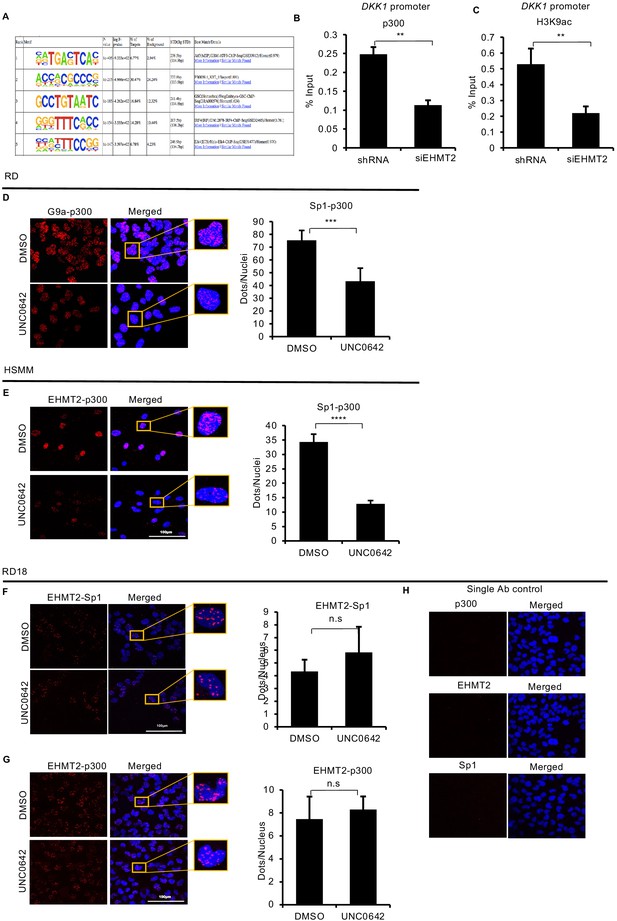
EHMT2 regulates Sp1 and p300 occupancy at the DKK1 promoter.
(A) Top DNA motifs at EHMT2-predicted binding sites by HOMER analysis. (B and C) ChIP-PCR analysis showed decrease in p300 occupancy and H3K9ac enrichment at the DKK1 promoter in shEHMT2 RD cells as compared to control (n = 2). Error bars indicate the mean ± SD. (D) Proximity ligation assay (PLA) was done to examine Sp1 and p300 interaction in control and 2.5 μM of UNC0642-treated RD cells. Sp1–p300 interaction showed a significant decrease upon UNC0642 treatment. Images were captured using confocal microscopy. Data shown is representative of two independent experiments. Error bars indicate the mean ± SD. (E) PLA was done to examine Sp1 and p300 interaction in control and 2.5 μM of UNC0642-treated human skeletal muscle myoblast cells. Sp1-p300 interaction showed a significant decrease upon UNC0642 treatment. Images were captured using confocal microscopy. Data shown is representative of two independent experiments. Error bars indicate the mean ± SD. (F) PLA was used to determine EHMT2-Sp1 interaction which did not change upon UNC0642 treatment. Data shown is representative of two independent experiments. Error bars indicate the mean ± SD. (G) EHMT2-p300 interaction was not significantly altered by UNC0642 treatment. Data shown is representative of two independent experiments. Error bars indicate the mean ± SD. (H) Single antibody controls for p300, EHMT2, and Sp1 were done for PLA in RD18 cells. In (B–G) the bar graphs are plotted for one biological representative each with three technical replicates of two biological experiments. Statistical significance in (B–G) was calculated as unpaired two-tailed t-test. **p≤0.001, ns = not significant.
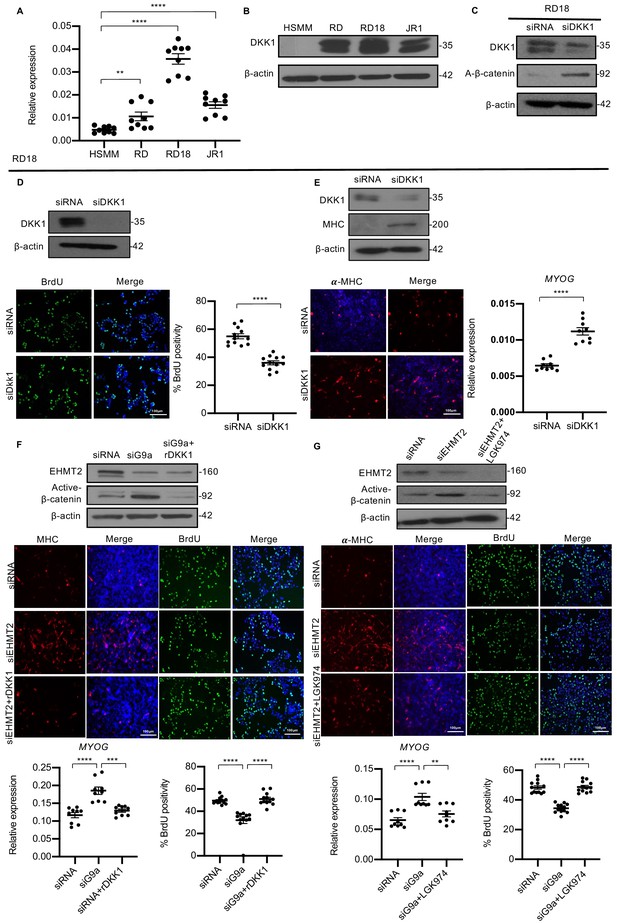
DKK1 is a downstream effector of EHMT2 function.
(A) DKK1 mRNA was examined by qPCR (n = 3) in human skeletal muscle myoblast (HSMM), RD, RD18, and JR1 cells. Values correspond to the average ± SEM. (B) DKK1 protein levels were analyzed by western blotting in HSMM, RD, RD18, and JR1. A representative image from three different experiments is shown. (C) Effect on canonical Wnt signaling upon knockdown of DKK1 was examined by analyzing active-β-catenin protein levels in control and siDKK1 RD18 cells. (D) DKK1 knockdown was analyzed in control and siDKK1 RD18 cells by western blot. Proliferation was analyzed in RD18 control and siDKK1 cells (n = 3) with anti-BrdU antibody. The dot plot shows the percentage of BrdU+ cells. Values correspond to the average ± SEM. (E) Differentiation was analyzed in control and siDKK1 RD18 cells that were cultured for 5 days in DM. Cells were analyzed by western blot and immunofluorescence and using anti-MHC antibody as indicated. A representative image of three different experiments is shown. MYOG expression was analyzed by qPCR (n = 3) at day 2 of differentiation. Values correspond to the average ± SEM. (F) Control, siEHMT2 cells, and siEHMT2 RD18 cells treated with rDKK1 for 24 hr and tested for active-β-catenin levels. Differentiation and proliferation were analyzed (lower panels) by MHC+ cells and BrdU+ cells as indicated. Representative images of three different experiments are shown. MYOG expression was analyzed by qPCR (n = 3) and the percentage of BrdU+ cells is shown in the dot plots. Values correspond to the average ± SEM. (G) Western blot showing active-β-catenin levels in control, siEHMT2 cells, and siEHMT2 RD18 cells treated with LGK974 for 24 hr. A representative image of three different experiments is shown. MHC+ and BrdU+ cells were analyzed. A representative image of three different experiments is shown. MYOG expression in control, siEHMT2, and siEHMT2 RD18 cells treated with LGK974 was analyzed by qPCR (n = 3). Values correspond to the average ± SEM. Statistical significance in (A) and (D–G) was calculated by unpaired two-tailed t-test. **p≤0.01, ***p≤0.001, ****p≤0.0001.
-
Figure 5—source data 1
qPCR data for endogenous DKK1 expression in ERMS cell lines.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig5-data1-v3.xlsx
-
Figure 5—source data 2
BrdU quantification data in RD18 cells upon DKK1 knockdown.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig5-data2-v3.xlsx
-
Figure 5—source data 3
qPCR data for day 2 myogenin expression in RD18 cells upon DKK1 knockdown.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig5-data3-v3.xlsx
-
Figure 5—source data 4
qPCR data for day 2 myogenin expression in RD18 cells upon rDKK1 treatment in G9a knockdown cells.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig5-data4-v3.xlsx
-
Figure 5—source data 5
BrdU quantification data in RD18 cells upon rDKK1 treatment in G9a knockdown cells.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig5-data5-v3.xlsx
-
Figure 5—source data 6
qPCR data for day 2 myogenin expression in RD18 cells upon LGK974 treatment in G9a knockdown cells.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig5-data6-v3.xlsx
-
Figure 5—source data 7
BrdU quantification data in RD18 cells upon LGK974 treatment in G9a knockdown cells.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig5-data7-v3.xlsx
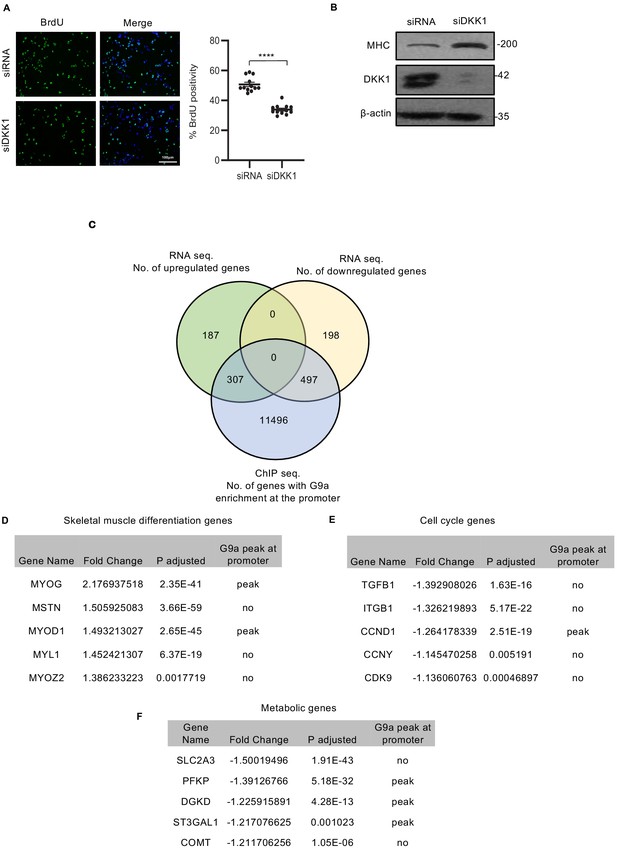
Effect of DKK1 knockdown in RD cells and integration of RNA-seq and ChIP-seq data.
(A) DKK1 knockdown was analyzed in control and siDKK1 cells by western blot. Proliferation was analyzed in control and siDKK1 cells (n = 3) with anti-BrdU antibody. The dot plot shows the percentage of BrdU+ cells. Values correspond to the average ± SEM. (B) Differentiation was analyzed in control and siDKK1 cells that were cultured for 5 days in DM. Cells were analyzed by western blot using anti-MHC antibody as indicated. (C) Venn diagram showing an overlap between number of genes that are significantly upregulated or significantly downregulated in RNA-seq analysis upon EHMT2 knockdown with a fold-change ≥ 1.2 and number of genes with significant EHMT2 occupancy on its promoter region (±3000 bp from TSS) in ChIP-seq analysis. (D–F) Lists of skeletal muscle differentiation genes (D), cell cycle genes (E), and metabolic genes (F) that were significantly altered upon EHMT2 knockdown with or without significant EHMT2 enrichment at the promoter. In (A) data from three independent biological replicates each with four technical replicates were plotted. Statistical significance in (A) was calculated by unpaired two-tailed t-test. ****p≤0.0001.
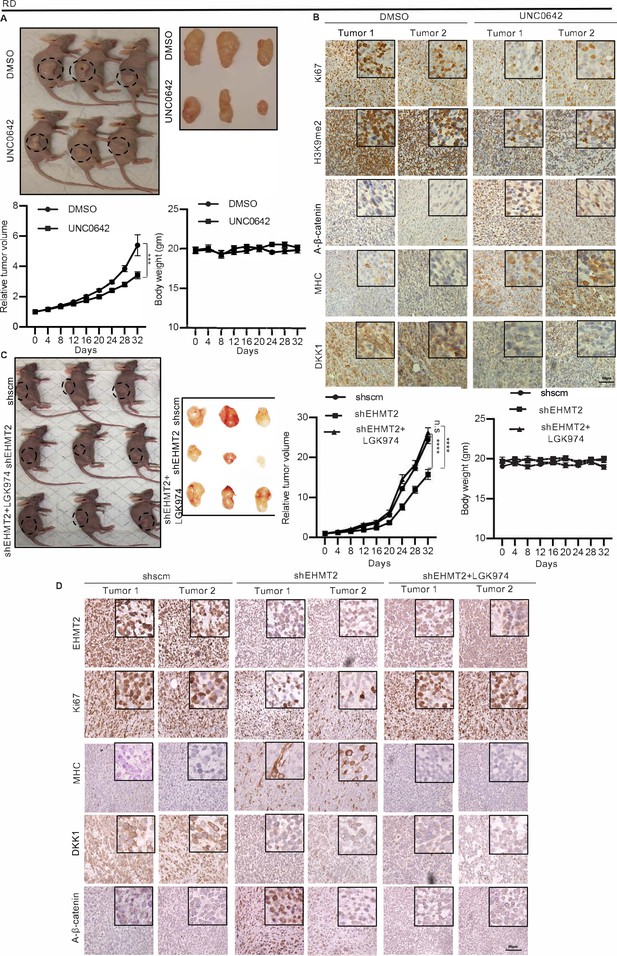
EHMT2 regulates tumor growth by regulation of DKK1 and Wnt signaling.
(A) Nude mice were injected with RD cells. Once tumors were palpable, mice were treated with DMSO (n = 10) or UNC0642 (n = 10). Representative images of three mice in each group (left panel) and resected tumors (right panel) are shown. The relative tumor volume in UNC0642-treated group showed a significant decrease compared to controls although the body weight of mice did not show any significant change. Statistical significance was calculated using repeated-measure two-way ANOVA where ***p≤0.001. Values correspond to the average ± SEM. (B) Tumors from two control and two UNC0642-treated mice were analyzed by immunohistochemistry (IHC) using anti-Ki67, anti-H3K9me2, anti-active-ß-catenin, anti-MHC, and anti-DKK1 antibodies. Scale bar: 50 μm. Inset shows zoomed in images. (C and D) Mice were injected with shRNA RD cells (shscm) (n = 10) or shEHMT2 RD cells (n = 20). Once tumors were palpable, half of shEHMT2 injected mice were treated with vehicle and the rest with LGK974. (C) Representative images of mice (left panel) injected with shscm control cells, shEHMT2 cells, and shEHMT2 cells + LGK974 are shown. Representative images of the tumors (right panel) isolated from the three cohorts are shown. The relative tumor volume and the body weight of mice in each group were determined. Statistical significance was calculated using repeated-measure two-way ANOVA where ****p≤0.0001. Values correspond to the average ± SEM. (D) Tumors from two different mice in each group were analyzed by IHC for Ki67, H3K9me2, DKK1, MHC, and active-ß-catenin staining as described above. Scale bar: 50 μm. Inset shows zoomed in images.
-
Figure 6—source data 1
Relative tumor volume and body weight of mice upon G9a activity inhibition by UNC0642.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig6-data1-v3.xlsx
-
Figure 6—source data 2
Relative tumor volume and body weight of mice upon G9a knockdown and treatment of G9a knockdown tumors with LGK974.
- https://cdn.elifesciences.org/articles/57683/elife-57683-fig6-data2-v3.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus female) | Nude mice | In Vivos | C.Cg/AnNTac-Foxn1nuNE9 BALB/c RRID:IMSR_TAC:balbnu | BALB/c inbred model |
| Cell line (Homo sapiens) | HSMM | Lonza Inc | #: CC-2580 | Isolated from upper arm or leg muscle tissue of normal donors and sold at second passage |
| Cell line (H. sapiens) | RD18 | Peter Houghton and Rosella Rota | RRID:CVCL_IU87 | Clone cells derived from RD cells |
| Cell line (H. sapiens) | RD | Peter Houghton and Rosella Rota | RRID:CVCL_1649 | Patient-derived cell line from pelvic mass of 7-year-old female |
| Cell line (H. sapiens) | JR1 | Peter Houghton and Rosella Rota | RRID:CVCL_J063 | Patient-derived cell line from lung metastasis of 7-year-old female |
| Cell line (H. sapiens) | RD shscm | This study | Transfected with shRNA control lentivirus particles | |
| Cell line (H. sapiens) | RD shEHMT2 | This study | Transfected with shEHMT2 lentivirus particles | |
| Transfected construct (H. sapiens) | SmartPool non-targeting siRNA | Dharmacon | D-001810-10-20 | Negative control of four siRNAs designed for minimal targeting |
| Transfected construct (H. sapiens) | SmartPool siRNA against EHMT2 | Dharmacon | L-006937-00-0010 | A mixture of four siRNA provided as a single reagent |
| Transfected construct (H. sapiens) | SmartPool siRNA against DKK1 | Dharmacon | L-003843-01-0010 | A mixture of four siRNA provided as a single reagent |
| Transfected construct (H. sapiens) | shRNA control lentivirus particles | Santa Cruz Biotechnology Inc | sc-108080 | Negative control. 200 μl viral stock containing 1 × 106IFU. Encodes nonspecific scrambled shRNA |
| Transfected construct (H. sapiens) | shEHMT2 control lentivirus particles | Santa Cruz Biotechnology Inc | sc-43–777V | 200 μl of viral stock containing 1 × 106 IFU. Pools of three to five target-specific sequences |
| Antibody | Rabbit monoclonal EHMT2 | Cell Signalling | #3306S | 1:300, western blot |
| Antibody | Mouse monoclonal MHC | Santa Cruz Biotechnology | Sc-32732 | 1:300, western blot |
| Antibody | Mouse monoclonal Myogenin | Santa Cruz Biotechnology | Sc-12732 | 1:250, western blot |
| Antibody | Mouse monoclonal Dkk1 | Santa Cruz Biotechnology | Sc-374574 | 1:300, western blot, 1:200 for IHC |
| Antibody | Mouse monoclonal active-ß-catenin | Merk Millipore | 05–665 | 1:500, western blot, 1:300 for IHC |
| Antibody | Rabbit polyclonal H3K9me2 | Cell Signaling | 9753S | 1:1000, western blot, 1:200 for IHC |
| Antibody | Mouse monoclonal ß-actin | Sigma-Aldrich | A2228 | 1:10,000, western blot |
| Antibody | Rabbit polyclonal H3 | Abcam | Ab-1791 | 1:10,000, western blot |
| Antibody | Mouse monoclonal Sp1 | Santa Cruz | Sc-17824 | 1:50 for PLA |
| Antibody | Rabbit Polyclonal Sp1 Rabbit | Merck Millipore | 07–645 | 1:100 for PLA, 3 μg was used for ChIP. 2 μg was used for IP pull down. 1:500 dilution for immunoblotting |
| Antibody | Mouse monoclonal p300 | Abcam | Ab14984 | 1:1000 for PLA, 2 μg was used for ChIP. 1:500 dilution for immunoblotting |
| Antibody | Rabbit polyclonal H3K9ac | Abcam | Ab4441 | 2 μg was used for ChIP |
| Antibody | Rabbit polyclonal EHMT2 | Abcam | Ab40542 | 2 μg was used for ChIP, 1:200 for IHC |
| Antibody | Mouse monoclonal Ki67 | Leica Biosystems | PA0118 | 1:100 for IHC |
| Antibody | Mouse monoclonal MHC | Sigma Aldrich | M4276 | 1:200 for IHC and IF |
| Sequence-based reagent | Primers | This study | As mentioned in Materials and methods | |
| Peptide, recombinant protein | Human DKK1 | R and D Systems | 5439-dk-010 | 100 ng/ml, Sf21(baculovirus)-derived human DKK1 protein |
| Commercial assay or kit | PLA kit (Duolink in situ- fluorescence) | Sigma | DUO92101 | |
| Commercial assay or kit | Lipofectamine RNAiMax | Thermo Fisher Scientific | 13778150 | |
| Chemical compound | LGK974 | Selleck Chemicals | S7143 | 200 nM, porcupine inhibitor |
| Chemical compound | Polybrene | Sigma Aldrich | TR-1003 | 2 Ul of 8 mg/ml |
| Chemical compound | Puromycin dihydrochloride | Sigma Aldrich | P8833 | 1 µg/ml |
| Software, algorithm | GraphPad prism | V9.0 | https://www.graphpad.com/ |
Additional files
-
Source data 1
Raw data for western blots.
- https://cdn.elifesciences.org/articles/57683/elife-57683-data1-v3.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57683/elife-57683-transrepform-v3.docx





