Cryo-EM structure of VASH1-SVBP bound to microtubules
Figures
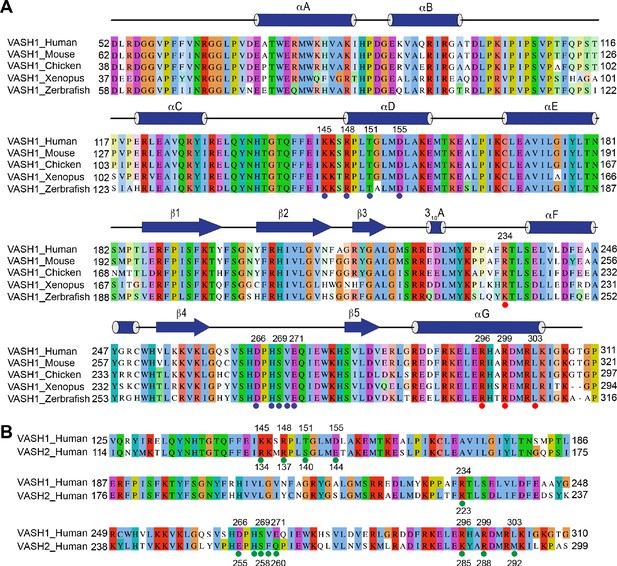
VASH1-SVBP efficiently detyrosinates and binds microtubules.
(A,B) Tubulin detyrosination assays of VASH1-SVBP in human cells. HeLa Tet-On cells were co-transfected with VASH1 and SVBP plasmids, and treated with 5 µM nocodazole (A) or 100 nM Taxol (B) for indicated times at 24 hr post-transfection. The cell lysates were blotted with the indicated antibodies. deY-tubulin, detyrosinated α-tubulin. Experiments were repeated three times with similar results. (C) Quantification of the relative detyrosination levels of α-tubulin in (A) and (B) (mean ± s.d., n = 3 independent experiments). Significance calculated using two-tailed student’s t-test; between control cells and cells treated with nocodazole or Taxol for the indicated time; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (D) Microtubule pelleting assays showing the binding of VASH1-SVBP to recombinant human microtubules. S, supernatant; P, pellet. (E) Cryo-EM map of 14-protofilament, GMPCPP-stabilized microtubules decorated by the VASH152-310-SVBP complex. The catalytically inactive C169S mutant of VASH1 was used in the complex. The map is lowpass filtered to 4 Å. The microtubule seam is indicated by a red dashed line. α-tubulin, β-tubulin, VASH1, and SVBP are colored in green, cyan, blue, and orange, respectively. The same color scheme is used for all figures. The inset shows a close-up view of the boxed region. (F) Close-up view of the cryo-EM map in (E), viewed from the lumen. The α- and β-tubulin molecules can be distinguished by the length of the S9-S10 loop (boxed with red dashed lines), with the loop in α-tubulin being longer.

Structure determination of VASH1-SVBP-decorated GMPCPP-microtubules.
(A) A representative micrograph of VASH1-SVBP-decorated GMPCPP-microtubules. Scale bar, 50 nm. (B) Processing workflow for cryo-EM structure determination of VASH1-SVBP-decorated GMPCPP-microtubules. The 2D classes of poorly decorated microtubules (red outlines) were discarded whereas the classes belonging to efficiently decorated microtubules (green outlines) were selected for subsequent 3D classification. (C) Fourier shell correlation (FSC) curves of GMPCPP-microtubules decorated with VASH1-SVBP. The FSC curves of microtubules (top) and VASH1-SVBP (bottom) were calculated separately. The final resolution for the reconstruction was estimated by calculating the Fourier shell correlation (FSC) of a single tubulin heterodimer in a ‘good’ protofilament after pseudo-helical averaging, using the FSC = 0.143 criterion.
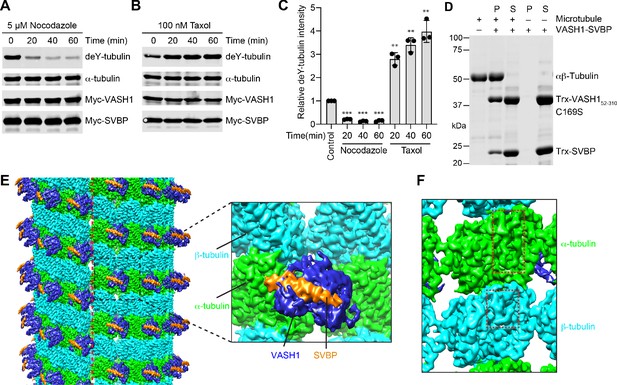
Cryo-EM structure of VASH1-SVBP bound to GMPCPP-stabilized microtubules.
(A) Models of VASH1-SVBP (VASH1, blue; SVBP, orange) and tubulin (α-tubulin, green; β-tubulin, cyan) were docked into the cryo-EM density (lowpass-filtered to 4 Å) and refined. (B) Cryo-EM density map of VASH1-SVBP lowpass-filtered to 6 Å. (C) Ribbon diagram of the cryo-EM structure of VASH1-SVBP bound to microtubules. (D) Two views of the electron density map (generated by Phenix.auto_sharpen, with local B factor sharpening and resolution cutoff at 7 Å) showing an unfitted, continuous density that belonged to the α-tubulin C-terminal tail (CTα).

Cryo-EM map of VASH1-SVBP-decorated GMPCPP-microtubules.
(A) Density corresponding to the αβ-tubulin heterodimer and the VASH1-SVBP complex colored by local resolution as determined in relion_postprocess. (B) Electron density map of nucleotides in the N-site (left) and E-site (right) of microtubules. (C) Representative regions of the cryo-EM map of α-tubulin, β-tubulin, and VASH1, highlighting the density of key residues at VASH1-microtubule interfaces.

Interactions between microtubules and VASH1-SBVP.
(A) Overlay of the ribbon diagrams of the cryo-EM structure of microtubule-bound VASH1-SVBP (colored blue and orange) and the crystal structure of VASH1-SVBP alone (PDB: 6OCG) (colored gray). (B) Surface drawing of the structure of VASH1-SVBP bound to two neighboring αβ-tubulin heterodimers in two different views. The C-terminal tail of α-tubulin (CTα) is indicated by a dashed green line. (C) Solvent-accessible surface of the VASH1-SVBP complex (PDB: 6OCG) colored by electrostatic potential (blue, positive; red, negative).
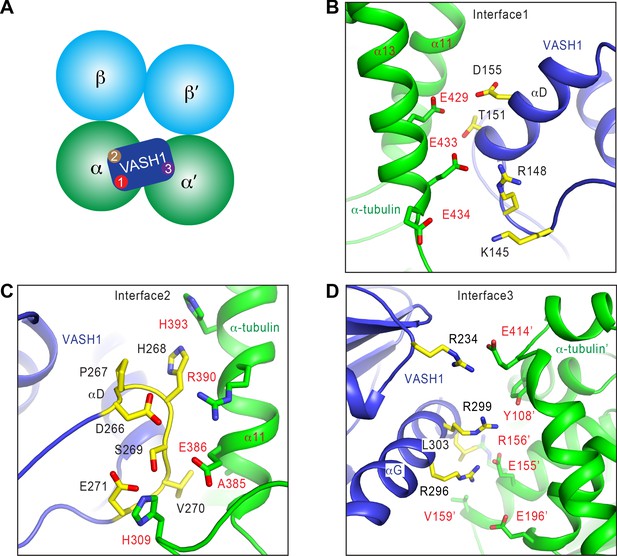
Interactions between VASH1 and microtubules.
(A) Schematic drawing of the VASH1-microtubule complex, with the three main interfaces indicated. (B–D) Close-up views of the VASH1–microtubule interfaces 1 (B), 2 (C), and 3 (D), with interacting residues shown as sticks. VASH1 residues are colored yellow and labeled with black letters while α-tubulin residues are colored green and labeled with red letters.
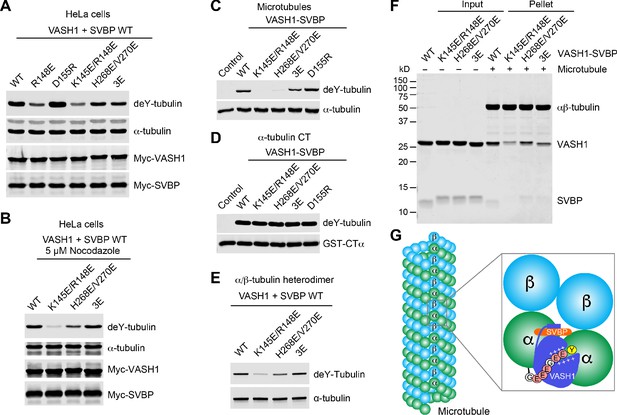
Requirement of VASH1-microtubule interactions in α-tubulin detyrosination.
(A,B) Tubulin detyrosination assays of VASH1-SVBP WT or mutants in human cells. HeLa Tet-On cells were co-transfected with Myc-VASH1 WT or mutants and Myc-SVBP WT plasmids. At 24 hr post-transfection, the cells were treated without (A) or with 5 µM Nocodazole (B) for 1 hr. The cell lysates were blotted with the indicated antibodies. Compared with VASH1 WT, VASH1 mutants with multiple glutamate substitutions had slightly slower mobilities. The mobility shift is likely caused by the introduction of multiple negative charges, akin to protein phosphorylation, which also sometimes retards gel mobility. deY-tubulin, detyrosinated α-tubulin. 3E, R234E/R299E/L303E. Experiments were repeated three times with similar results. (C–E) In vitro detyrosination of GMPCPP-stabilized human microtubules (C), the C-terminal peptide of α-tubulin (CTα) fused to GST (D), or free αβ-tubulin heterodimers (E) by the indicated recombinant VASH1–SVBP WT or mutant complexes. Experiments were repeated at least three times with similar results. (F) Coomassie-stained gel of microtubule pelleting assays of VASH1-SVBP WT and mutant complexes. (G) Model of microtubule lattice binding, substrate recognition, and tubulin detyrosination by VASH1-SVBP. The ‘+’ signs indicate positive charges.

Requirement of microtubule-VASH1 interactions in α-tubulin detyrosination.
(A–E) Quantification of the relative detyrosination levels of α-tubulin in Figure 4A (A), 4B (B), 4C (C), 4D (D), and 4E (E). Mean ± s.d.; n = 3 independent experiments. (F) Quantification of the percentage of VASH1-SVBP bound to microtubules in Figure 4F. Mean ± s.d.; n = 3 independent experiments. Significance calculated using two-tailed student’s t-test; between WT and mutants; *p < 0.05, **p < 0.01 ***p < 0.001, and ****p < 0.0001.
Tables
Data collection and refinement statistics.
| VASH1-SVBP-microtubule | |
|---|---|
| Data collection and processing | |
| Magnification | 105,000 |
| Voltage (kV) | 300 |
| Electron exposure (e-Å−2) | 50 |
| Defocus range (μm) | −0.9 to −2.5 |
| Pixel size (Å) | 0.83 |
| Symmetry imposed | Pseudo-Helical |
| Initial particle images (no.) | 156,525 |
| Final particle images (no.) | 46,999 |
| Map resolution (Å)/FSC threshold | 3.1/0.143 |
| Map sharpening B factor (Å−2) | −60 |
| Refinement | |
| Initial model used | PDB: 6OCG, 6DPU |
| Model resolution (Å)/FSC threshold | 3.9/0.5 |
| Model composition | |
| Nonhydrogen atoms | 18,196 |
| Protein residues | 2296 |
| Ligands | 4 |
| B factors (Å−2) | |
| Protein | 143.68 |
| Ligands | 101.97 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.931 |
| Validation | |
| MolProbity score | 1.81 |
| Clashscore | 6.2 |
| Poor rotamers (%) | 0.36 |
| Ramachandran plot | |
| Favored (%) | 92.56 |
| Allowed (%) | 7.35 |
| Disallowed (%) | 0.09 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) | Novagen | Cat.#: 69450 | competent cells |
| Cell line (Homo-sapiens) | HeLa Tet-On cells | Takara Bio | Cat.#: 631183 RRID:CVCL_V353 | Cell based detyrosination assay |
| Transfected construct (Homo-sapiens) | pCS2-MYC-human VASH1 WT | This paper | Cell based detyrosination assay | |
| Transfected construct (Homo-sapiens) | pCS2-MYC-human VASH1 R148E | This paper | Cell based detyrosination assay | |
| transfected construct (Homo-sapiens) | pCS2-MYC-human VASH1 D155R | This paper | Cell based detyrosination assay | |
| Transfected construct (Homo-sapiens) | pCS2-MYC-human VASH1 K145E/R148E | This paper | Cell based detyrosination assay | |
| Transfected construct (Homo-sapiens) | pCS2-MYC-human VASH1 H268E/V270E | This paper | Cell based detyrosination assay | |
| Transfected construct (Homo-sapiens) | pCS2-MYC-human VASH1 R234E/R299E/L303E | This paper | Cell based detyrosination assay | |
| Transfected construct (Homo-sapiens) | pCS2-MYC-human SVBP | This paper | Cell based detyrosination assay | |
| Antibody | anti-Myc (Mouse monoclonal) | Roche | Cat.#: 11667203001, RRID:AB_390911 | WB (1:5000) |
| Antibody | anti-α-tubulin (Mouse monoclonal) | Sigma-Aldrich | Cat.#: T6199, RRID:AB_477583 | WB (1:2000) |
| Antibody | anti-detyrosinated tubulin (rabbit Polyclonal) | EMD Millipore | Cat.#: AB320, RRID:AB_177350 | WB (1:2000) |
| Antibody | anti-GST (Mouse monoclonal) | Sigma-Aldrich | Cat.#: SAB4200237, RRID:AB_2858197 | WB (1:2000) |
| Antibody | anti-rabbit IgG (H+L) (Dylight 800 conjugates) | Cell signaling | Cat.#:5151 RRID:AB_10697505 | WB (1:5000) |
| Antibody | anti-mouse IgG (H+L) (Dylight 680 conjugates) | Cell signaling | Cat.#: 5470 RRID:AB_10696895 | WB (1:5000) |
| Recombinant DNA reagent | pRSF-32M-3C-VASH152–310 WT | This paper | See Materials and methods, Section: Protein expression and purification | |
| Recombinant DNA reagent | pRSF-32M-3C-VASH152–310 C169S | This paper | See Materials and methods, Section: Protein expression and purification | |
| Recombinant DNA reagent | pRSF-32M-3C-VASH152–310K145E/R148E | This paper | See Materials and methods, Section: Protein expression and purification | |
| Recombinant DNA reagent | pRSF-32M-3C-VASH152–310H268E/V270E | This paper | See Materials and methods, Section: Protein expression and purification | |
| Recombinant DNA reagent | pRSF-32M-3C-VASH152–310R234E/R299E/L303E | This paper | See Materials and methods, Section: Protein expression and purification | |
| Recombinant DNA reagent | pRSF-32M-3C-VASH152–310 D155R | This paper | See Materials and methods, Section: Protein expression and purification | |
| Recombinant DNA reagent | pET-32M-3C-SVBP | This paper | See Materials and methods, Section: Protein expression and purification | |
| Recombinant DNA reagent | pET-21b-SVBP | This paper | See Materials and methods, Section: Protein expression and purification purification | |
| Sequence-based reagent | VASH1_1up Fse1 sense | This paper | PCR primers | GGAGGCCGGCCAATGCCAGGGGGGAAGAAG |
| Sequence-based reagent | VASH1_52 up Fse1 sense | This paper | PCR primers | CGAGGCCGGCCAGACCTGCGAGACGGAGGC |
| Sequence-based reagent | VASH1_310 down Asc1 anti-ense | This paper | PCR primers | CCAGGCGCGCCCTAGACCCGGATCTGGTACCC |
| Sequence-based reagent | VASH1_365 down Asc1 anti-ense | This Paper | PCR primers | CCAGGCGCGCCCTAGACCCGGATCTGGTACCC |
| Sequence-based reagent | SVBP_1up Fse1 sense | This Paper | PCR primers | TGCGGCCGGCCAATGGATCCACCTGCACGT |
| Sequence-based reagent | SVBP_66 down Asc1 anti-sense | This Paper | PCR primers | CGTGGCGCGCCTCATTCTCCAGGAGGCTGC |
| Sequence-based reagent | VASH1_C169S anti-sense | This Paper | PCR primers | TTTGATTGGCAGGGCCTCT |
| Sequence-based reagent | VASH1_C169S sense | This Paper | PCR primers | AGCCTGGAAGCCGTGATCC |
| Sequence-based reagent | VASH1 K145E anti-sense | This Paper | PCR primers | AATTTCAAAGAACTGTGTCCCTGT |
| Sequence-based reagent | VASH1 K145E sense | This Paper | PCR primers | GAGAAGAGCAGACCTCTGACAGG |
| Sequence-based reagent | VASH1 R148E anti-sense | This Paper | PCR primers | GCTCTTCTTAATTTCAAAGAACTGT |
| Sequence-based reagent | VASH1 R148E sense also used for K145E/R148E mutation | This Paper | PCR primers | GAACCTCTGACAGGGCTGATG |
| Sequence-based reagent | VASH1 K145E/R148E anti-sense | This Paper | PCR primers | GCTCTTCTCAATTTCAAAGAACTGT |
| Sequence-based reagent | VASH1_D155R sense | This Paper | PCR primers | AGGGCTGATGCGCCTGGCCAAGG |
| Sequence-based reagent | VASH1_D155R anti-sense | This Paper | PCR primers | GTCAGAGGTCTGCTCTTC |
| Sequence-based reagent | VASH1_R234E sense | This Paper | PCR primers | GCCCGCCTTCGAGACGCTCAGCG |
| Sequence-based reagent | VSH1_R234E anti-sense | This Paper | PCR primers | GGCTTGTACATCAGGTCC |
| Sequence-based reagent | VASH1_268/270E sense | This Paper | PCR primers | CGAGGAGCAGATCGAGTGGAAGCAC |
| Sequence-based reagent | VASH1_268/270E anti-sense | This Paper | PCR primers | CTCTCCGGGTCGTGTGACACGCT |
| Sequence-based reagent | VASH1_R299E sense | This Paper | PCR primers | GCGCCACGCCGAGGACATGCGGC |
| Sequence-based reagent | VASH1_R299E anti-sense | This Paper | PCR primers | TCCAGCTCCTTGCGGAAG |
| Sequence-based reagent | VASH1_L303E sense | This Paper | PCR primers | CGACATGCGGGAGAAGATTGGCAAAGGGACGGGC |
| Sequence-based reagent | VASH1_L303E anti-sense | This Paper | PCR primers | CGGGCGTGGCGCTCCAGC |
| Peptide, recombinant protein | VASH152-310 WT in complex with SVBP | This paper | In vitro detyrosination and pelleting assay | |
| Peptide, recombinant protein | VASH152-310 D155R in complex with SVBP | This paper | In vitro detyrosination | |
| Peptide, recombinant protein | VASH152-310 K145E/R148E in complex with SVBP | This paper | In vitro detyrosination and pelleting assay | |
| Peptide, recombinant protein | VASH152-310 H268E/V270E in complex with SVBP | This paper | In vitro detyrosination and pelleting assay | |
| Peptide, recombinant protein | VASH152-310 R234E/R299E/L303E in complex with SVBP | This paper | In vitro detyrosination and pelleting assay | |
| chemical compound, drug | GMPCPP | Jena Bioscience | Cat.#: NC0641143 | |
| chemical compound, drug | Taxol | Cytoskeleton | Cat.#: TXD01 | |
| chemical compound, drug | Nocodazole | Sigma-Aldrich | Cat. #: M1404 | |
| chemical compound, drug | Isopropyl-beta-D-thiogalactoside (IPTG) | Gold Biotechnology | Cat. #: 12481C100 | Induce protein expression |
| Software, algorithm | UCSF Chimera | Pettersen et al., 2004 | RRID:SCR_004097 | https://www.cgl.ucsf.edu/chimera/ |
| Software, algorithm | MotionCorr2 | Zheng et al., 2017 | RRID:SCR_016499 | http://msg.ucsf.edu/em/software/motioncor2.html |
| Software, algorithm | GCTF | Zhang, 2016 | RRID:SCR_016500 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| Software, algorithm | RELION3 | Zivanov et al., 2018 | RRID:SCR_016274 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Download_%26_install |
| Software, algorithm | Coot | Emsley and Cowtan, 2004 | RRID:SCR_014222 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Software, algorithm | Phenix.refine | Adams et al., 2010 | RRID:SCR_014224 | https://www.phenix-online.org/documentation/reference/refinement.html |
| Software, algorithm | Graphpad prism 8.30 | Graphpad | RRID:SCR_002798 | https://www.graphpad.com/scientific-software/prism/ |
| Software, algorithm | Frealign | Grigorieff, 2016 | RRID:SCR_016733 | https://grigoriefflab.umassmed.edu/frealign |
| Other | Ni-NTA Agarose | Qiagen | Cat. #: 30230 | Recombinant protein purification |
| Other | Lipofectamine 2000 | Thermo Fisher Scientific | Cat. #: 11668019 | Mammalia cell transfection |