An analog to digital converter controls bistable transfer competence development of a widespread bacterial integrative and conjugative element
Figures
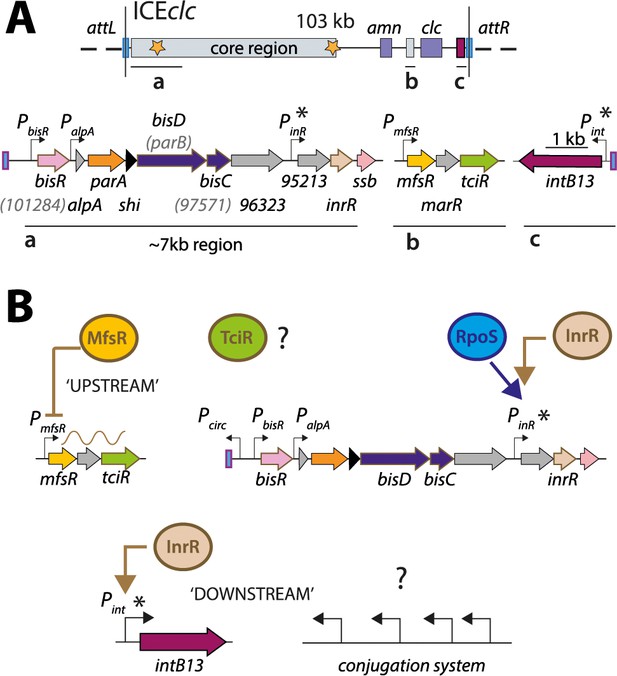
ICEclc and postulated regulation network for transfer competence formation.
(A) Schematic representation of the genetic organization of ICEclc (GenBank accession number AJ617740.2). Loci of interest (a, b and c) are detailed below the general map and drawn to scale. Note the ~7 kb left-end region, which is the major focus of the study. Genes are represented by coloured arrows with their name below (former names shown in lighter font inside brackets). Promoters are represented by hooked arrows pointing towards the transcription orientation. Those marked with an asterisk are known to be expressed only in the subpopulation of transfer competent cells. attL and attR, attachment sites; clc genes: 3-chlorocatechol degradation, amn genes: 2-aminophenol degradation. (B) Known steps in ICEclc transfer competence regulation. An ‘upstream’ cascade, with MfsR autorepressing its own transcription and that of tciR; TciR overexpression leading to transfer competence in almost all cells (Pradervand et al., 2014). Bistable expression of ‘downstream’ genes from PinR and Pint in the subpopulation of transfer competent cells, and further roles of additional factors RpoS (Miyazaki et al., 2012) and InrR (Minoia et al., 2008).
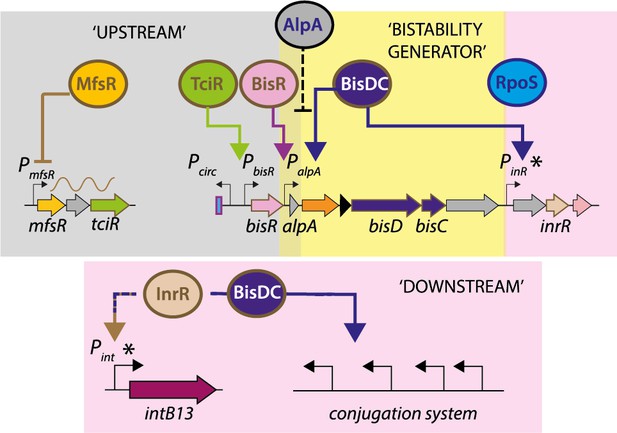
Postulated model of bistability generation of ICEclc.
An upstream cascade of sequential transcription factors MfsR, TciR and BisR, each acting on a single target promoter.
A bistability generator involving a feedback loop by BisDC activating PalpA and potential modulatory repressive role of AlpA. The feedback loop regenerates sufficient BisDC to activate downstream promoters, in conjunction with RpoS and InrR (whose role remains uncharacterized).
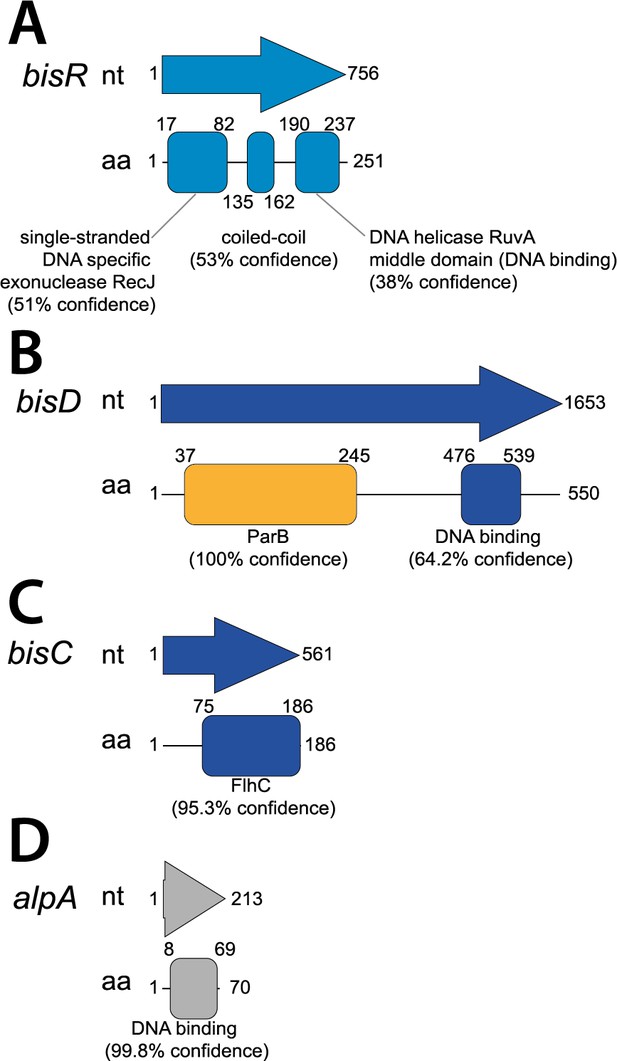
Protein domain predictions in the ICEclc bistability regulators BisR, BisD and BisC, and AlpA.
Genes are schematically represented by arrows with indicated size (nucleotides, nt). Corresponding protein domains predicted using Phyre2 (prediction confidence within brackets) are shown below the respective genes, with amino acid positions (aa) indicated.
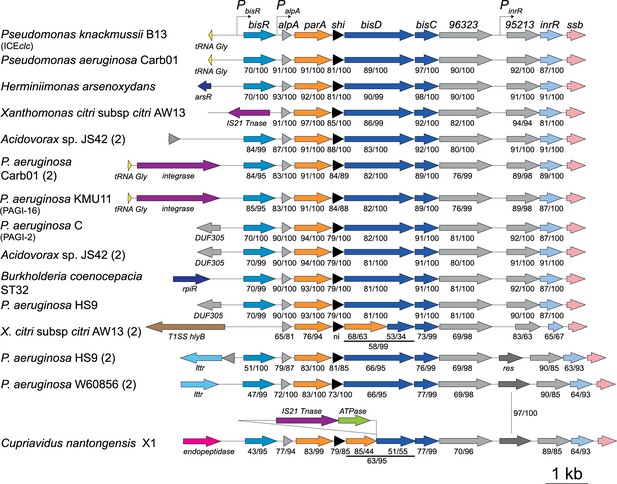
Gene organization of ICEclc-related regulatory loci in different bacterial species.
Diagrams show gene organization (coloured arrows, to scale) of the ICEclc left-end regulatory loci (Pseudomonas knackmussii B13, Accession number HG322950.1), and homologs in Pseudomonas aeruginosa Carb01_63 (CP011317.1), Herminiimonas arsenicoxydans (CU207211.1); Acidovorax sp. JS42 (CP000539.1); P. aeruginosa KMU11 (PAGI-16) (KX196167); P. aeruginosa C (PAGI-2) (AF440523.1); Burkholderia cenocepacia ST32 (CP011917.1); Pseudomonas aeruginosa HS9 (CP030861.1); Xanthomonas citri subsp. citri AW13 (CP009031.1); Pseudomonas aeruginosa W60856 (CP008864.2); and Cupriavidus nantongensis X1 (CP014844.1). Genes of unknown function (dark grey) inserted between orfs96323 and 95213 in strains AW13, HS9 (2) and W60856. Numbers below genes indicate the percentage of amino acid identity and coverage to the corresponding one from ICEclc. ni: no identity.
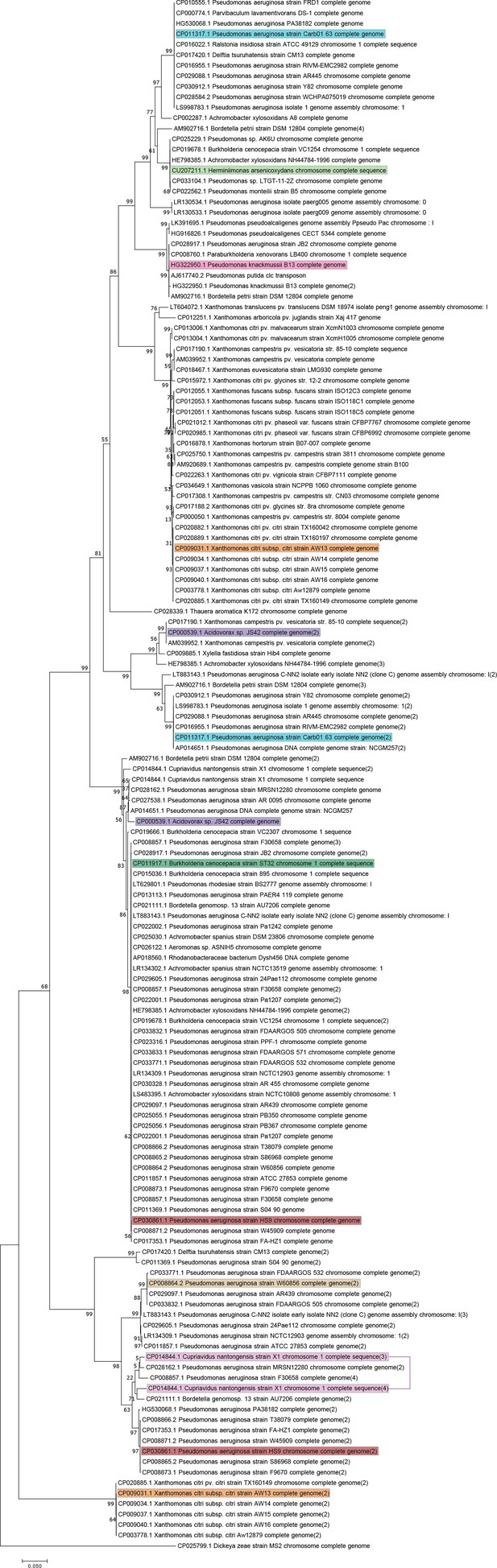
BisDC homologs among Proteobacteria.
Maximum Likelihood phylogenetic tree of BisDC homologs among Proteobacteria based on amino acid alignment using the Tamura-Nei model. Numbers next to branching points indicate the percentage of trees with clustering as shown among 100 bootstraps. BisDC representatives displayed in Figure 1—figure supplement 2 are highlighted. C. nantogensis X1 sequences were linked from its two separate genes.
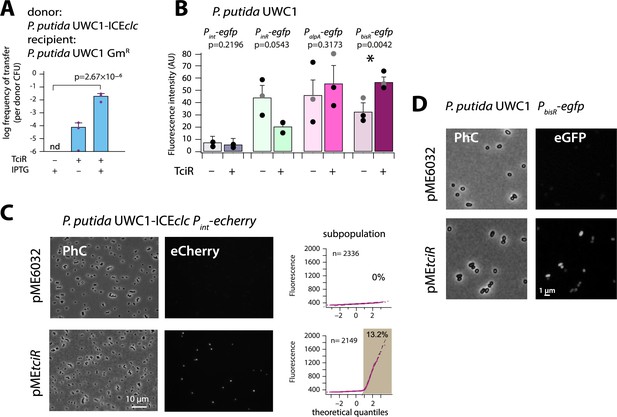
The LysR-type regulator TciR links to a single node in the regulation network.
(A) Ectopic overexpression of tciR induces ICEclc wild-type conjugative transfer under non-permissive conditions. Bars show the means (+ one standard deviation) of transconjugant formation after 48 hr in triplicate matings using P. putida UWC1 donors carrying the indicated ICEclc or plasmids, in absence (-) or presence (+) of 0.1 mM IPTG, and with a GmR-derivative of P. putida as recipient. Dots represent individual transfer; nd: not detected (<10−7 for the three replicates). p-value derives from one-sided t-test comparison (n = 3). (B) Reporter expression from single copy chromomosomal PbisR, PinR, Pint, or PalpA transcriptional egfp fusions in P. putida UWC1 without ICEclc as a function of ectopically expressed TciR, in comparison to strains carrying the empty vector pME6032. Bars show means of the 75th percentile fluorescence of 500–1000 individual cells each per triplicate culture grown on succinate, induced with 0.05 mM IPTG. Error bars denote standard deviation from the means from biological triplicates (dots show individual 75th percentiles). AU, arbitrary units of brightness at 500 ms exposure. p-values derive from pair-wise comparisons in t-tests between cultures expressing TciR and not. (C) Proportion of cells expressing eCherry from a single-copy chromosomal insertion of Pint in P. putida with ICEclc in presence of induced TciR (pMEtciR, 0.05 mM IPTG) or with empty vector (pME6032). Fluorescence images scaled to same brightness (300–2000). Diagrams show quantile-quantile plots of individual cell fluorescence levels, with n denoting the number of analysed cells and the shaded part indicating the subpopulation size expressing Pint-echerry. (D) Fluorescence images of P. putida without ICEclc with a single-copy chromosomal PbisR-egfp fusion in presence of empty vector or of induced TciR. Images scaled to same brightness (300–1200).
-
Figure 2—source data 1
Figure 2 panel A: ICEclc transfer data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Figure 2 panel B: 75th percentile fluorescence data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Figure 2 panel C: qq-plot single cell fluorescence data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig2-data3-v2.xlsx
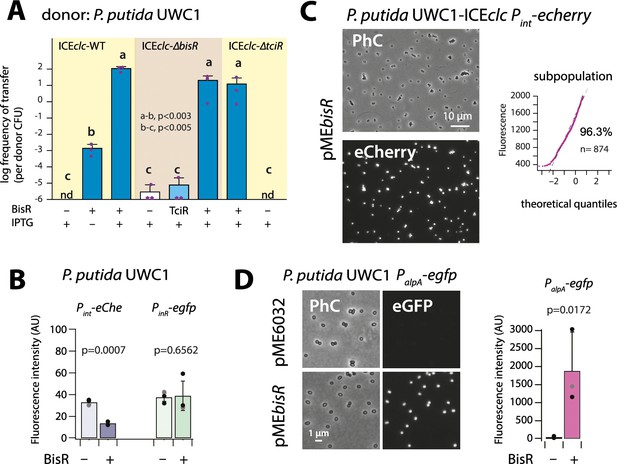
Identification of BisR as a new intermediary regulator for PalpA activation.
(A) Ectopic overexpression of bisR induces ICEclc conjugative transfer under non-permissive conditions and from ICEclc deleted of key regulatory genes. For explanation of bar diagram meaning, see Figure 2A legend. BisR (+), plasmid with bisR; TciR, pMEtciR; –, empty vector pME6032. nd: not detected (<10−7 for the three replicates). Letters indicate significance groups in ANOVA followed by post-hoc Tukey testing (e.g., a-b: p-values between groups a and b; b-c: p-values between groups b and c). (B) Absence of direct induction by BisR of Pint or PinR fluorescence reporters in P. putida without ICE. For explanation of bars, see Figure 2B legend. (C) Population-wide expression of Pint-echerry in P. putida with ICEclc upon ectopic induction of plasmid-located BisR (pMEbisR, 0.05 mM IPTG). Image brightness scale: 300–2000. For vector control, see Figure 2C. (D) Induced BisR from plasmid leads to reporter expression from the alpA-promoter in all cells of P. putida without ICEclc. Image brightness scales: 300–1200. Bars show means and standard deviation from median fluorescence intensity of single cells (n = 500–1000, summed from 6 to 12 images per replicate) of biological triplicates. p-value derives from pair-wise t-test between cultures with empty vector (–) and those with induced BisR (+).
-
Figure 3—source data 1
Figure 3 panel A: ICEclc transfer frequencies.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Figure 3 panel C: qq plot single cell fluorescence values.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Figure 3 panel B and panel D: 75th percentile fluorescence data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig3-data3-v2.xlsx
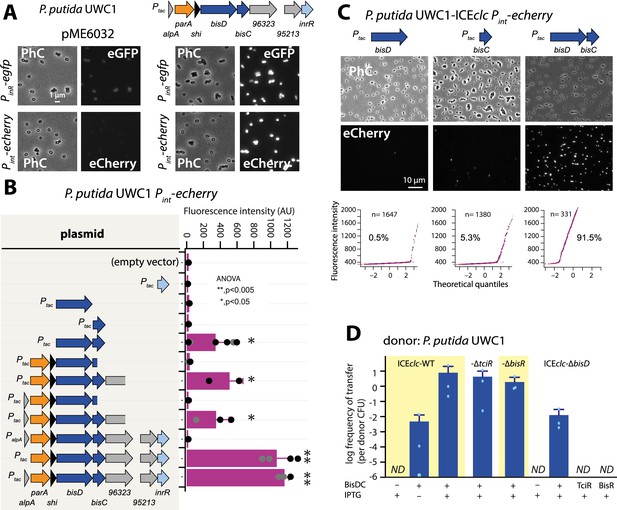
A new regulatory factor BisDC for activation of downstream ICEclc functions.
(A) IPTG (0.05 mM) induction of a plasmid with the cloned ICEclc left-end gene region (as depicted on top) leads to reporter expression from the ‘downstream’ PinR- and Pint-promoters in P. putida without ICEclc. Fluorescence images scaled to same brightness (300–1200). (B) Pint-echerry reporter expression upon IPTG induction (0.05 mM) of different plasmid-subcloned left-end region fragments (grey shaded area on the left) in P. putida without ICEclc. Bars show means of median cell fluorescence levels with one standard deviation, from triplicate biological cultures (n = 500–1000 cells, summed from 6 to 12 images per replicate). Asterisks denote significance groups in ANOVA followed by post-hoc Tukey testing. (C) Population response of Pint-echerry induction in P. putida with ICEclc in presence of plasmid constructs expressing bisD, bisC or both (fluorescence images scaled to 300–2000 brightness). Quantile-quantile plots (n = number of cells) below show the estimated size of the responding subpopulation. (D) Effect of bisDC induction from cloned plasmid (0.05 mM IPTG) on conjugative transfer of ICEclc wild-type or mutant derivatives. Transfer assays as in legend to Figure 2A. ND, below detection limit (10–7).
-
Figure 4—source data 1
Figure 4 panel B: median fluorescence values.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Figure 4 panel C: qq plot single cell fluorescence data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Figure 4 panel D: ICEclc transfer data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig4-data3-v2.xlsx
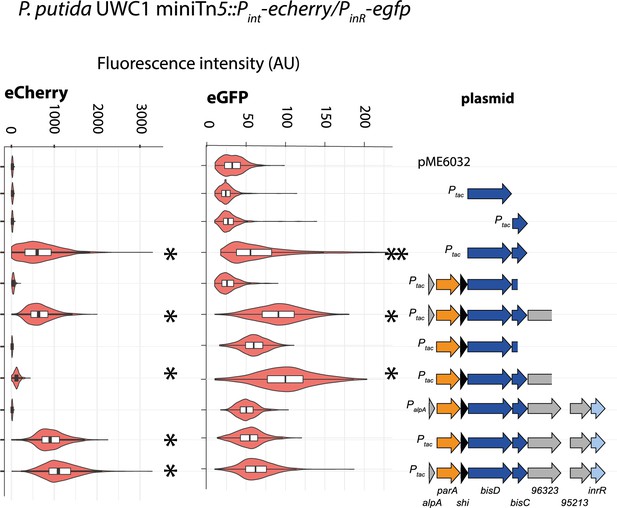
Effect of bisDC containing plasmid constructs on fluorescent protein expression from a single copy dual Pint-PinR reporter in P. putida without ICEclc.
Diagrams show distributions of individual cell fluorescence of P. putida from Pint (eCherry) and PinR (eGFP), complemented with the indicated plasmid constructs (on the right), upon induction with 0.05 mM IPTG. Distributions represented by box plots (black on white, with first quartile, median and second quartile), underlaid with a violin distribution of all data (salmon red). Single asterisks indicate statistically significant differences of median values compared to P. putida carrying the empty vector in pair-wise t-test of biological triplicates (p<0.05), whereas double asterisks indicate statistically significant differences of the measured subpopulations in quantile-quantile plots.
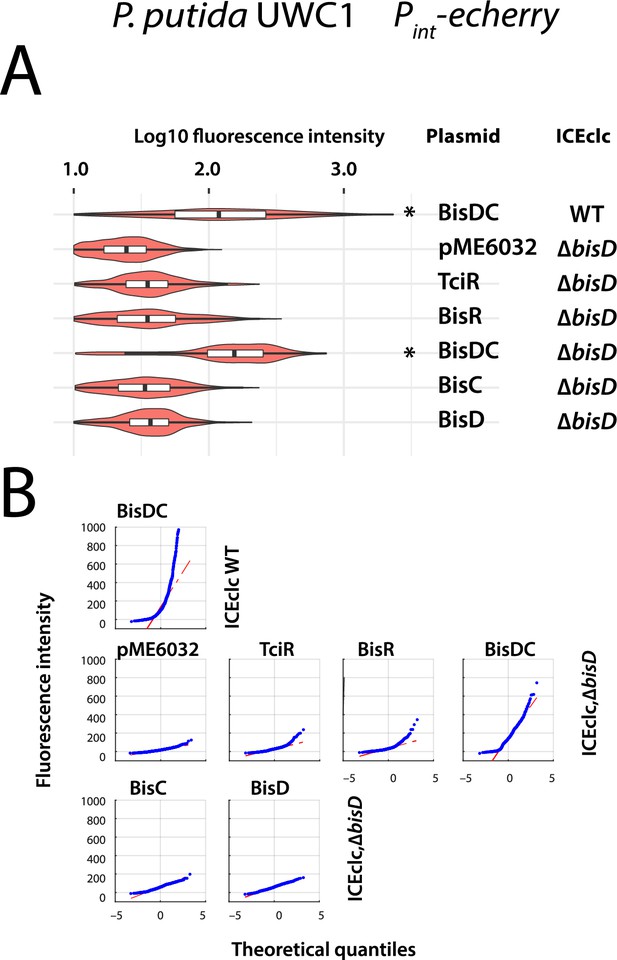
Effect of bisDC expression on reporter expression from Pint in P. putida carrying ICEclc with a bisD deletion.
(A) Violin/box plots (as in legend to Figure 4—figure supplement 1) show distributions of log10 individual cell fluorescence from Pint (eCherry) of P. putida with ICEclc–∆bisD or wild-type, complemented with the indicated plasmid constructs (in the middle), upon induction with 0.05 mM IPTG. (B) Corresponding quantile-quantile plots to identify subpopulations of reporter-expressing cells. Single asterisks indicate statistically significant differences of median values compared to P. putida carrying the empty vector in pair-wise t-test of biological triplicates (p<0.05).
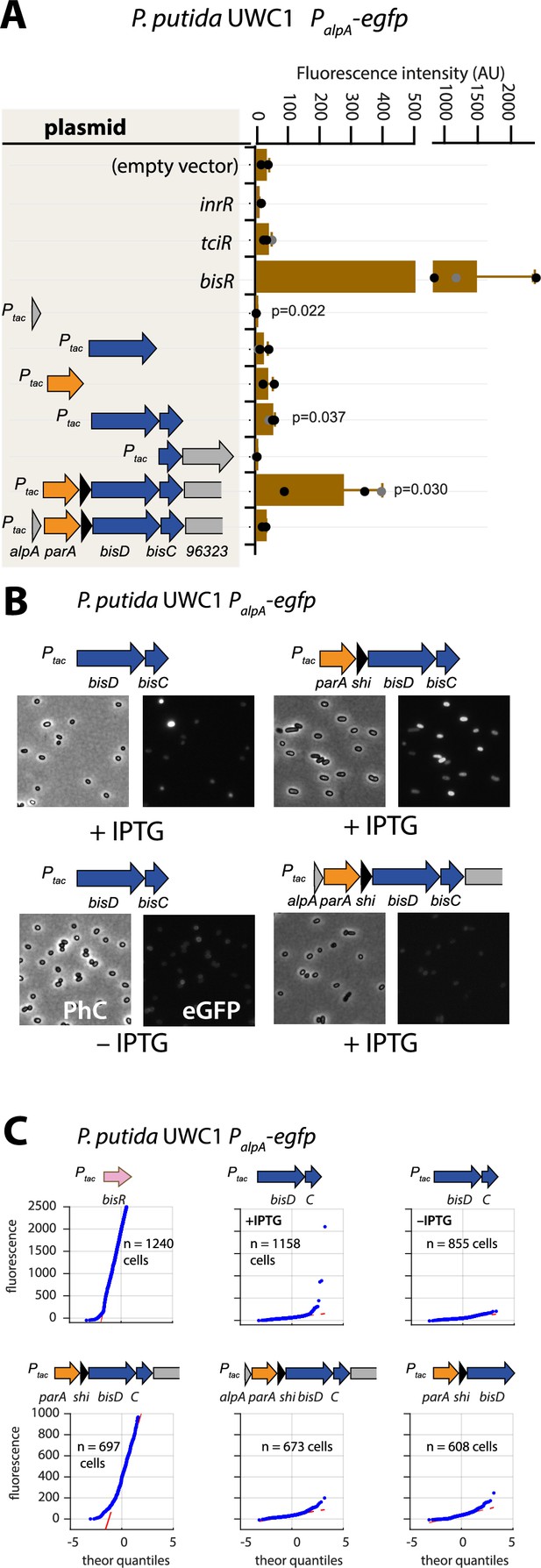
Autoregulatory feedbacks on the alpA promoter.
(A) PalpA-reporter expression upon IPTG induction (0.05 mM) of plasmid-cloned individual genes or gene combinations (as depicted in the shaded area on the left) in P. putida without ICEclc. Bars represent means of median cell fluorescence plus one standard deviation, as in legend to Figure 2B. p-values stem from pair-wise comparisons between triplicate cultures carrying the empty vector pME6032 and the indicated plasmid-cloned gene(s). (B) Cell images of P. putida PalpA-egfp without ICEclc expressing plasmid-cloned combinations with bisDC (fluorescence brightness scaled to 300–1200, 0.05 mM IPTG). (C) Quantile-quantile estimation of subpopulation expression of the PalpA-egfp reporter, showing the sufficiency of bisDC induction for autoregulatory feedback and the reinforcement from upstream elements (n denotes the number of cells used for the quantile-quantile plot, summed from 6 to 12 images of a single replicate culture).
-
Figure 5—source data 1
Figure 5 panel A: Median fluorescence values.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Figure 5 panel C : qq plot single cell fluorescence values.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig5-data2-v2.xlsx
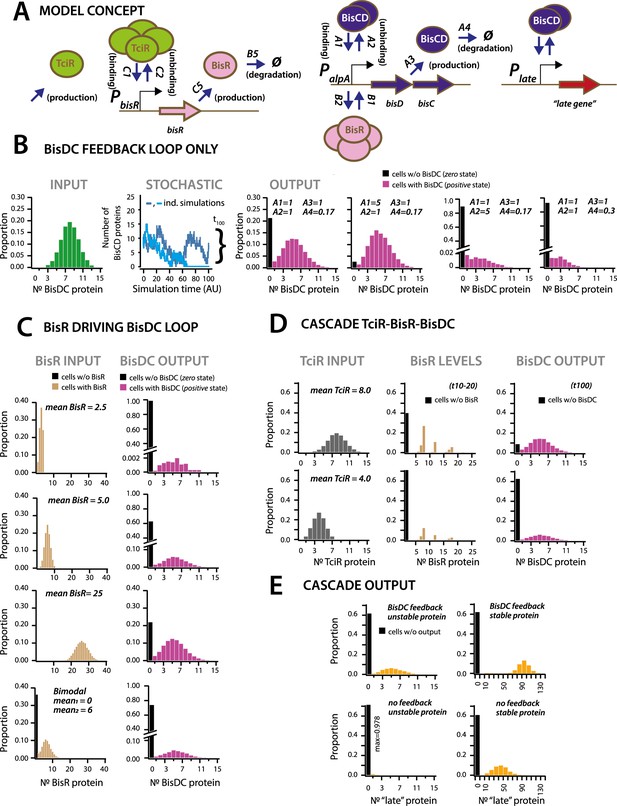
Stochastic simulations of ICEclc regulatory network configurations.
(A) Conceptual model of the ICEclc regulatory cascade producing bistable output. Ellipses indicate the three major regulatory factors (TciR, BisR and BisDC) interacting with their target promoters (PbisR, PalpA), and BisDC-regulated downstream output (here schematically as Plate and a late gene). Relevant simulated processes include: production (combination of transcription and translation, with corresponding rates: C5, A3), oligomerization (assumed number of protein monomers in the binding complex), binding and unbinding to the target promoter, and degradation. All processes are simulated as stochastic events across 100 time steps, and protein output levels are summarized from 10,000 individual stochastic simulations (curly bracket in B; detailed parametrization in Supplementary file 4; one simulation being equivalent to an individual cell). (B) Behaviour of a BisDC autoregulatory feedback loop on the distribution of BisDC protein levels per cell (histograms, n = 10,000 simulated cells) as a function of different binding (A1), unbinding (A2), production (A3) and degradation (A4) rate constants, starting from a uniformly distributed set of BisDC levels (input, in green). Black bar indicates the proportion of cells with zero output (i.e., non-activated circuit). Light blue: simulation example where BisDC levels go to zero and loop would die out. Dark blue: BisDC levels remain positive. (C) As for A, but for an architecture of BisR initiating bisDC expression, with different input distributions (uniformly low to high mean, or bimodal BisR input). Note how higher or bimodal BisR input is not expected to change the median BisDC quantity in active cells, but only the proportion of ‘cells’ with positive (magenta, bars) and zero state (black bars; n = 10,000 simulated cells; note different ordinate scales). (D) As for A, but for the complete cascade starting with TciR. Shown are regulatory factor level distributions from two different TciR starting distributions across 10,000 simulations; for BisR integrated between time points 10 and 20 (t10-20), and for BisDC after 100 time steps (t100). Bimodal expression of zero and positive states arises at the bisR node, but is further maintained to constant BisDC output as a result of the feedback loop. (E) Importance of the BisDC-feedback on the output of a downstream (‘late’) BisDC-dependent expressed protein, for a case of a stable and an unstable protein (n = 10,000 simulated cells).
-
Figure 6—source data 1
Figure 6 panels B–E: Model simulations and histogram data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig6-data1-v2.xlsx
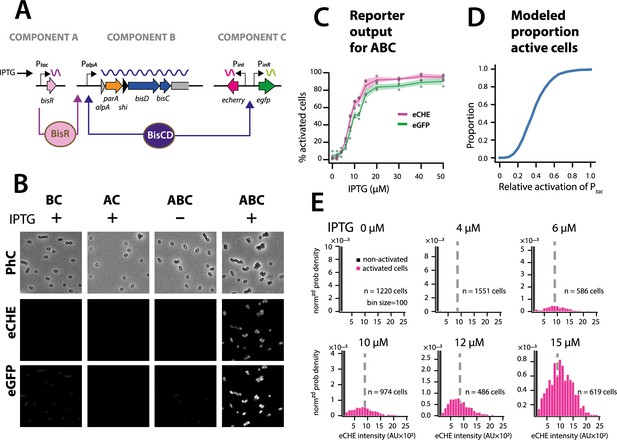
The ICEclc bistability generator is a scalable analog-digital converter.
(A) Schematic representation of the three ICEclc components used to generate scalable bistable output in P. putida UWC1 without ICE. (B) Cell images of P. putida with the different bistability-generator components as indicated, induced in presence or absence of IPTG (0.1 mM). Fluorescence brightness scaled to between 300–1200. (C) Proportion of active cells (estimated from quantile-quantile plotting as in Figure 2C) as a function of IPTG concentration (same induction time for all). Lines correspond to the means from three biological replicates with transparent areas representing the standard deviation. (D) Modelled proportion of cells with positive output in the architecture of Figure 6C as a function of the relative BisR starting levels from Ptac. (E) Measured distributions (as normalized probability density) of eCherry fluorescence among the subpopulations of activated (magenta bars) and non-activated cells (black bars) at different IPTG concentrations, showing same subpopulation fluorescence median (dotted grey lines), as predicted in the stochastic model. AU, arbitrary units of fluorescence brightness at 500 ms exposure. n denotes the number of cells used to produce the histograms, summed from 6 to 12 images from a single replicate culture. Panels autoscaled to maximum ordinate.
-
Figure 7—source data 1
Figure 7 panel C: Mean fluorescence data.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Figure 7 panel E: Single cell and histogram fluorescence data of IPTG induction.
- https://cdn.elifesciences.org/articles/57915/elife-57915-fig7-data2-v2.xlsx
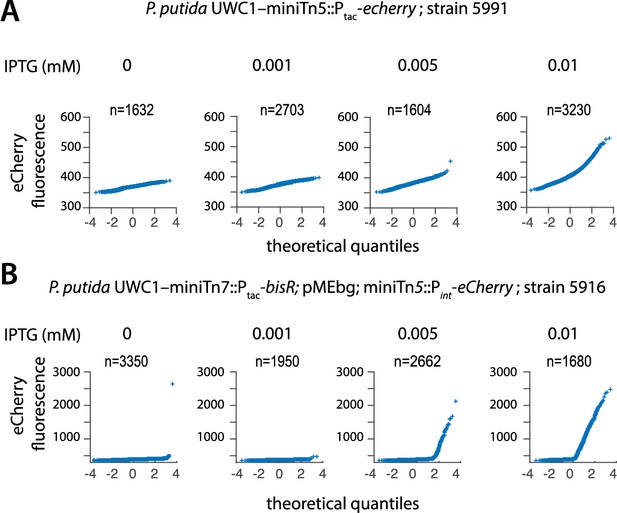
Scalable analog (Ptac) to digital bimodal (Pint) expression in P. putida with ICEclc bistability generator.
(A) Unimodal eCherry expression among individual P. putida cells from a single copy Ptac-echerry chromosomal insertion, as a function of increasing IPTG concentration. (B) Bimodal eCherry fluorescence from Pint among individual P. putida cells expressing BisR from Ptac at different IPTG concentrations and carrying the plasmid cloned PalpA-inrR gene region (bistability generator). Shown are quantile-quantile plots of the observed versus expected distribution of fluorescence levels (n = number of observed cells summed from 10 to 12 technical replicates).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Pseudomonas knackmussii ICEclc) | ICEclc | PMID:16484212 | GenBank: AJ617740.2 | Full ICEclc sequence |
| Gene (Pseudomonas knackmussii ICEclc) | tciR | PMID:24945944 | GenBank: CAE92867.2 | Transcriptional regulator of ICEclc |
| Gene (Pseudomonas knackmussii ICEclc) | bisR; orf101284 | this paper | GenBank: CAE92957.1 | Transcriptional regulator of ICEclc |
| Gene (Pseudomonas knackmussii ICEclc) | bisD; parB; orf98147 | this paper | GenBank: CAE92953.1 | Subunit D of the transcriptional regulator complex BisCD of ICEclc |
| Gene (Pseudomonas knackmussii ICEclc) | bisC; orf97571 | this paper | GenBank: CAE92952.1 | Subunit C of the transcriptional regulator complex BisCD of ICEclc |
| Gene (plasmid pZS2FUNR) | echerry | PMID:19098098 | Red fluorescent protein gene used for the miniTn7:Ptac-echerry reporter | |
| Strain, strain background (Pseudomonas putida) | UWC1 | PMID:2604401 | NCBI: txid1407054 | Background strain for ICEclc transfer and mutagenesis experiments |
| Strain, strain background (Pseudomonas putida) | 2737 | PMID:21255116 | UWC1 carrying ICEclc in tRNAGly | |
| Strain, strain background (Pseudomonas putida) | UWCGC | PMID:21255116 | Recipient strain used for conjugation transfer experiments | |
| Strain, strain background (Escherichia coli) | DH5αλpir | PMID:10610816 | Cloning strain | |
| Recombinant DNA reagent | pME6032; pME (plasmid) | PMID:11807065 | GenBank: DQ645594.1 | Broad-host range cloning vector, lacIq-Ptac expression |
| Recombinant DNA reagent | miniTn5 (plasmid; transposon) | PMID:21342504 (RRID:Addgene_60487) | GenBank: HQ908071.1 | Transposon suicide vector for gene reporter constructs |
| Recombinant DNA reagent | miniTn7 (plasmid; transposon) | PMID:15908923 (RRID:Addgene_63121) | GenBank: AY619004.1 | Transposon suicide vector for single copy insertions |
| Recombinant DNA reagent | miniTn7:Ptac (plasmid; transposon) | PMID:15908923 | GenBank: AY599234.1 | Transposon vector used for miniTn7:Ptac-echerry reporter |
| Recombinant DNA reagent | miniTn5:PinR-egfp/Pint-echerry | PMID:19098098 | Dual single copy insertion reporter system for ICEclc bistable activity | |
| Sequence-based reagent | DNA fragment containingPalpA, alpA, parA, shi and the 5’ part of bisD | ThermoFisher Scientific | Synthetic DNA fragment | |
| Commercial assay or kit | Nucleospin Plasmid Kit | Macherey-Nagel | Macherey-Nagel: 740588.50 | Plasmid purification |
| Commercial assay or kit | Nucleospin Gel and PCR Clean-up Kit | Macherey-Nagel | Macherey-Nagel: 740609.50 | PCR fragment purification |
| Commercial assay or kit | Quick-Fusion Cloning Kit | Bimake | Bimake: B22611 | Generation of recombinant vectors |
| Chemical compound, drug | IPTG; isopropyl β-D-1-thiogalactopyranoside | Sigma-Aldrich | Sigma-Aldrich: I5502 | Used for induction of Ptac promoter |
| Chemical compound, drug | 3-CBA; 3-chlorobenzoate (3-chlorobenzoic acid) | Sigma-Aldrich | Sigma-Aldrich: C24604 | Specific carbon source for selection of ICEclc in Pseudomonas |
| Chemical compound, drug | Sodium succinate; succinate (Sodium succinate dibasic hexahydrate) | Sigma-Aldrich | Sigma-Aldrich:14170 | General carbon source for growth of Pseudomonas |
| Software, algorithm | MEGA7 | PMID:27004904 | Phylogenetic analysis | |
| Software, algorithm | Visiview | Visitron systems GMbH | https://www.visitron.de/products/visiviewr-software.html | Microscopy images acquisition |
| Software, algorithm | ImageJ | PMID:22930834 | Image processing | |
| Software, algorithm | MatLab (v 2016a) | Mathworks | Data treatment | |
| Software, algorithm | R ggplot | Hadley Wickham | https://ggplot2.tidyverse.org/ | Data visualization |
| Software, algorithm | Julia; DifferentialEquations.jl package | DOI:http://doi.org/10.5334/jors.151 | Mathematical model | |
| Other | Minimum media; MM (culture media) | PMCID:PMC494262 | Supplementary file 2 | Carbon source-free base for minimal media |
| Other | Bio-Rad GenePulser Xcell | Biorad | Biorad: 165–2660 | Device used for electro-transformation of bacterial strains |
| Other | Zeiss Axioplan II microscope (Carl Zeiss); EC ‘Plan-Neofluar’ 100x/1.3 Oil Pol Ph3 M27 objective lens (Carl Zeiss); SOLA SE light engine (Lumencor); SPOT Xplorer slow-can charge coupled device camera 1.4 Megapixels monochrome w/o IR (Diagnostic Instruments) | Carl Zeiss; Lumencor; Diagnostic instruments | Carl Zeiss: Axioplan II;Carl Zeiss: 420491-9910-000; Lumencor: SOLA 6-LCR-SC; Diagnostic instrument: XP2400 | Microscope for single cell phase contrast and epifluorescence imaging |
| Other | 0.2–μm cellulose acetate filters | Sartorius | Sartorius:11107–25 N | filters for conjugative transfer |
Strains and plasmids used in this study.
| Strains or plasmids | Relevant genotype or phenotype | References |
|---|---|---|
| E. coli | ||
| DH5αλpir | endA1 hsdR17 glnV44 (=supE44) thi-1 recA1 gyrA96 relA1 φ80dlacΔ(lacZ)M15 Δ(lacZYA-argF)U169 zdg-232::Tn10 uidA::pir+ | Platt et al., 2000 |
| P. putida UWC1 | plasmid-free derivative ofP. putida KT2440 (Rif) | McClure et al., 1989 |
| UWCGC | Single copy integration of lacI-less Ptac promoter controlling echerry expression (Gm) | Miyazaki and van der Meer, 2011b |
| ICEclc | ICEclc copy integrated into tRNAgly-5 (3-CBA) | Miyazaki and van der Meer, 2011b |
| ICEclc ∆tciR | tciR (orf17162) derivative mutant of ICEclc (3-CBA) | Pradervand et al., 2014 |
| ICEclc ∆bisR | bisR (orf101284) derivative mutant of ICEclc (3-CBA) | This work |
| ICEclc ∆bisD | bisD (orf98147) derivative mutant of ICEclc (3-CBA) | This work |
| miniTn7::PinR-egfp | Single copy chromosomal integration of PinR promoter fused to egfp (Gm) | This work |
| miniTn7::PalpA-egfp | Single copy chromosomal integration of PalpA promoter fused to egfp (Gm) | This work |
| miniTn5:: PbisR-egfp | Single copy chromosomal integration of PbisR promoter fused to egfp (Kn) | This work |
| miniTn5:: Pint-echerry/PinR egfp (C) | Single copy chromosomal integration of a dual reporter Pint and PinR promoter fused to echerry and egfp, respectively (Kn) | Minoia et al., 2008 |
| miniTn7::Ptac-bisR (A) | Single copy chromosomal integration of Ptac promoter fused to bisR (Gm) | This work |
| miniTn7::Ptac-echerry | Single copy chromosomal integration of Ptac promoter fused to echerry (Gm) | This work |
| Plasmids | ||
| pME6032 | pVS1-p15A shuttle vector carrying the lacIq-Ptac expression system (Tc) | Heeb et al., 2000 |
| pMEtciR | pME6032 derivative allowing IPTG-controlled expression of tciR (Tc) | This work |
| pMEbisR | pME6032 derivative allowing IPTG-controlled expression of bisR (Tc) | This work |
| pMEbisC | pME6032 derivative allowing IPTG-controlled expression of bisC (Tc) | This work |
| pMEbisD | pME6032 derivative allowing IPTG-controlled expression of bisD (Tc) | Reinhard et al., 2013 |
| pMEbisDC | pME6032 derivative allowing IPTG-controlled expression of bisCD (Tc) | This work |
| pMEparA | pME6032 derivative allowing IPTG-controlled expression of parA (Tc) | Reinhard et al., 2013 |
| pMEparA-shi-bisD | pME6032 derivative allowing IPTG-controlled expression ofparA, shi and bisD (Tc) | Reinhard et al., 2013 |
| pMEbisC96 | pME6032 derivative allowing IPTG-controlled expression of bisC and 96323 (Tc) | This work |
| pMEalpA | pME6032 derivative allowing IPTG-controlled expression of alpA (Tc) | This work |
| pMEinrR | pME6032 derivative allowing IPTG-controlled expression of inrR (Tc) | This work |
| pMEreg | pME6032 derivative allowing IPTG-controlled expression of the alpA-inrR loci (Tc) | This work |
| pMEreg∆alpA | pME6032 derivative allowing IPTG-controlled expression of the parA-inrR loci (Tc) | This work |
| pMEreg∆alpA∆P | pMEreg∆alpA derivative lacking 3’ half of 96323, 95213 and inrR (Tc) | This work |
| pMEreg∆alpA∆A | pMEreg∆alpA derivative lacking the bisC, 96323, 95213 and inrR (Tc) | This work |
| pMEreg∆P | pMEreg derivative lacking 3’ half of 96323, 95213 and inrR (Tc) | This work |
| pMEreg∆A | pMEreg derivative lacking the bisC, 96323, 95213 and inrR (Tc) | This work |
| pMEbg | lacIq-Ptac-less pME6032 derivative carrying the PalpA-inrR loci (Tc) | This work |
| pMEbg_short (comp B) | pMEbg derivative lacking 96323, 95213 and inrR (Tc) | This work |
| pUX-BF13 | helper plasmid for integration of Tn7 (Ap) | Heeb et al., 2000 |
-
3-chlorobenzoate (3-CBA); Ampicillin (Ap); gentamycin (Gm); kanamycin (Kn); rifampicin (Rf); tetracycline (Tc).
(A), (B) and (C) refer to components of the reconstituted bistability generator.
-
For strain numbers, see Supplementary file 1.
Additional files
-
Source code 1
Julia code for the stochastic modelling of bistability.
- https://cdn.elifesciences.org/articles/57915/elife-57915-code1-v2.zip
-
Supplementary file 1
Full strain list.
- https://cdn.elifesciences.org/articles/57915/elife-57915-supp1-v2.docx
-
Supplementary file 2
Composition of minimal growth medium.
- https://cdn.elifesciences.org/articles/57915/elife-57915-supp2-v2.docx
-
Supplementary file 3
List of Primers.
- https://cdn.elifesciences.org/articles/57915/elife-57915-supp3-v2.docx
-
Supplementary file 4
Stochastic model for bistability architectures.
- https://cdn.elifesciences.org/articles/57915/elife-57915-supp4-v2.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57915/elife-57915-transrepform-v2.docx





