The push-to-open mechanism of the tethered mechanosensitive ion channel NompC
Figures
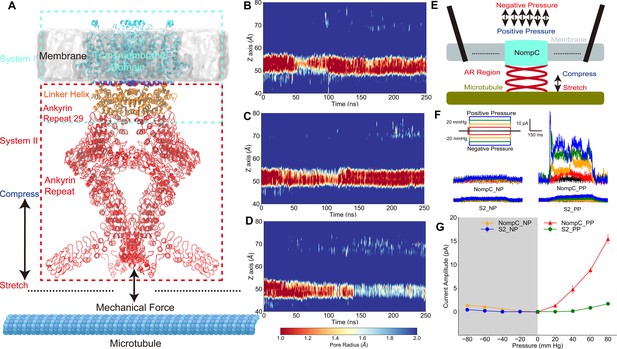
The tethered NompC channel was opened by compression of the intracellular ankyrin repeat domain.
(A) The simulation systems. The NompC was divided into two subsystems, denoted by the cyan and red rectangular boxes, for the molecular dynamics (MD) simulations. (B−D) The transmembrane (TM) pore size evolution for the force-free (B), pulling/stretch (C), and pushing/compression (D) simulations, calculated from the MD trajectories FI0, SI0, and CI0 (Supplementary file 1a), respectively. (E) A schematic figure of the cell-attached patch-clamp electrophysiological experiment for NompC. (F) Representative traces of the electrophysiological measurements for the S2 blank cell and NompC-expressed cell, showing that there are significantly larger signals under positive pressure (PP) in the presence of NompC. (G) The mean and standard deviation (SD) of the mechano-gated currents in the S2 blank and NompC expressing cells under positive (PP) and negative pressure (NP) in the cell-attached patch-clamp experiments (S2_NP: n = 6; S2_PP: n = 6; NompC_NP: n = 5; NompC_PP: n = 7). All of the error bars denote ± SD.
The sequence of NompC used for the molecular dynamics (MD) simulations (highlighted).
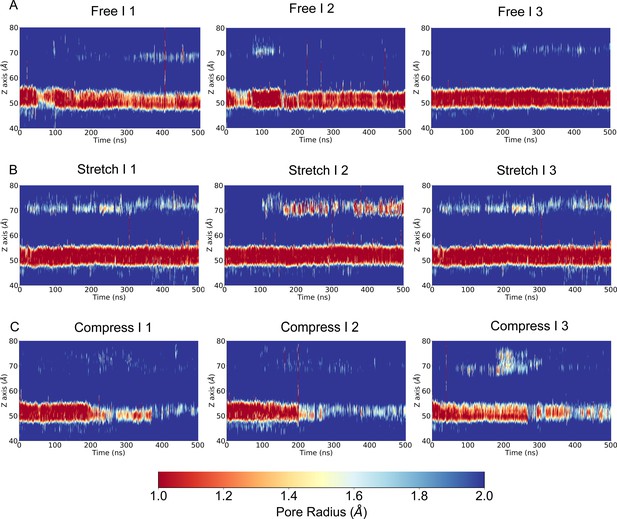
The transmembrane (TM) pore size evolution of multiple replicate simulations with a slower pulling/pushing speed, for the force-free, pulling/stretch, and pushing/compress simulations, calculated from the molecular dynamics (MD) trajectories FI1-3, SI1-3, and CI1-3 (Supplementary file 1a), respectively.

This figure is similar to Figure 1B–D except that the pore radius was calculated with the structure of the transmembrane (TM) backbone (side chain removed) in the molecular dynamics (MD) trajectories FI0, SI0, and CI0 (Supplementary file 1a),respectively.
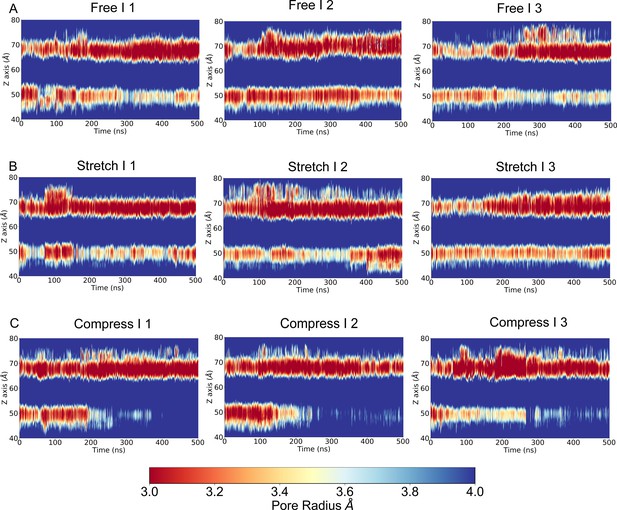
This figure is similar to Figure 1—figure supplement 2 except that the pore radius was calculated with the structure of the transmembrane (TM) backbone only (side chain removed) in the molecular dynamics (MD) trajectories FI1-3, SI1-3, and CI1-3, respectively.
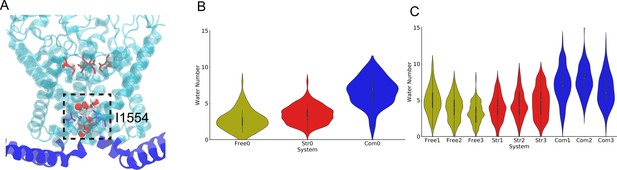
The number of water molecules around the gate region of NompC in the molecular dynamics (MD) simulations.
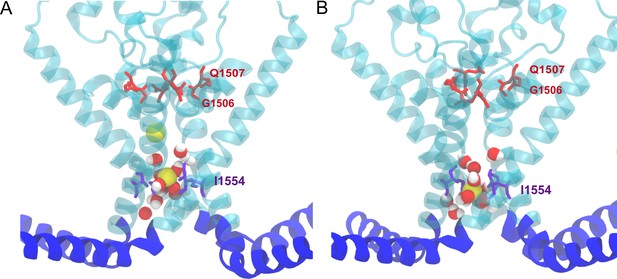
Sodium ions spontaneously permeated through the partially opened gate in the ‘pushing’ molecular dynamics (MD) simulations in the absence of a transmembrane potential in the trajectories CI1 and CI2, respectively.
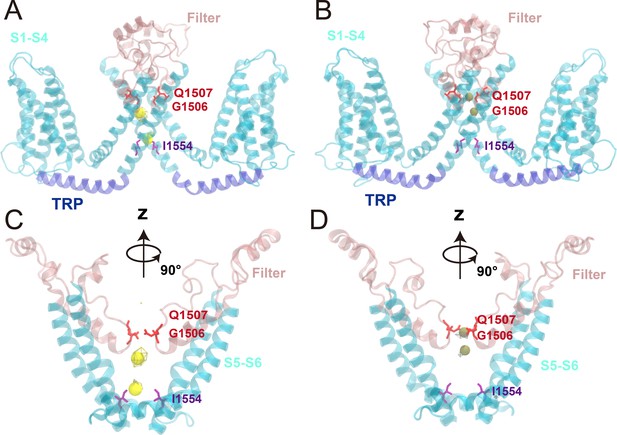
Ion density maps from the ion permeation trajectories.
The ion density maps were obtained from the simulation trajectories with a −300 mV transmembrane potential for Na+ ions and a 300 mV transmembrane potential for K+ ions, calculated from the molecular dynamics (MD) trajectories PI1-3 and PII1-3 (Supplementary file 1c), respectively.

The sodium ion and potassium ion permeation count through the partially opened structure of NompC in the permeation molecular dynamics (MD) trajectories PI1-3 and PII1-3, respectively.

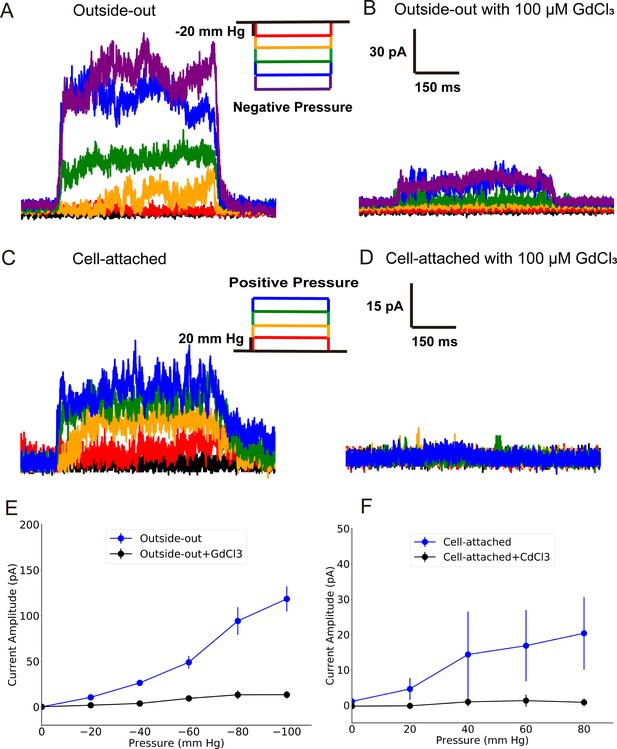
Experiment results of the inside-out (IO) and outside-out (OO) patch clamp.
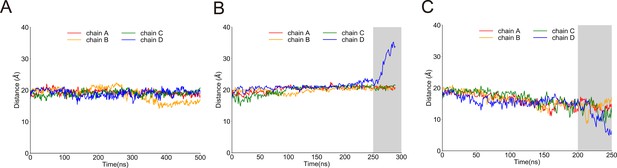
Distances between the centers of AR29 and the transmembrane (TM) domain of NompC in the 250 ns molecular dynamics/steered molecular dynamics (MD/SMD) simulations of system I, for the free, pulling, and pushing simulations, calculated from the MD trajectories FI0, SI0, and CI0, respectively.

Distances between the centers of AR29 and the transmembrane (TM) domain of NompC in the 500 ns molecular dynamics/steered molecular dynamics (MD/SMD) simulations of system I, for the free, pulling, and pushing simulations, calculated from the MD trajectories FI1-3, SI1-3, and CI1-3, respectively.
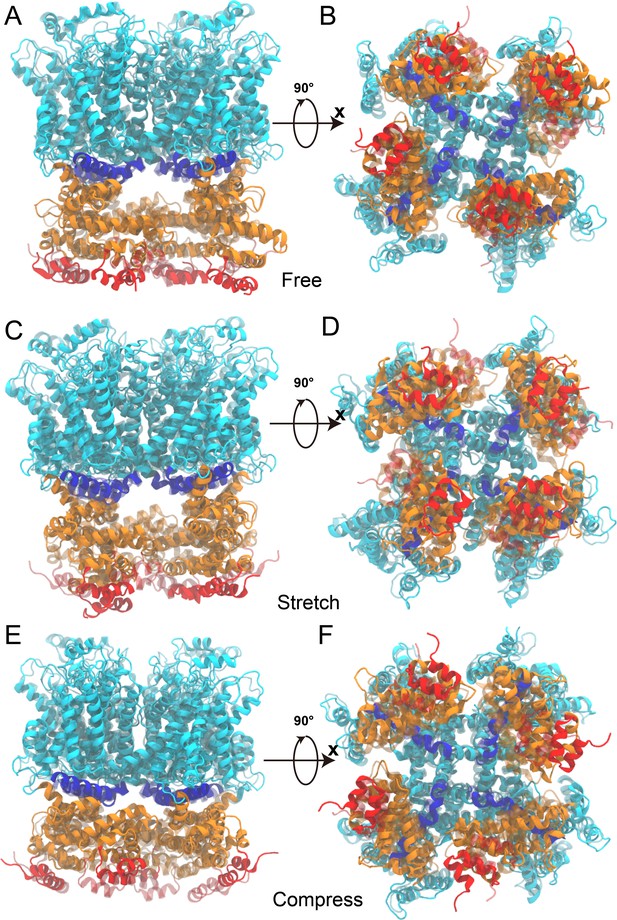
The overlaid initial and 200 ns conformations of the simulation system I in the free, pulling/stretch, and pushing/compress simulations, from the molecular dynamics (MD) trajectories FI0, SI0, and CI0, respectively.
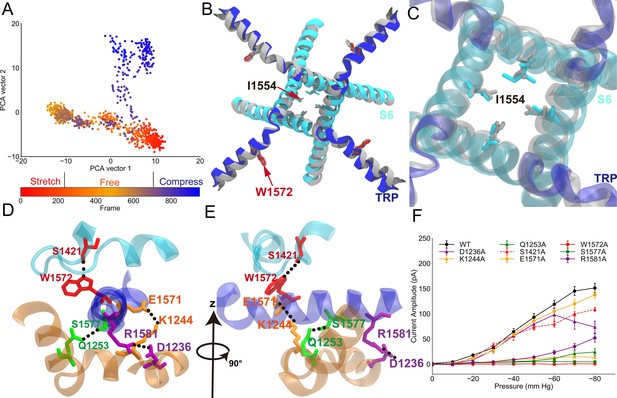
Conformational changes of the transient receptor potential (TRP) and transmembrane (TM) domains during gating.
(A) Principal component analysis (PCA) of the molecular dynamics (MD) simulation trajectories (FI0, SI0, and CI0 in Supplementary file 1a). The projections on the second eigenvector can distinguish the conformations under pulling (red) or pushing (blue). (B) The overlaid extreme structures along the second eigenvector of the PCA. The most closed conformation (silver) and open conformation (cyan) showed the global changes of the TRP domain during gating: a clockwise rotation. (C) The orientation and position of the gate residue, I1554, in the most closed (silver) and open (cyan) conformations in the simulations. (D, E) The residues forming four stable hydrogen bonds between the TRP and LH domains. (F) The mean and standard deviation (SD) of the mechano-gated current of the wild-type NompC, as well as the mutants W1572A, S1421A, Q1253A, S1577A, K1244A, E1571A, D1236A, and R1581A, under negative pressure in the outside-out patch-clamp experiments (wild type: n = 13; W1572A: n = 7; S1421A: n = 6; Q1253A: n = 6; S1577A: n = 5; K1244A: n = 6; E1571A: n = 6; D1236A: n = 9; R1581A: n = 6). All of the error bars denote ± SD. Hydrogen bonds are indicated by dashed lines (D, E).
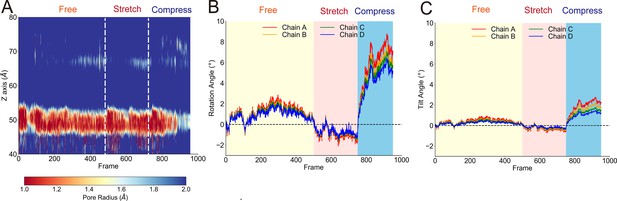
The transmembrane (TM) pore size evolution and rotation angle evolution of the transient receptor potential (TRP) domain from principal component analysis (PCA).

The conformational change of the transient receptor potential (TRP) domain in the steered molecular dynamics (SMD) simulations.
The side and bottom views of the S6 and TRP domains before and after pulling or pushing.
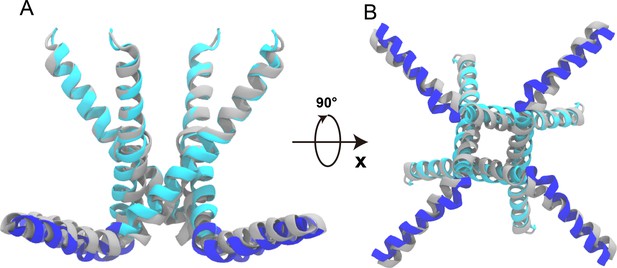
The overlaid closed-state and open-state structures of TRPV1, obtained in lipid nanodisc.
The schematic figure of pUAST-NOMPC_EGFP (del-miniwhite) used in the experiment.
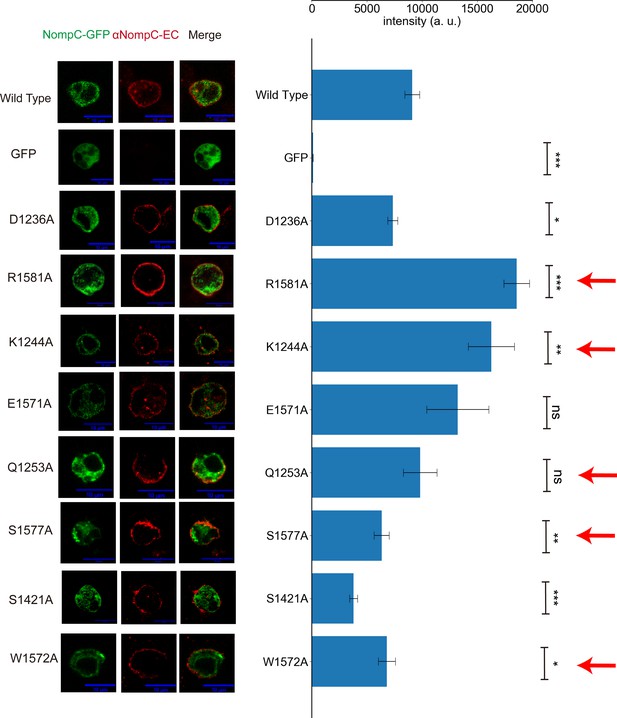
The mutants of residues listed in Figure 2D–F showed normal membrane targeting.
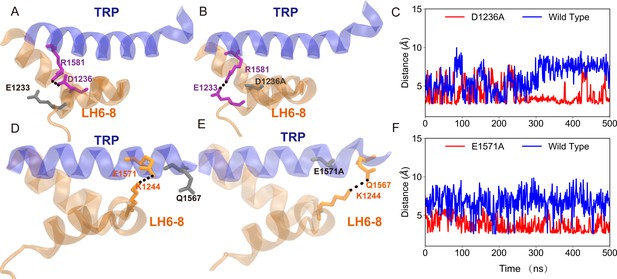
Formation of the alternative hydrogen bonds in the mutant, as identified in the molecular dynamics (MD) simulations (from chain A in the trajectories D1236A and E1571A in Supplementary file 1a).
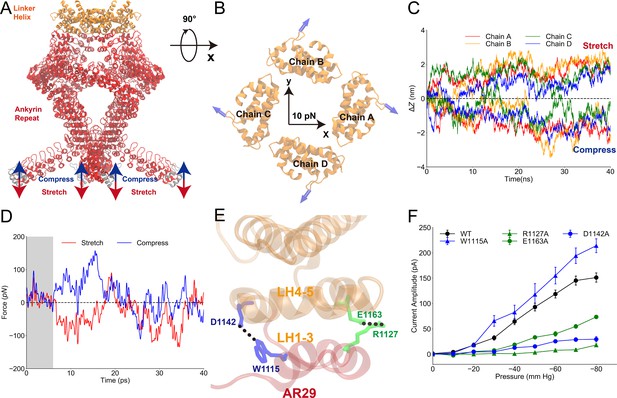
Mechanical properties of the ankyrin repeat (AR) region.
(A) The simulation system in which the linker helix (LH) domain (orange) was restrained and a pushing or pulling force was applied to the first AR (gray). (B) Projection of the reaction forces of the restraints on the LH domain (same as the forces exerted on the transient receptor potential [TRP] domain by the LH domain) on the plane parallel to the membrane surface, showing that a torque is generated that will drive the LH and TRP domain to rotate clockwise (looking from the intracellular side). The calculation was based on the molecular dynamics (MD) trajectories CII1 and symmetrized from the original data as shown in Figure 3—figure supplement 1. (C) The AR region was compressed/stretched by a pushing/pulling force of 5 pN and reached its equilibrium length within 40 ns simulations (from MD trajectories SII1 and CII1 with respect to FII1). (D) The evolution of the net average reaction forces of the restraints on the LH domain when pushing (blue) or pulling forces (red) were applied to AR1, calculated from the MD trajectories FII2-6, CII2-6, and SII2-6 (Supplementary file 1b). A clear deviation occurred at around 7-8 ps during the simulation time, indicating that the forces applied to AR1 have reached LH at the time. (E) The residues forming two stable hydrogen bonds between the LH domain and AR29. (F) The mean and standard deviation (SD) of the mechano-gated current of the wild-type NompC and the mutants W1115A, D1142A, R1127A, and E1163A, under negative pressure in the outside-out patch-clamp experiments (wild type: n = 13; W1115A: n = 5; D1142A: n = 4; R1127A: n = 5; E1163A: n = 6). All of the error bars denote ± SD. Hydrogen bonds are indicated by dashed lines (E).

The forces exerted on the linker helix (LH) domain when AR1 was being pushed/pulled.
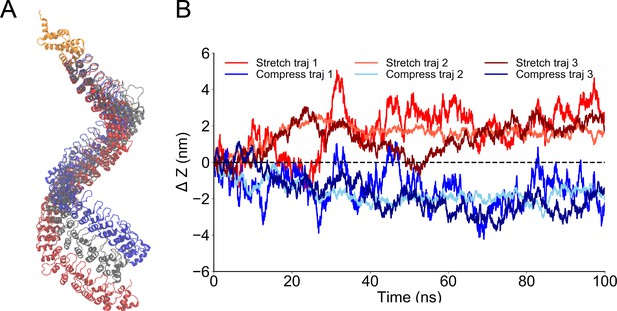
Steered molecular dynamics (MD) of the single ankyrin repeat (AR) chain of NompC.

The force constant of one ankyrin repeat (AR) chain in the AR bundle calculated from the molecular dynamics (MD) simulations with various pulling or pushing forces.

The reaction forces to the restraints on the linker helix (LH) domain after applying a force on the AR1.
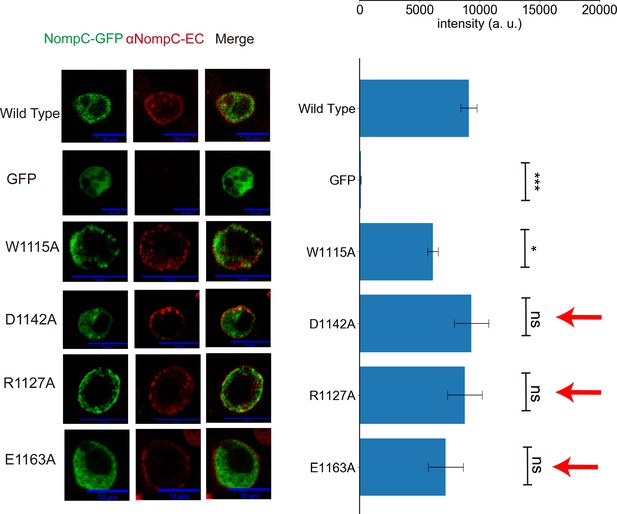
The mutants of residues listed in Figure 3F showed normal membrane targeting.
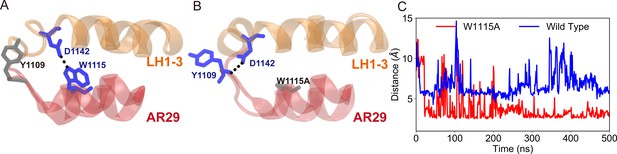
Formation of the alternative hydrogen bonds in the mutant, as identified in the molecular dynamics (MD) simulations (from chain A in the trajectories W1115A in Supplementary file 1a).

Formation of the two hydrogen bonds that were not observed in the cryo-EM structure.
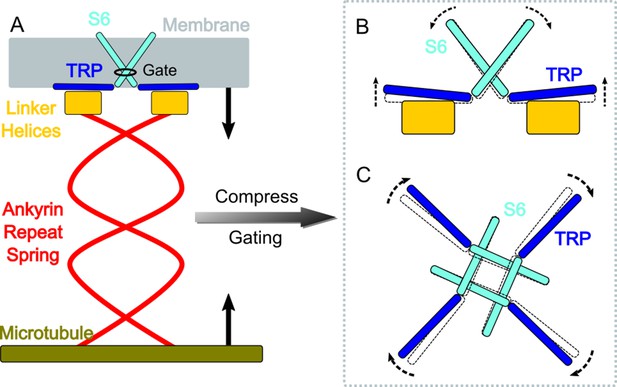
A gating model of NompC.
(A) The compression of the ankyrin repeat (AR) region will generate a pushing force and a torque on the linker helix (LH) domain, pointing to the extracellular side. (B) The LH domain further pushes the transient receptor potential (TRP) domain, leading to a tilt (side view), and (C) a clockwise rotation of the TRP domain (looking from the intracellular side). The motion of the TRP domain pulls the S6 helices to slightly tilt and rotate, which dilates the constriction site of the pore.
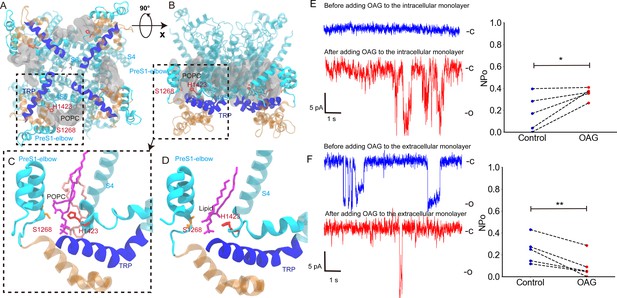
The interaction between H1423 and lipids and the effect of adding 1-oleoyl-2-acetyl-sn-glycerol (OAG) on the NompC opening.
(A) The bottom view and (B) the side view of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) molecules moving around H1423. The transient receptor potential (TRP) domain is shown in blue, the S4S5 linker is shown in cyan, and the POPC density around H1423 is shown with silver transparent surfaces. The analysis was performed on the molecular dynamics (MD) trajectory FI0. (C) The initial (salmon) and final (violet) locations of a POPC molecule in the MD simulation trajectory FI0. (D) A lipid molecule was observed in the cryo-EM structure of NompC (PDB ID: 5vkq). The pocket between H1423 and S1268 can stably accommodate a lipid molecule in both the MD simulations and the cryo-EM structure. (E) The representative traces of the spontaneous NompC current before/after adding OAG to the intracellular monolayer of the membrane, and the corresponding average open probabilities (N = 5, paired Student’s t-test, p=0.0208). (F) The representative traces of the spontaneous NompC current before/after adding OAG to the extracellular monolayer of the membrane, and the corresponding average open probabilities (N = 5, paired Student’s t-test, p=0.0047).
Videos
The transmembrane (TM) pore size evolution during the 250 ns simulation trajectories (FI0, SI0, and CI0) as shown in Figure 1B–D.
This video shows the TM pore size evolution during the 250 ns ‘free’, ‘pulling’, and ‘pushing’ simulations.
Ion permeation through the partially opened NompC channel under transmembrane potential.
The rotation of the transient receptor potential (TRP) domain in the steered molecular dynamics (SMD) simulations (SI0 and CI0 in Supplementary file 1a).
The tilt of the transient receptor potential (TRP) domain in the steered molecular dynamics (SMD) simulations (SI0 and CI0 in Supplement 1a).
The conformational changes of the AR domain in the “free”, “pulling”, and “pushing” MD simulations as shown in Figure 3C.
The conformational changes of the AR domain in the “free”, “pulling”, and “pushing” MD simulations, in which a 2-pN force was applied to each AR chain.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (D. mel) | Schneider 2 (S2) cells | CCTCC (China Center for Type Culture Collection) | Serial# GDC0138 | Cell species report and Mycoplasma contamination test reports provided |
| Antibody | Rabbit anti-αNOMPC-EC (polyclonoal) | Ref. (Zhang et al., 2015) | Immunostaining dilution (1: 500), primary antibody | |
| Antibody | Alexa Fluor 594 AffiniPure Donkey Anti Rabbit IgG(H + L) | Yeason | Cat# 34212ES60 | Immunostaining dilution (1: 100), secondary antibody |
| Recombinant DNA reagent | pActin-Gal4 (plasmid) | Ref. (Yan et al., 2013) | Plasmid for driving Gal4 expression under actin promoter in S2 cells | |
| Recombinant DNA reagent | pUAST-NOMPC-EGFP (plasmid) | Ref. (Yan et al., 2013) | Plamid for Gal4-driven NompC expression in S2 cells | |
| Recombinant DNA reagent | pUAST-NOMPC-EGFP (del-miniwhite,dm) (plasmid) | This paper | Plamid for Gal4-driven WT NompC expression in S2 cells, no miniwhite sequence | |
| Recombinant DNA reagent | pUAST-NOMPC(D1236A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on D1236 | |
| Recombinant DNA reagent | pUAST-NOMPC(R1581A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on R1581 | |
| Recombinant DNA reagent | pUAST-NOMPC(K1244A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on K1244 | |
| Recombinant DNA reagent | pUAST-NOMPC(E1571A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on E1571 | |
| Recombinant DNA reagent | pUAST-NOMPC(Q1253A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on Q1253 | |
| Recombinant DNA reagent | pUAST-NOMPC(S1577A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on S1577 | |
| Recombinant DNA reagent | pUAST-NOMPC(S1421A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on S1421 | |
| Recombinant DNA reagent | pUAST-NOMPC(W1572A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on W1572 | |
| Recombinant DNA reagent | pUAST-NOMPC(W1115A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on W1115 | |
| Recombinant DNA reagent | pUAST-NOMPC(D1142A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on D1142 | |
| Recombinant DNA reagent | pUAST-NOMPC(R1127A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on R1127 | |
| Recombinant DNA reagent | pUAST-NOMPC(E1163A)-EGFP(dm) (plasmid) | This paper | Contains Drosophila NOMPC CDS with alanine substitution on E1163 | |
| Chemical compound, drug | 1-Oleoyl-2-acetyl-sn-glycerol (OAG) | Sigma-Aldrich | Cat# O6754 | DAG analogue |
| Chemical compound, drug | GdCl3 | Sigma-Aldrich | Cat# 439770 | NOMPC blocker |
| Chemical compound, drug | Concanavalin A (Con A) | Sigma-Aldrich | Cat# C5275 | Cell adhesion |
| Chemical compound, drug | ClonExpress II One-step Cloning Kit | Vazyme | Serial# C112 | Site-directed mutagenesis |
| Chemical compound, drug | TransIT-Insect Transfection Reagent | Mirus | Cat# MIR 6100 | S2 cell transfection |
Additional files
-
Supplementary file 1
Tables.
(a) Molecular dynamics/steered molecular dynamics (MD/SMD) trajectories of system I. (b) MD/SMD trajectories of system II. (c) Ion permeation simulations of the partially opened NompC. (d) Stable hydrogen bonds and their occupancies in the MD/SMD trajectories. (e) Different domains/components used in the MD simulations. (f) The primers used for the alanine substitution.
- https://cdn.elifesciences.org/articles/58388/elife-58388-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58388/elife-58388-transrepform-v1.docx