Atg1 kinase in fission yeast is activated by Atg11-mediated dimerization and cis-autophosphorylation
Figures
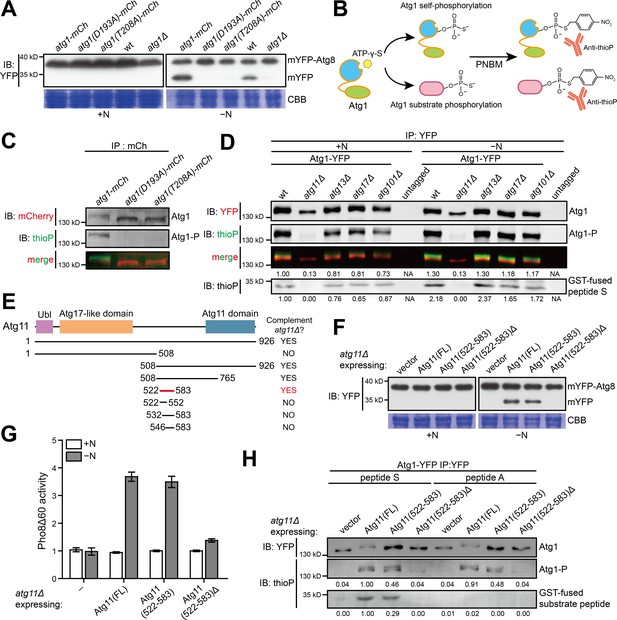
A 62 amino acid region in S. pombe Atg11 is necessary and sufficient for autophagy and for supporting the kinase activity of Atg1.
(A) Mutations predicted to inactivate the kinase activity of S. pombe Atg1 abolished its autophagy function. The processing of endogenously mYFP-tagged Atg8 was used as a readout for autophagy. Cells were collected before (+N) and after shifting to a nitrogen-free medium for 12 hr (−N), and lysates were analyzed by immunoblotting using anti-YFP antibody. Coomassie brilliant blue (CBB) staining of PVDF membrane after immunodetection was used as a control for protein loading (Welinder and Ekblad, 2011). wt, wild type; mCh, mCherry; IB, immunoblotting. (B) Schematic of the non-radioactive in vitro Atg1 kinase assay. (C) In vitro Atg1 kinase assay confirmed that the kinase activity of S. pombe Atg1 was abolished by either D193A or T208A mutation. Endogenously mCherry-tagged Atg1 was immunopurified from cells growing in nutrient-rich medium, and subjected to the in vitro Atg1 kinase assay depicted in B. The immunoblotting signals were detected using the LI-COR Biosciences Odyssey infrared imaging system. IP: immunoprecipitation; Atg1-P, phosphorylated Atg1. (D) Atg11 but not Atg13, Atg17, or Atg101 is required for S. pombe Atg1 kinase activity in cells grown in a nutrient-rich medium (+N) and in cells shifted to a nitrogen-free medium for 1 hr (−N). Endogenously YFP-tagged Atg1 was immunopurified and analyzed by the in vitro kinase assay. Immunoblotting signal generated using anti-thioP antibody was quantified by densitometry and normalized to the immunoblotting signal of Atg1 protein, and reported as values relative to that of the wild-type sample under the +N condition. Results shown are the representative of three independent experiments. See Figure 1—figure supplement 1A for quantitation of data from three independent experiments. GST-fused peptide S is a non-self substrate (see Figure 1—figure supplement 1C). (E) Truncation analysis to identify the minimal region of Atg11 sufficient for its autophagy function. Conserved domains in S. pombe Atg11 are depicted at the top (see Figure 1—figure supplement 2A for domain conservation in Atg11/FIP200 proteins from representative eukaryotes). Starvation-induced vacuolar entry of mYFP-Atg8 was used as an autophagy readout to assess the ability of truncated Atg11 fragments to complement the autophagy defect of atg11Δ (see Figure 1—figure supplement 2B for imaging data). (F) Atg11(522-583) is necessary and sufficient for the autophagy function of Atg11. mYFP-Atg8 processing assay was performed as in A. FL, full length. (G) Pho8Δ60 assay confirmed that Atg11(522-583) is necessary and sufficient for the autophagy function of Atg11. pho8Δ cells expressing S. cerevisiae Pho8Δ60 were collected before (+N) and after nitrogen starvation for 4 hr (−N). The activity of Pho8Δ60 was normalized to the average activity of all samples under the +N condition. Data shown represent mean ± SEM (n = 3). (H) Atg11(522-583) is necessary and sufficient for the ability of Atg11 to support the ability of Atg1 to phosphorylate itself and the non-self substrate GST-fused peptide S. Endogenously YFP-tagged Atg1 was immunopurified and analyzed by the in vitro kinase assay. Results shown are the representative of three independent experiments. Quantitation was performed as in D. Values are relative to that of the reaction using Atg1 immunopurified from Atg11(FL)-expressing cells and using peptide S as the non-self substrate.
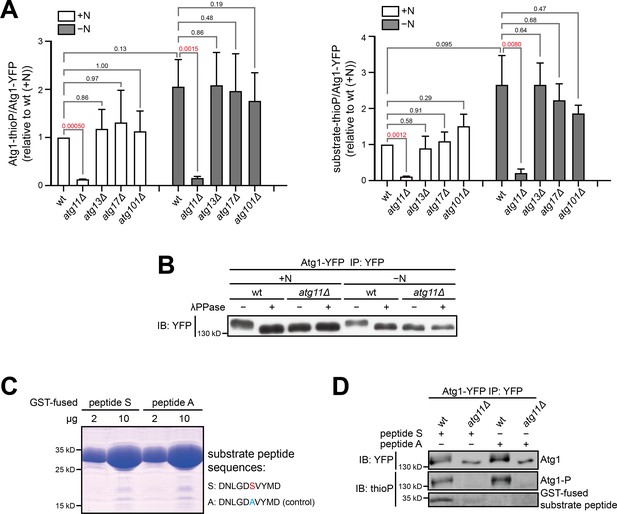
Atg11 is required for Atg1 kinase activity.
(A) Quantitation of phosphorylation signals from three independent experiments including the one shown in Figure 1D. Data shown represent mean ± SEM. P values calculated using the ratio paired t-test are shown. Significant P values are highlighted in red. Left, Atg1 autophosphorylation; right, phosphorylation of the non-self substrate peptide S. (B) The slower migration of Atg1 immunopurified from wild type compared to Atg1 immunopurified from atg11Δ is owing to the phosphorylation of Atg1. Immunopurified Atg1-YFP was treated with or without lambda protein phosphatase (λPPase), and analyzed by immunoblotting with anti-YFP antibody. (C) Atg1 substrate peptide (peptide S) and a negative control peptide (peptide A) were expressed and purified from E. coli as GST-fusions. Left: coomassie-stained SDS-PAGE gel showed the purity of these proteins. Right: the amino acid sequences of peptide S and peptide A. The only phosphorylatable residue in peptide S is highlighted in red, and the alanine substitution in peptide A is highlighted in blue. (D) Atg1 immunopurified from wild type can phosphorylate peptide S but Atg1 immunopurified from atg11Δ cannot.
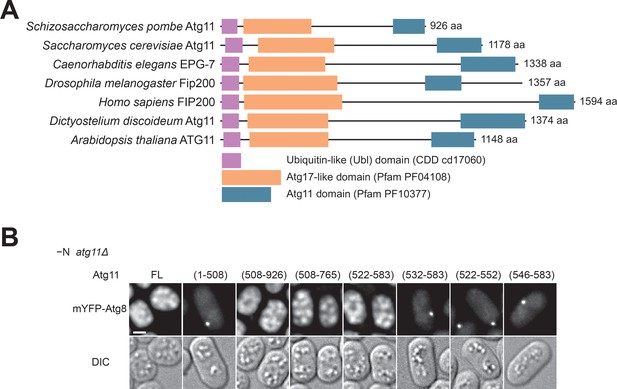
Atg11(522-583) is sufficient for autophagy.
(A) Domain organization of Atg11/FIP200-family proteins. Atg11/FIP200-family proteins from seven representative species all contain three conserved domains: a ubiquitin-like (Ubl) domain (NCBI Conserved Domain Database accession cd17060), an Atg17-like domain (Pfam accession PF04108), and an Atg11 domain (Pfam accession PF10377). Domain boundaries are defined based on our sequence homology analysis. Accession numbers of these protein sequences are: gi|19114767 (Schizosaccharomyces pombe), gi|6325306 (Saccharomyces cerevisiae), gi|1061385443 (Caenorhabditis elegans), gi|45553257 (Drosophila melanogaster), gi|134304842 (Homo sapiens), gi|66808869 (Dictyostelium discoideum), and gi|15234869 (Arabidopsis thaliana). (B) The ability of truncated Atg11 to complement the autophagy defect of atg11Δ was assessed using the starvation-induced vacuolar entry of mYFP-Atg8 as an autophagy readout. Scale bar, 2 μm.
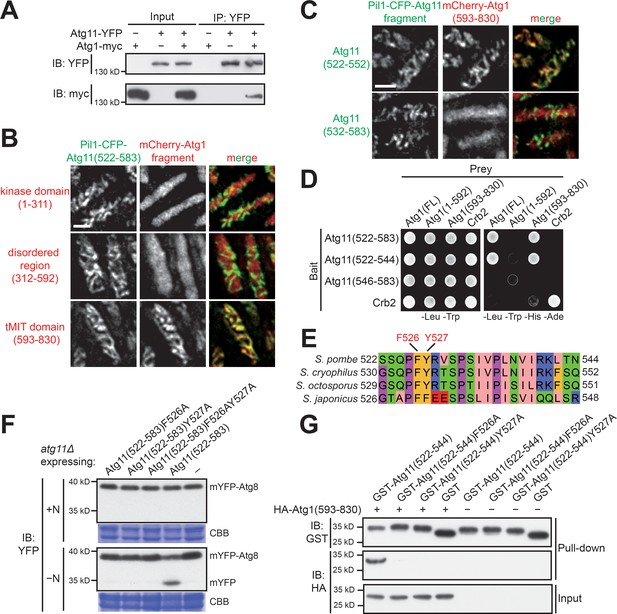
Atg11(522-544) mediates a specific and direct interaction with the tMIT domain of Atg1.
(A) Coimmunoprecipitation assay showed that Atg11 physically interacts with Atg1. Atg11 and Atg1 were endogenously tagged with YFP and myc, respectively. (B) Pil1 co-tethering assay identified an interaction between Atg11(522-583) and the tMIT domain of Atg1. Log-phase cells co-expressing Pil1-CFP-Atg11(522-583) and an mCherry-tagged Atg1 fragment were examined by fluorescence microscopy. Scale bar, 3 μm. (C) Atg11(522-552) but not Atg11(532-583) interacted with the tMIT domain of Atg1 in the Pil1 co-tethering assay. Scale bar, 3 μm. (D) Y2H assay showed that Atg11(522-544) is sufficient for interacting with Atg1. Crb2, which can self-interact, served both as a positive control and a specificity control. (E) The sequence of S. pombe Atg11(522-544) was aligned to the corresponding regions of Atg11 proteins from three other fission yeast species. (F) Mutating either F526 or Y527 to alanine disrupted the autophagy function of Atg11(522-583). mYFP-Atg8 processing assay was performed as in Figure 1A. (G) In vitro GST pull-down assay using recombinant proteins demonstrated a direct interaction between Atg11(522-544) and the tMIT domain of Atg1.
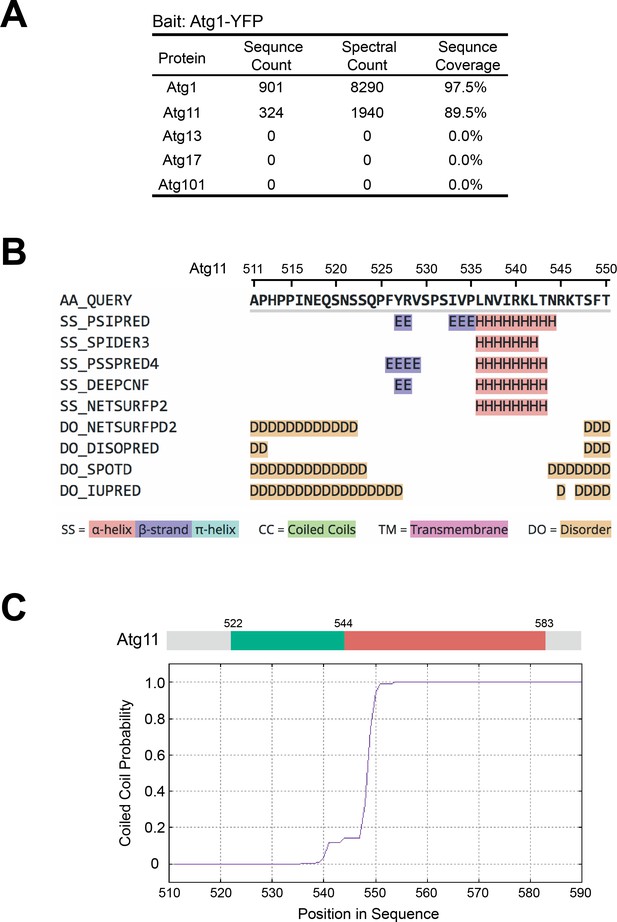
AP-MS analysis of Atg1-associated proteins and secondary structure prediction analysis on Atg11.
(A) AP-MS analysis showed that Atg11, but not Atg13, Atg17, or Atg101, co-purified with Atg1. Endogenously YFP-tagged Atg1 was immunopurified, and purified proteins were identified using mass spectrometry. (B) The sequence of Atg11(511-550) was subjected to secondary structure prediction using the Quick2D tool in the MPI Bioinformatics Toolkit (https://toolkit.tuebingen.mpg.de; Zimmermann et al., 2018). (C) The sequence of Atg11(511-590) was subjected to coiled coil prediction using the MARCOIL tool in the MPI Bioinformatics Toolkit (Delorenzi and Speed, 2002; Zimmermann et al., 2018).
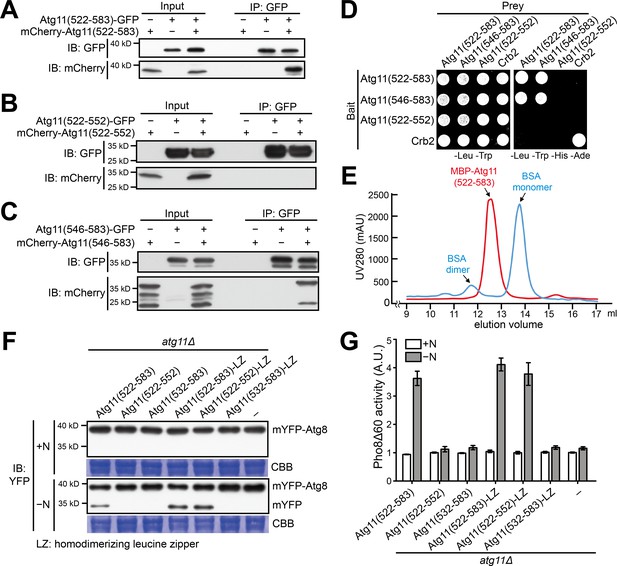
Atg11(546-583) mediates homodimerization.
(A–C) Coimmunoprecipitation assay showed that Atg11(522-583) can self-interact (A), and Atg11(546-583) but not Atg11(522-552) is sufficient for this self-interaction (B and C). To examine whether an Atg11 fragment can self-interact, its fusions with GFP and mCherry were co-expressed and the GFP-fused version was immunoprecipitated. (D) Y2H assay showed that Atg11(522-583) and Atg11(546-583) but not Atg11(522-552) can self-interact. (E) Atg11(522-583) forms a dimer. MBP-tagged Atg11(522-583) purified from E. coli was analyzed by gel filtration chromatography. Absorbance of eluates was monitored at 280 nm. Bovine serum albumin (BSA) monomer (66.5 kD) and dimer (133 kD) served as molecular weight markers. mAU, milli-absorbance units. (F) mYFP-Atg8 processing assay showed that fusing a heterologous dimerization motif (LZ) to Atg11(522-552) restored its ability to rescue the autophagy defect of atg11Δ. (G) Pho8Δ60 assay confirmed that Atg11(522-552)-LZ but not Atg11(522-552) can rescue the autophagy defect of atg11Δ. Data shown represent mean ± SEM (n = 3).
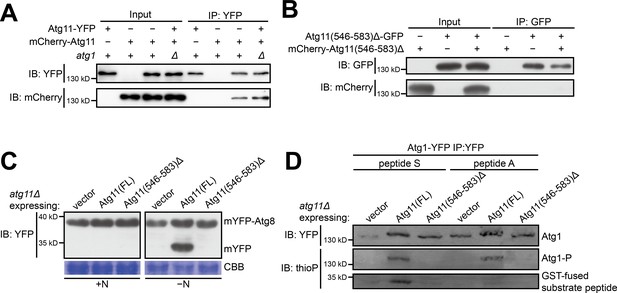
The self-interaction of Atg11 is important for the autophagy function of Atg11 and for supporting the kinase activity of Atg1.
(A) Coimmunoprecipitation assay showed that full-length Atg11 can self-interact independently of Atg1. Atg11 tagged with YFP and mCherry respectively were co-expressed in wt and atg1Δ cells, and the YFP-tagged version was immunoprecipitated. (B) Coimmunoprecipitation assay showed that Atg11(546-583)Δ, which lacks the 38-amin-acid region (546-583) that mediates homodimerization, cannot self-interact. Atg11(546-583)Δ tagged with GFP and mCherry respectively were co-expressed and the GFP-tagged version was immunoprecipitated. (C) mYFP-Atg8 processing assay showed that Atg11(546-583)Δ cannot rescue the autophagy defect of atg11Δ. FL, full length. (D) Atg11(546-583)Δ failed to support the kinase activity of Atg1.
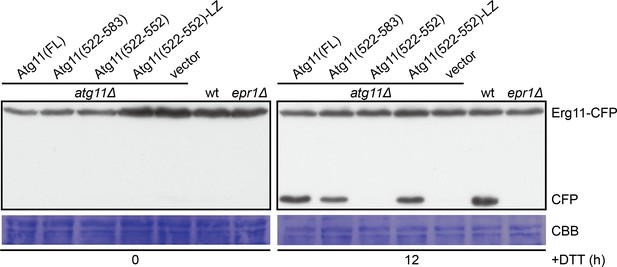
Atg11(522-583) and Atg11(522-552)-LZ but not Atg11(522-552) can support DTT-induced selective ER-phagy.
The processing of CFP-tagged Erg11, an ER membrane protein, was used as a readout for ER stress-induced selective ER-phagy. epr1Δ mutant lacking the ER-phagy receptor required for DTT-induced ER-phagy was used as a control. Cells were collected before and after DTT treatment for 12 hr, and lysates were analyzed by immunoblotting using an antibody that can recognize CFP.
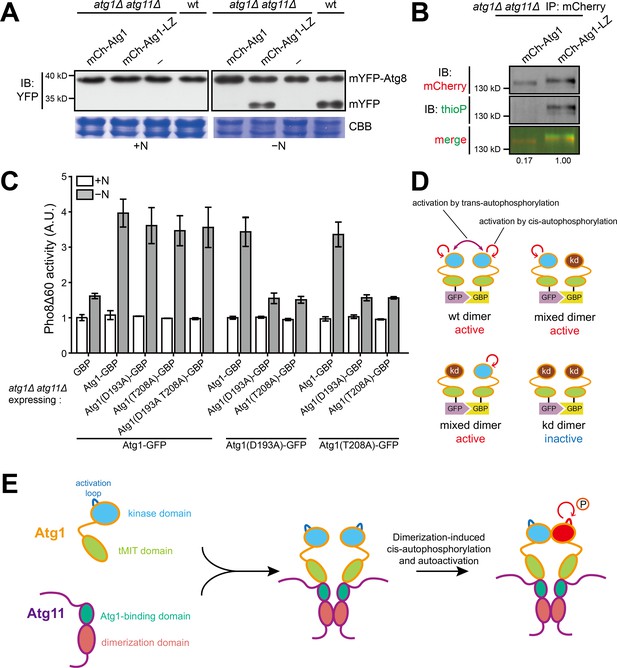
Dimerization of Atg1 activates its kinase activity through cis-autophosphorylation.
(A) mYFP-Atg8 processing assay showed that fusing a heterologous dimerization motif (LZ) to Atg1 bypassed the requirement of Atg11 for autophagy. (B) In vitro Atg1 kinase assay showed that LZ-mediated dimerization of Atg1 bypassed the requirement of Atg11 for Atg1 kinase activity. Quantification was performed as in Figure 1D, and reported values are relative to that of the Atg1-LZ sample. Results shown are the representative of three independent experiments. (C) Pho8Δ60 assay showed that artificial heterodimerization of Atg1 mediated by GFP and GFP-binding protein (GBP) resulted in the bypass of Atg11 even when one Atg1 molecule in a dimer is inactive. GFP-tagged Atg1 was co-expressed with GBP-tagged Atg1 in atg1Δ atg11Δ cells. Data shown represent mean ± SEM (n = 3). (D) Diagram explaining the results in C. kd, kinase-dead. (E) Model depicting how Atg11 promotes Atg1 activation through dimerization-induced cis-autophosphorylation of Atg1.
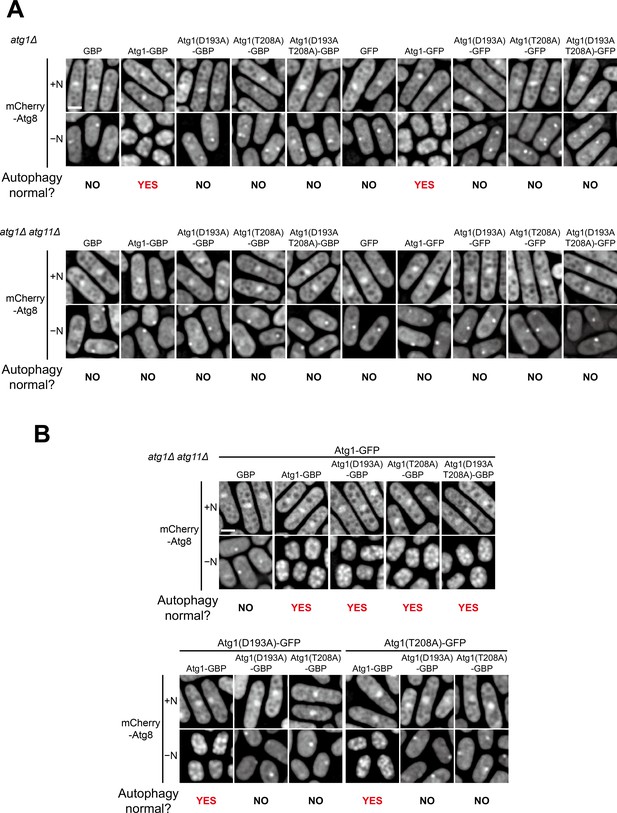
Atg1-GBP or Atg1-GFP alone rescued atg1Δ but not atg1Δ atg11Δ, and their combination rescued atg1Δ atg11Δ even when one of them was kinase-dead.
(A) GBP, GBP-tagged Atg1, GFP, or GFP-tagged Atg1 was separately expressed in atg1Δ and atg1Δ atg11Δ cells. Starvation-induced vacuolar entry of mCherry-Atg8 was used as an autophagy readout. Cells were collected before (+N) and after nitrogen starvation for 8 hr (−N), and examined by fluorescence microscopy. Scale bar, 3 μm. (B) Artificial heterodimerization of Atg1 mediated by the interaction between GFP and GBP rescued the autophagy defect of atg1Δ atg11Δ when one but not both subunits of the heterodimer was kinase-dead. GFP-tagged Atg1 was co-expressed with GBP-tagged Atg1 in atg1Δ atg11Δ cells. Starvation-induced vacuolar entry of mCherry-Atg8 was used as an autophagy readout. Cells were collected before (+N) and after nitrogen starvation for 8 hr (−N), and examined by fluorescence microscopy. Scale bar, 3 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Schizosaccharomyces pombe) | atg1 | PomBase | SPCC63.08C | |
| Gene (Schizosaccharomyces pombe) | atg11 | PomBase | SPAC7D4.04 | |
| Genetic reagent (Schizosaccharomyces pombe) | Fission yeast strains used in this study | This paper | See Supplementary file 1 | |
| Antibody | Anti-GFP (mouse monoclonal) | Roche | Cat# 11814460001; RRID:AB_390913 | 1:3000 dilution |
| Antibody | Anti-mCherry (mouse monoclonal) | Huaxingbio | Cat# HX1810 | 1:1000 dilution |
| Antibody | Anti-thiophosphate ester (rabbit monoclonal) | Abcam | Cat# ab92570; RRID:AB_10562142 | 1:5000 dilution |
| Antibody | Anti-myc (mouse monoclonal) | Abmart | Cat# M200002L | 1:3000 dilution |
| Antibody | Anti-GST (mouse monoclonal) | Abmart | Cat# M200007 | 1:3000 dilution |
| Antibody | Anti-HA (mouse monoclonal) | MBL | Cat# M180-3; RRID:AB_10951811 | 1:3000 dilution |
| Recombinant DNA reagent | Plasmids used for this study | This paper | See Supplementary file 1 | |
| Chemical compound, drug | ATPγS | Sigma-Aldrich | Cat# A1388 | |
| Chemical compound, drug | p-Nitrobenzyl mesylate (PNBM) | Abcam | Cat# ab138910 | |
| Commercial assay or kit | GFP-Trap agarose beads | ChromoTek | Cat# gta-20; RRID:AB_2631357 | |
| Commercial assay or kit | RFP-Trap agarose beads | ChromoTek | Cat# rta-20; RRID:AB_2631362 | |
| Commercial assay or kit | Glutathione Sepharose 4 Fast Flow | GE Healthcare | Cat# 17-0756-01 | |
| Commercial assay or kit | Ni-NTA Agarose | QIAGEN | Cat# 30210 |
Additional files
-
Supplementary file 1
Fission yeast strains and plasmids used in this study.
- https://cdn.elifesciences.org/articles/58073/elife-58073-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58073/elife-58073-transrepform-v2.doc





