Heterogeneity of murine periosteum progenitors involved in fracture healing
Figures
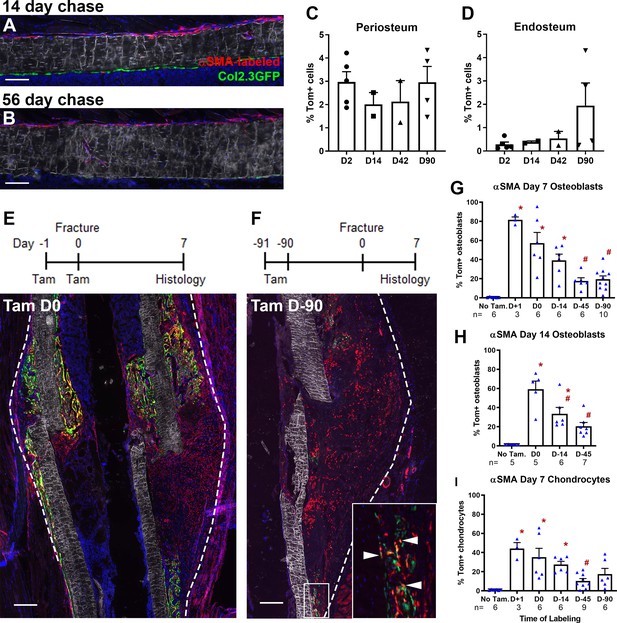
Alpha smooth muscle actin (αSMA) identifies long-term progenitor cells in the periosteum.
Representative images of tibia mid-shaft periosteum: (A) 14 days or (B) 56 days after tamoxifen with the periosteal surface near the top and endosteal surface near the bottom of the image. Percentage of αSMA-labeled (Tom+) cells within the CD45- population at different time points in (C) periosteum and (D) endosteum as measured by flow cytometry (n = 2–5). Fracture histology of representative day 7 tibial fracture calluses in different groups. Diagrams indicate the experimental design. (E) αSMA-labeled with tamoxifen (Tam) at day −1, 0; (F) αSMA-labeled with tamoxifen at day −91, –90. Quantification of Col2.3GFP+ osteoblasts derived from αSMA-labeled cells in the fracture callus based on image analysis (Tom+GFP+ cells/GFP+ cells) in (G) day 7 fracture callus and (H) day 14 fracture callus at different times after tamoxifen administration. (I) Quantification of Sox9+ chondrocytes derived from αSMA-labeled cells in day 7 fracture callus. All images show mineralized bone by dark field (gray) and DAPI+ nuclei in blue. Scale bars are 100 µm (A, B) or 250 µm (E–F). In F, the boxed area is magnified and αSMA-labeled osteoblasts as quantified in G-H are indicated with arrowheads. Approximate callus area used for analysis is indicated by the dashed lines in E-F, cortical bone surface defines the inner boundary. *p<0.05 compared to no tamoxifen control with ANOVA followed by Dunnett’s posttest. #p<0.05 compared to tamoxifen day 0 group.
-
Figure 1—source data 1
Image analysis processed data and statistical analysis tables (from Graphpad Prism) for graphs in Figure 1G–I and supplement 3B-C.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig1-data1-v2.xlsx
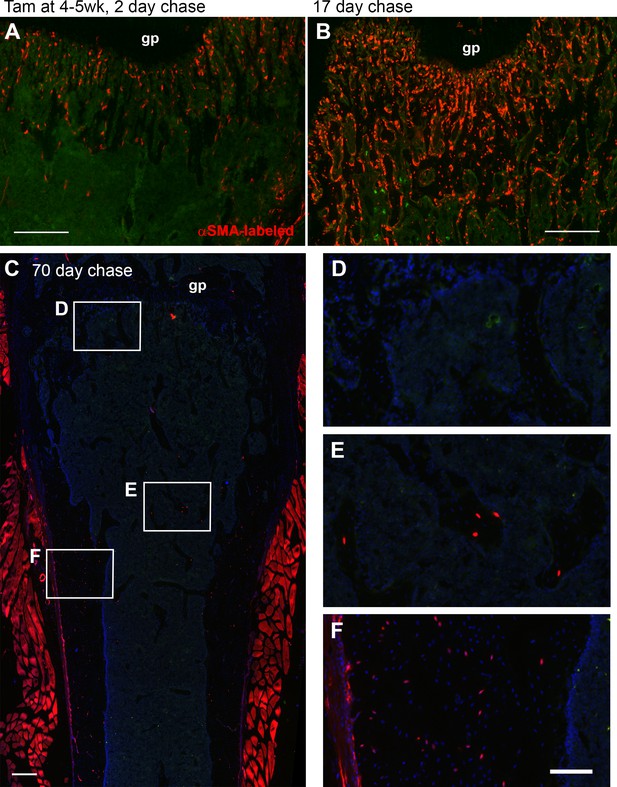
Alpha smooth muscle actin (αSMA) lineage tracing in the bone marrow compartment of juvenile mice.
Lineage tracing in the distal femur of αSMACreER/Ai9 mice at different time points after tamoxifen treatment at 4–5 weeks of age. (A) and (B) are replicates of data shown in Grcevic et al., 2012. Both show background signal in bone marrow in green and (C–F) have DAPI staining in blue. After 17 days, many osteoblasts and osteocytes in trabeculae are labeled, but at day 70 magnified areas show that few labeled cells are present in the primary spongiosa (D), but labeled osteocytes are retained in more proximal trabeculae (E) and in some cortical regions (F). Muscle is labeled due to targeting of some satellite cells by αSMACreER (Matthews et al., 2016). Scale bars are 250 µm (A–C) and 100 µm (D–F). gp, growth plate.
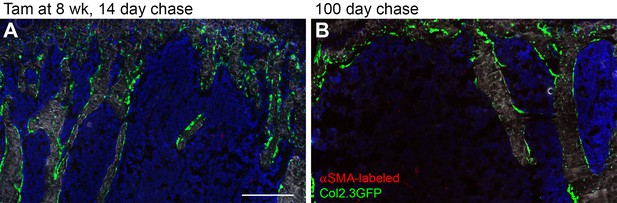
Alpha smooth muscle actin (αSMA) lineage tracing in the bone marrow compartment of adult mice.
Proximal tibia trabecular histology in αSMACreER/Ai9/Col2.3GFP mice 14 (A) and 100 days (B) after tamoxifen treatment during adulthood (at least 8 weeks of age). Note that there are few red cells and almost no contribution to osteoblasts or osteocytes. Scale bars are 250 µm. gp, growth plate.
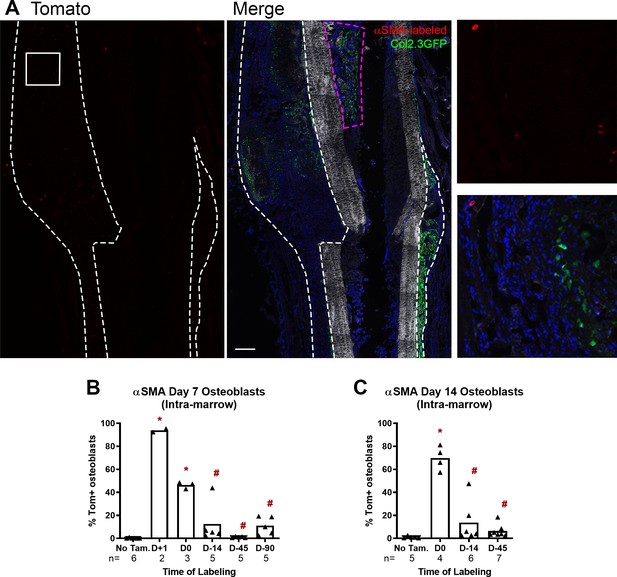
Minimal αSMACreER leakiness in fracture callus and contribution to endosteal bone formation.
(A) Fracture histology of representative day 7 callus in an αSMACreER/Ai9/Col2.3GFP mouse never exposed to tamoxifen showing the red channel only and merged image. The magnified images show the boxed area. A small number of red cells are visible; however, very few colocalize with Col2.3GFP. Images show mineralized bone by dark field (gray) and DAPI+ nuclei in blue. Callus area is outlined by white dashed lines. Scale bar is 250 µm. (B) Contribution of αSMA-labeled cells to new Col2.3GFP+ osteoblasts in the bone marrow compartment (as indicated by pink box in A) in day 7 fractures and (C) in day 14 fractures. *p<0.05 compared to no tamoxifen control with ANOVA followed by Dunnett’s posttest. #p<0.05 compared to tamoxifen day 0 group. αSMA, alpha smooth muscle actin.
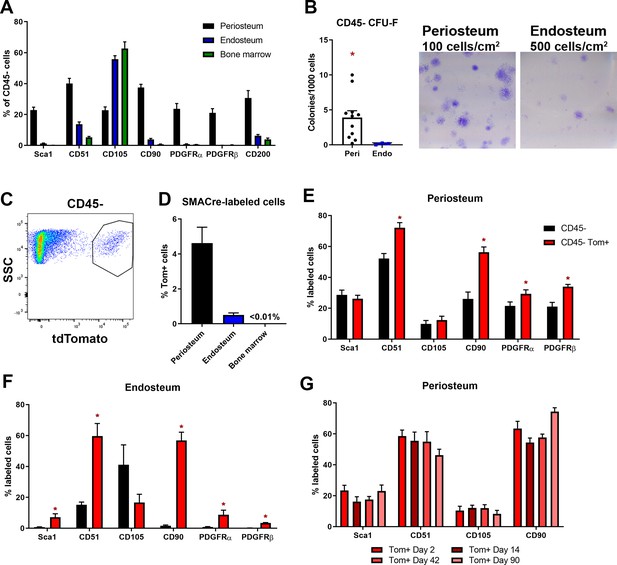
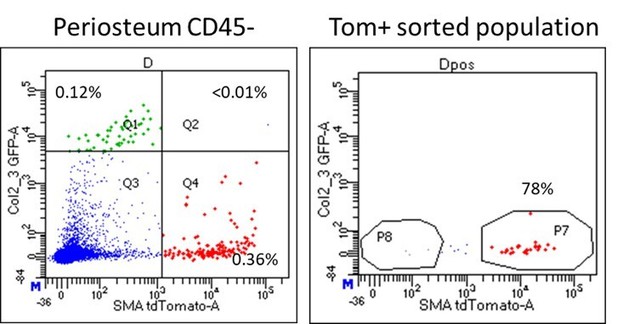
Mesenchymal stem cell markers are enriched in periosteum.
Flow cytometry and cell sorting was used to evaluate marker expression in tissue from adult long bones. (A) Expression of cell surface markers in periosteum, endosteum, and bone marrow samples as a percentage of the CD45/Ter119/CD31- fraction, n = 4–50. Expression in periosteum is significantly different from both bone marrow and endosteum for all markers (two-way ANOVA with posttest comparing cell types). (B) Colony-forming unit fibroblast (CFU-F) formation in the total CD45/Ter119/CD31- fraction. (C) Representative dot plot showing gating of alpha smooth muscle actin (αSMA)-labeled cells. (D) Proportion of αSMA-labeled cells within the CD45/Ter119/CD31- gate 2–3 days after tamoxifen in different tissue compartments (n = 17). Cell surface marker expression in the Tom+ population 2–3 days after tamoxifen in (E) periosteum and (F) endosteum (n = 4–17). (G) Cell surface profile of αSMA-labeled periosteum cells at different time points (n = 2–7). *p<0.05 compared to CD45- population.
-
Figure 2—source data 1
Data tables and statistical analysis tables (from Graphpad Prism) for graphs shown in Figure 2 and figure supplement 2.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig2-data1-v2.xlsx
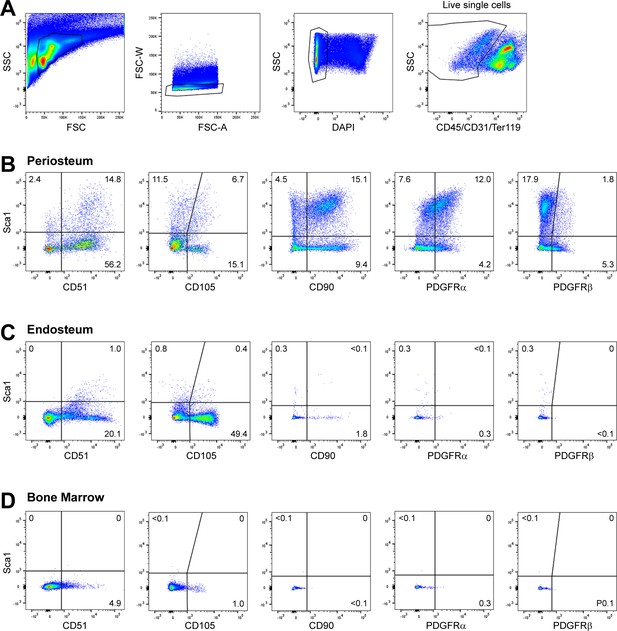
Flow cytometry plots of mesenchymal progenitor marker expression.
Representative plots showing (A) the initial gating scheme to enable analysis of CD45/CD31/Ter119- mesenchymal cell populations in periosteum and expression of indicated markers in (B) periosteum, (C) endosteum, and (D) bone marrow of representative samples in relation to Sca1. Quadrant percentages are specific to the sample that is shown. The periosteum, endosteum, and bone marrow samples in each column come from the same sample (obtained from two animals). FSC, forward scatter; SSC, side scatter.
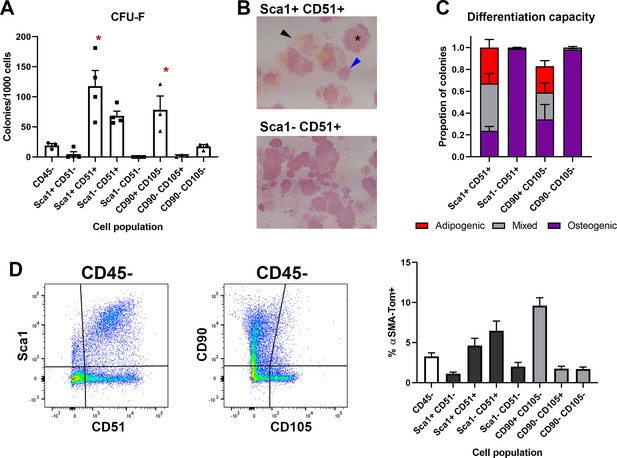
Sca1+CD51+ and CD90+ periosteal mesenchymal cells are enriched for multipotent progenitor cells.
(A) Colony-forming unit fibroblast (CFU-F) formation of different CD45- periosteal populations (n = 3–4). *p<0.05 compared to total CD45- based on one-way ANOVA with Dunnett’s posttest. (B) Differentiation potential of cell populations grown in mixed osteogenic adipogenic conditions. Pink is ALP stain, red is Oil Red O, colonies indicated show osteogenic only (blue arrowhead), adipogenic only (black arrowhead), and mixed (*) colonies. (C) Quantification of colony differentiation from different populations. (D) Dot plots indicating different populations sorted and analyzed and percentage of αSMA+ cells (Tom+ cells 2–3 days after tamoxifen injection) within each of the CD45- cell populations indicated (n = 10–16).
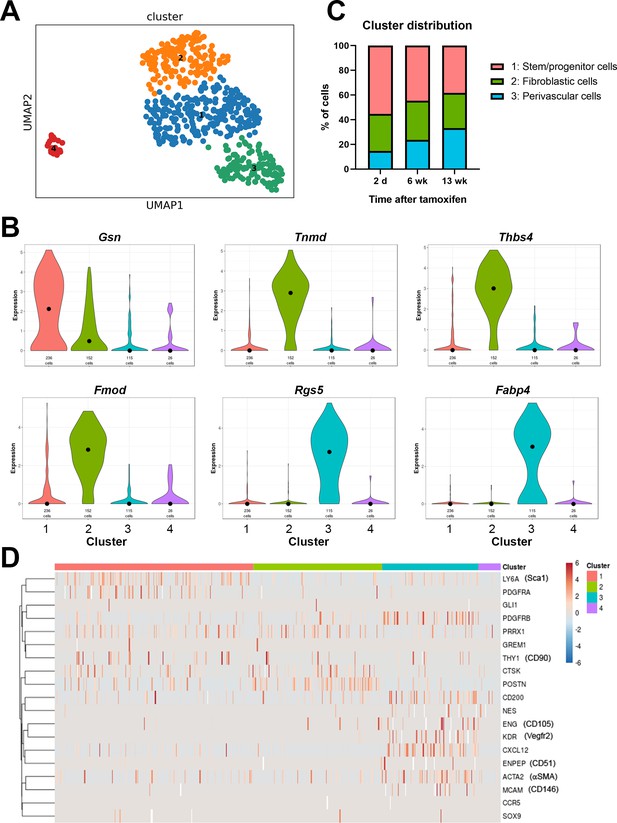
Single cell RNAseq analysis of αSMA-Tom+ cells.
Plate-based RNAseq was performed on alpha smooth muscle actin (αSMA)-Tom+ cells at up to 13 weeks after tamoxifen delivery. (A) UMAP cluster plot indicating four clusters. (B) Violin plots of top markers for clusters 1–3. (C) Distribution of cells in clusters 1–3 at different time points following tamoxifen. (D) Heat map of expression of putative skeletal stem and periosteal progenitor markers.
-
Figure 3—source data 1
scRNAseq cluster makers with AUROC values >0.7.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig3-data1-v2.xlsx
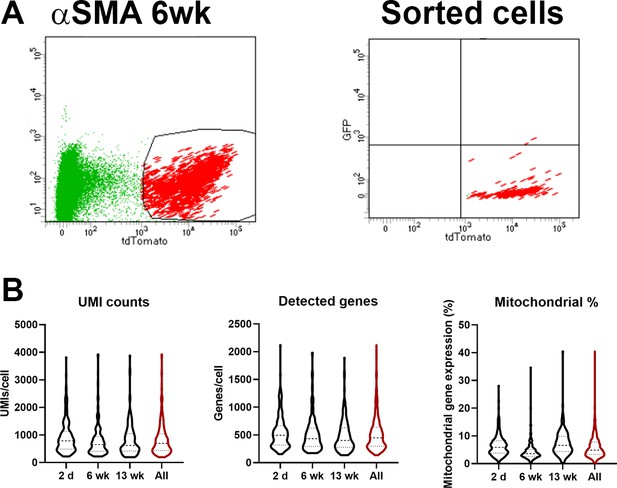
Quality control data for single-cell RNAseq dataset.
(A) Gating for cell sorting of the 6-week chase cells. (B) Violin plots showing summary data for unique molecular identifier (UMI) counts, number of genes detected, and proportion of mitochondrial transcripts within the dataset.
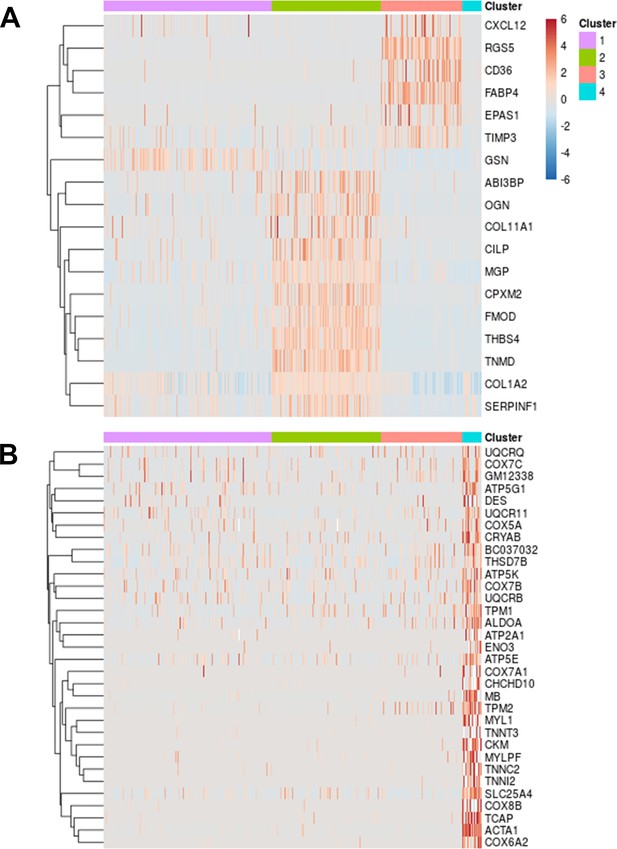
Heat maps of cluster markers.
Heat map indicating expression of cluster markers with area under the receiver operating characteristics curve (AUROC) values >0.7 for (A) clusters 1–3 which appear to be periosteal populations and (B) cluster 4 which shows expression of skeletal muscle lineage genes.
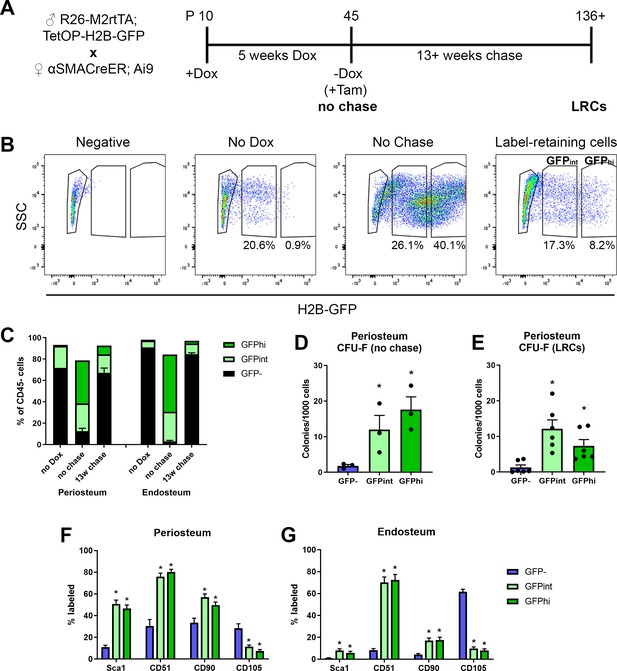
Label-retaining cells are enriched with most progenitor markers.
(A) Experimental design for label-retaining cell experiments. (B) Representative gating of the CD45- population in different periosteum samples to define three populations based on GFP expression. (C) Quantification of different GFP populations in a representative experiment for periosteum and endosteum (n = 1, no doxycycline [dox]; n = 4, no chase; n = 6, 13-week chase). (D) Colony-forming unit fibroblast (CFU-F) frequency in different CD45- GFP populations in periosteal no chase samples (n = 3) and (E) following chase (n = 6). Cell surface marker expression is shown in different CD45- GFP subsets following chase (populations equivalent to right-hand label-retaining cell plot in B) in (F) periosteum cells and (G) endosteal cells. Data is pooled from three separate experiments involving 13–22 weeks chase (n = 19). *p<0.05, repeated measures ANOVA with Dunnett’s posttest compared to GFP- population.
-
Figure 4—source data 1
Data tables and statistical analysis tables (from Graphpad Prism) for graphs shown in Figure 4F–G.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig4-data1-v2.xlsx
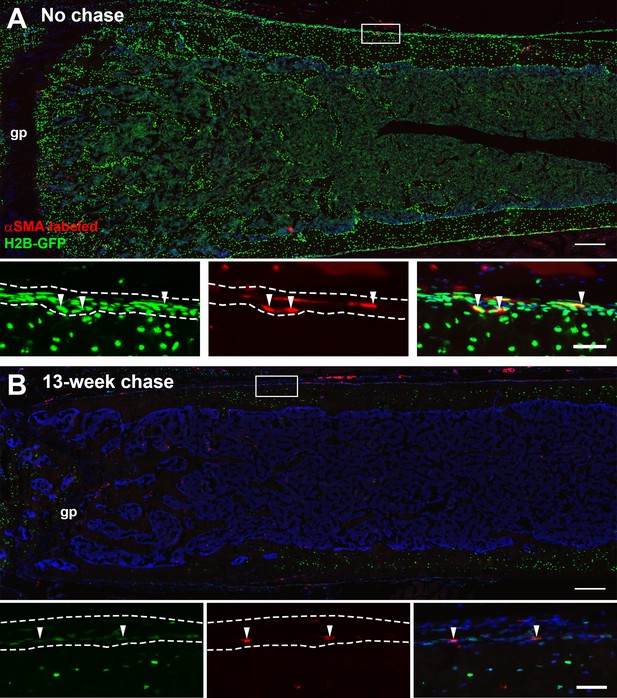
Histology of H2B-GFP/αSMACreER/Ai9 femurs.
(A) H2B-GFP expression in a distal femur following 5-week doxycycline (dox) and 2 days after tamoxifen. (B) H2B-GFP label-retaining cells after 13-week chase; tamoxifen was given at the time of dox withdrawal. Boxed areas are magnified. Dashed lines indicate the periosteum and arrowheads indicate GFP+ αSMA-Tom+ cells in the cambium layer of the periosteum. gp, growth plate. Scale bars are 250 or 100 µm (high magnification images).
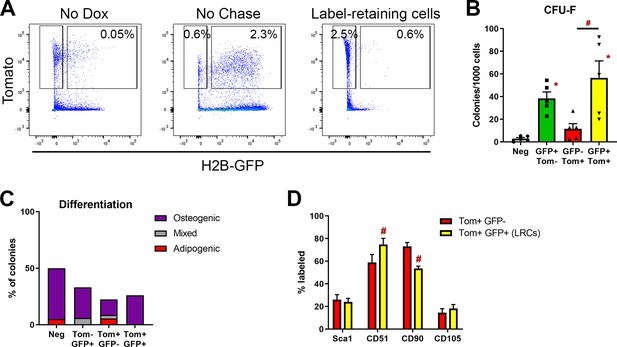
Alpha smooth muscle actin (αSMA)-labeled periosteal label-retaining cells are enriched with colony-forming unit fibroblast (CFU-F).
(A) Representative flow plots showing identification of αSMA-labeled label-retaining cells. Values represent an average of n = 3–10. (B) CFU-F formation and (C) differentiation of colonies under mixed osteogenic/adipogenic conditions of αSMA-labeled label-retaining cells and other subpopulations (n = 5 for CFU-F, n = 3 for differentiation). (D) Cell surface marker expression in αSMA-labeled subpopulations. *p<0.05 compared to negative population, #p<0.05 compared to Tom+GFP- population.
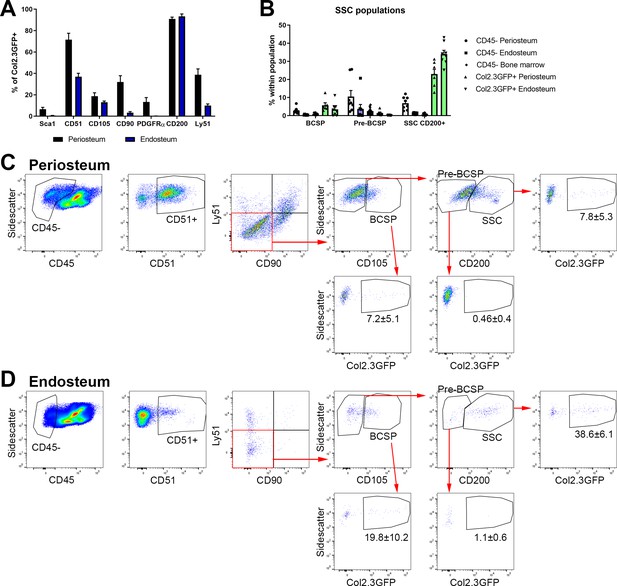
Bone, cartilage, and stromal progenitor (BCSP) and skeletal stem cell (SSC) populations in adult bone contain mature osteoblasts.
(A) Marker expression on Col2.3GFP+ osteoblasts, n = 4–32. (B) Frequency of BCSP and SSC populations in different tissue compartments and populations (n = 8–12). (C–D) Gating to identify BCSPs, pre-BCSPs, SSCs, and presence of Col2.3GFP+ osteoblasts as a major component of these populations in periosteum (C) and endosteum (D). Mean ± standard deviation for % GFP+ cells are shown.
-
Figure 5—source data 1
Data tables for graphs and plots shown in Figure 5.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig5-data1-v2.xlsx
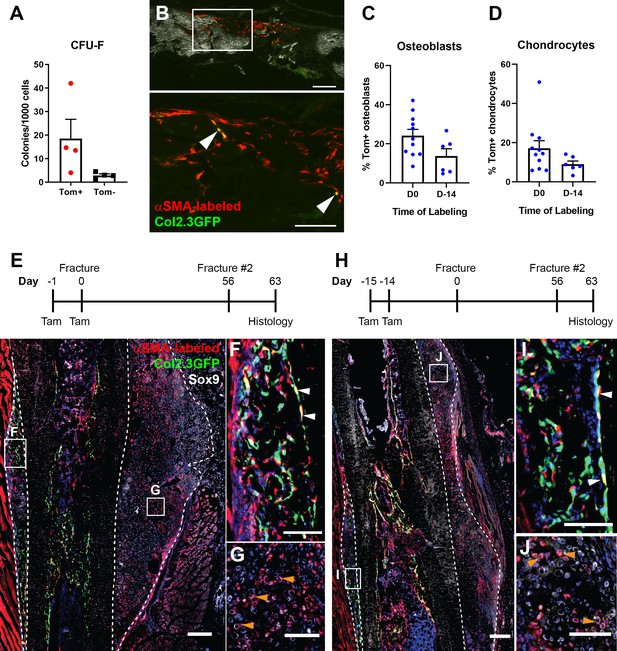
Alpha smooth muscle actin (αSMA)-labeled cells are transplantable self-renewing osteochondroprogenitors.
(A) Colony-forming unit fibroblast (CFU-F) frequency of periosteal αSMA-labeled cells sorted 2 days after tamoxifen. (B) Engraftment of transplanted αSMA-labeled cells in a calvarial defect 5 weeks after transplantation. Arrowheads indicate cells that are Col2.3GFP+. Quantification of αSMA-labeled cell contribution to (C) osteoblasts and (D) chondrocytes in day 7 secondary fracture calluses is shown. (E) Experimental design and fracture callus histology following a second tibia fracture in αSMACreER/Ai9/Col2.3GFP mice showing contribution of Tom+ cells to callus tissue, including osteoblasts (F) and Sox9+ chondrocytes (white) (G). (H–J) Histology of a similar secondary fracture where tamoxifen was given 14 days prior to the primary fracture. White arrowheads indicate αSMA-labeled Col2.3GFP+ osteoblasts, orange arrowheads indicate αSMA-labeled Sox9+ chondrocytes. DAPI stain is shown in blue. Approximate callus area used for analysis is indicated by the dashed lines in E and H. Scale bars indicate 250 µm (low magnification) and 100 µm (high magnification).
-
Figure 6—source data 1
Data tables and statistical analysis tables (from Graphpad Prism) for graphs shown in Figure 6C-D.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig6-data1-v2.xlsx
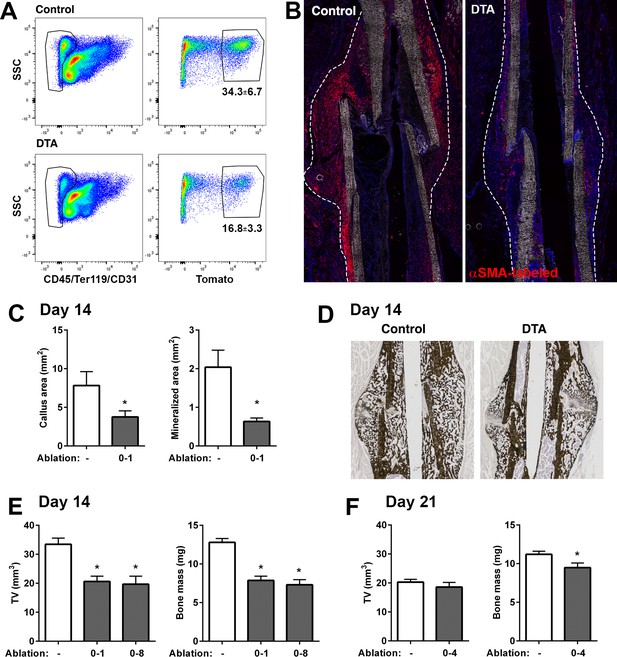
Ablation of alpha smooth muscle actin (αSMA)+ cells impairs fracture healing.
(A) Flow cytometry of day 4 fracture callus from control αSMACreER/Ai9 mice and DTA animals (αSMACreER/Rosa-DTA/Ai9) indicating significantly fewer (p<0.01) αSMA-labeled cells in the ablation model (mean ± SD shown, n = 4). (B) Representative histological sections from day 4 fracture callus in controls or with DTA-mediated ablation. The outline of the callus is indicated. (C) Histological analysis of callus area and mineralized area at day 14, and (D) representative images of von-Kossa staining (n = 4–7). (E) Micro-computed tomography (MicroCT) analysis of day 14 fracture callus in DTA- and DTA+ mice that had undergone two different tamoxifen regimens to ablate αSMA+ cells, day 0, 1 of fracture or day 0, 2, 4, 6, 8. Callus tissue volume and bone mass were measured. Cre-: n = 14, 8 male (M), 6 female (F); DTA D0-1: n = 10, 4M 6F; DTA D0-8: n = 8, 4M 4F. (F) MicroCT analysis of day 21 fracture callus in mice treated with tamoxifen on day 0, 2, and 4 (n = 14–15).
-
Figure 7—source data 1
Data tables and statistical analysis tables (from Graphpad Prism) for graphs shown in Figure 7C,E and F.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig7-data1-v2.xlsx
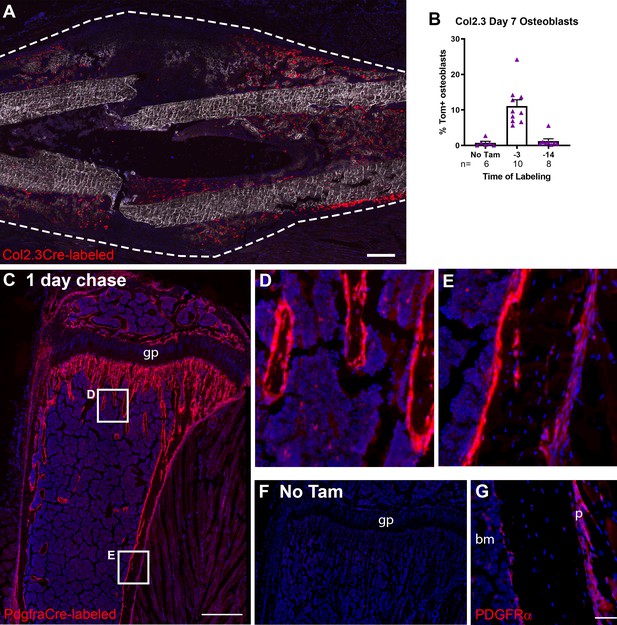
Contribution of mature osteoblasts to fracture healing and PDGFRα distribution.
(A) Fracture histology of a representative day 7 tibial fracture callus from a Col2.3CreER/Ai9 mouse treated with tamoxifen at days −4 and −3. Mineralized bone is shown by dark field (gray) and DAPI+ nuclei in blue. Scale bar is 250 µm. (B) Quantification of Col2.3-labeled cell contribution to Osx+ osteoblasts. *p<0.05 based on ANOVA with Dunnett’s posttest compared to no tamoxifen group. (C) Tibia histology of PdgfraCreER/Ai9 mice given three daily doses of tamoxifen and euthanized 1 day later. Scale bar is 200 µm. Magnified areas of (D) trabecular and (E) cortical bone are shown. (F) Tibial histology of a PdgfraCreER/Ai9 mouse that had not been exposed to tamoxifen showing minimal leakiness of the CreER. Representative images of n = 3 aged 5–8 weeks of age are shown. (G) PDGFRα immunostaining of bone. Scale bar is 50 µm. bm, bone marrow; gp, growth plate; p, periosteum.
-
Figure 8—source data 1
Data table and statistical analysis table (from Graphpad Prism) for graph shown in Figure 8B.
- https://cdn.elifesciences.org/articles/58534/elife-58534-fig8-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | αSMACreER (Tg(Acta2-cre/ERT2)1Ikal) | PMID:22083974 | MGI:5461154 | |
| Genetic reagent (Mus musculus) | Col2.3CreER (B6.Cg-Tg(Col1a1-cre/ERT2)1Crm/J) | Jackson Labs | Jax: 016241 | |
| Genetic reagent (Mus musculus) | Col2.3GFP (B6.Cg-Tg(Col1a1*2.3-GFP)1Rowe/J) | PMID:11771662 | Jax: 013134 | |
| Genetic reagent (Mus musculus) | Ai9 (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) | Jackson Labs | Jax: 007909 | |
| Genetic reagent (Mus musculus) | Rosa-DTA (B6.129P2-Gt(ROSA)26Sortm1(DTA)Lky/J) | Jackson Labs | Jax: 009669 | |
| Genetic reagent (Mus musculus) | R26-M2rtTA; TetOP-H2B-GFP (B6;129S4-Gt(ROSA)26Sortm1(rtTA*M2)JaeCol1a1tm7(tetO-HIST1H2BJ/GFP)Jae/J) | Jackson Labs | Jax: 016836 | |
| Genetic reagent (Mus musculus) | NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) | Jackson Labs | Jax: 005557 | |
| Genetic reagent (Mus musculus) | PdgfraCreER (B6.129S-Pdgfratm1.1(cre/ERT2)Blh/J) | Jackson Labs | Jax: 032770 | |
| Antibody | CD45 eFluor450 (rat monoclonal, clone 30-F11) | eBioscience | 48–0451 | Flow (1:400) |
| Antibody | Ter119 eFluor450 (rat monoclonal, clone TER-119) | eBioscience | 48–5921 | Flow (1:200) |
| Antibody | CD31 eFluor450 (rat monoclonal, clone 390) | eBioscience | 48–0311 | Flow (1:400) |
| Antibody | CD45 APC (rat monoclonal, clone 30-F11) | eBioscience | 17–0451 | Flow (1:400) |
| Antibody | Ter119 APC (rat monoclonal, clone TER-119) | eBioscience | 17–5921 | Flow (1:200) |
| Antibody | CD31 APC (rat monoclonal, clone 30-F11) | eBioscience | 17–0311 | Flow (1:400) |
| Antibody | CD140a APC (rat monoclonal, clone APA5) | eBioscience | 17–1401 | Flow (1:100) |
| Antibody | CD140a PE-Cy7 (rat monoclonal, clone APA5) | eBioscience | 25–1401 | Flow (1:400) |
| Antibody | CD140b APC (rat monoclonal, clone APB5) | eBioscience | 17–1402 | Flow (1:100) |
| Antibody | CD105 APC (rat monoclonal, clone MJ7/18) | eBioscience | 17–1051 | Flow (1:200) |
| Antibody | CD90.2 APC eFluor 780 (rat monoclonal, clone 53–2.1) | eBioscience | 47–0902 | Flow (1:200) |
| Antibody | CD90.2 BV605 (rat monoclonal, clone 30-H12) | BD Bioscience | 740334 | Flow (1:100) |
| Antibody | CD51 Biotin (rat monoclonal, clone RMV-7) | eBioscience | 13–0512 | Flow (1:100) |
| Antibody | Sca1 AlexaFluor700 (rat monoclonal, clone D7) | eBioscience | 56–5981 | Flow (1:100) |
| Antibody | Ly51 BV711 (rat monoclonal, clone 6C3) | BD Bioscience | 740691 | Flow (1:100) |
| Antibody | CD200 PE (rat monoclonal, clone OX-2) | Biolegend | 123807 | Flow (1:100) |
| Antibody | Sox9 (rabbit polyclonal) | EMD Millipore | ABE2868 | IHC (1:200) |
| Antibody | Osterix (rabbit monoclonal) | Abcam | ab209484 | IHC (1:400) |
| Antibody | PDGFRα (rabbit polyclonal) | R and D Systems | AF1062 | IHC (1:80) |
| Antibody | Goat anti rabbit AlexaFluor647 (polyclonal) | ThermoFisher | A21244 | IHC (1:300) |
| Chemical compound, drug | Streptavidin APC eFluor 780 | eBioscience | 47–4317 | Flow (1:400) |
| Chemical compound, drug | Streptavidin PE-Cy7 | eBioscience | 25–4317 | Flow (1:400) |
| Chemical compound, drug | Streptavidin APC | eBioscience | 17–4317 | Flow (1:400) |
| Chemical compound, drug | Tamoxifen | Sigma Aldrich | T5648 | 75 mg/kg i.p. |
| Chemical compound, drug | Doxycycline | Envigo | TD.01306 | 625 mg/kg in Teklad Custom Diet |
| Sequence-based reagent | Barcode oligo primer | IDT | 5’-AAGCAGTGGTATCAACGC AGAGTACJJJJJJJJJJJJNNNNN NNNTTTTTTTTTTTTTTTTTTT TTTTTTTTTTTVN-3’ | |
| Sequence-based reagent | Custom primer sequence | IDT | 5'-AATGATACGGCGACCACC GAGATCTACACGCCTGTCCG CGGAAGCAGTGGTATCAACG CAGAGT*A*C-3’ | |
| Sequence-based reagent | Custom read one sequence | IDT | 5’-CGGAAGCAGTGGTATCAA CGCAGAGTAC-3’ |