Striatal direct and indirect pathway neurons differentially control the encoding and updating of goal-directed learning
Figures
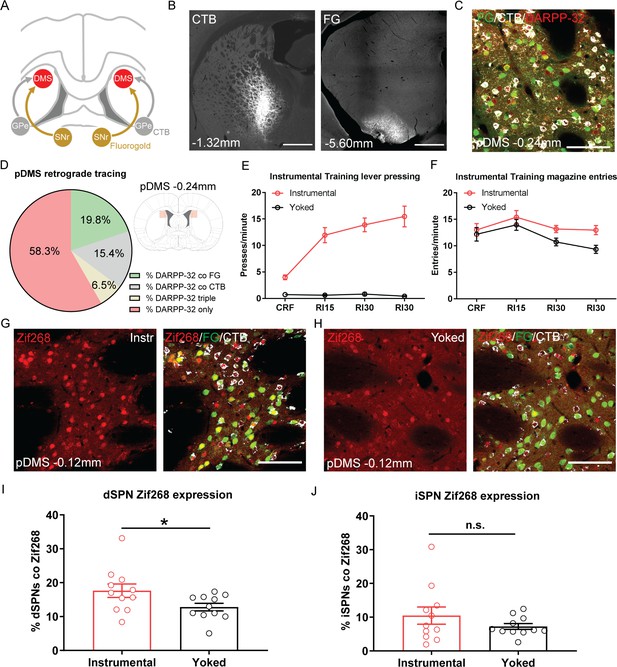
The acquisition of goal-directed actions induces response-specific expression of Zif268 in direct spiny projection neurons (dSPNs) in the posterior dorsomedial striatum (pDMS).
(A) Schematic illustrating the surgery design for retrograde tracing. Rats received bilateral infusions of the retrograde tracers fluorogold (FG) and cholera-toxin B (CTB) into the substantia nigra pars reticulata (SNr) and globus pallidus (GPe) respectively. (B) Fluorescent confocal images showing injection sites of CTB (left) and FG (right) in the GPe and SNr respectively; scale bars, 1000 µm. (C) Fluorescent confocal image taken from one 40 µm coronal section in the pDMS, illustrating fluorescence labelling of DARPP-32 (red), FG (green), and CTB (white); scale bar, 100 µm. (D) Percentages represent the proportion of total DARPP-32 positive SPNs that also express FG or CTB in the pDMS (imaging region indicated in pale red, right). Data presented are mean percentages for 22 rats, averaged across hemispheres. (E) Mean (± SEM) lever presses per minute averaged across each day of instrumental or yoked training for each group. (F) Mean (± SEM) magazine entries per minute averaged across each day of instrumental or yoked training for each group. (G) Fluorescent confocal image of a coronal 40 µm section for an instrumentally trained rat; left image shows Zif268 expression (red), and right image shows the same merged with FG labelled dSPNs (green) and CTB labelled indirect spiny projection neurons (iSPNs; white); scale bar, 100 µm. (H) Same as G but for a yoked trained rat. (I) Percentage of labelled dSPNs that were co-labelled with Zif268 for each rat in Group Instrumental and Group Yoked. Data presented are means of four pDMS sections per rat. Bars represent group means ± SEM. (J) Same as I but for iSPNs. *p<0.05.
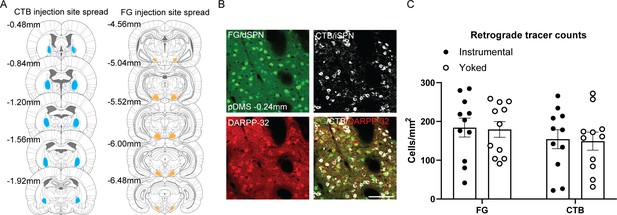
Projection target verification for the SPN tracing and immunofluorescence experiment.
(A) The extent of cholera-toxin B (CTB) spread at the globus pallidus injection site (left) and FG spread at the substantia nigra pars reticulata injection site (right), for all included rats, imaged and mapped at five different anteroposterior coordinates. (B) Confocal images (scale bar 100 µm) showing FG labelled cells (green), CTB labelled cells (white), DARPP-32 labelled cells (red), and a merged image of all three. (C) Total number of cells/mm2 labelled with FG and CTB for each rat in Group Instrumental and Group Yoked. Bars represent group means ± SEM.
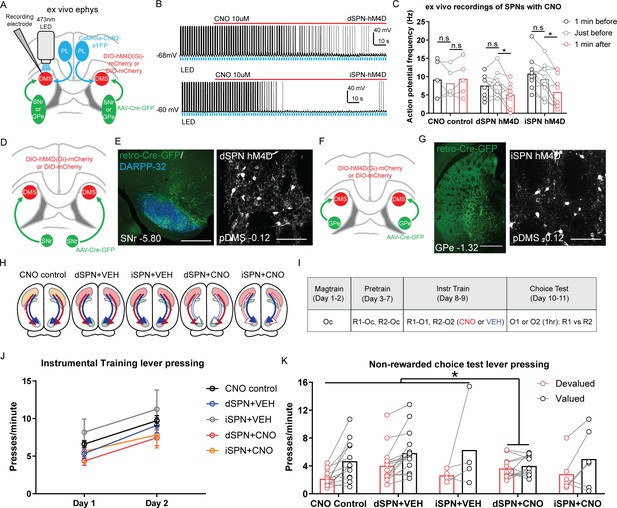
Goal-directed learning is disrupted by chemogenetic inhibition of direct spiny projection neurons (dSPNs) but not indirect spiny projection neurons (iSPNs) in the posterior dorsomedial striatum (pDMS).
(A) Schematic depicting the viral injections for ex vivo slice electrophysiology recordings; retro-Cre was injected into the substantia nigra pars reticulata (SNr) or globus pallidus (GPe), DIO-hM4D or DIO-mCherry was injected into the pDMS and ChR2-eYFP injected into the PL; direction of arrows indicates retrograde or anterograde transport of the virus. (B) Example trace from one recorded dSPN (top) and one recorded iSPN (bottom) expressing DIO-hM4D. Upward deflection black lines represent action potentials, which were elicited by the pulsing of a 473 nm wavelength LED light (blue) onto corticostriatal terminals (see Materials and methods). The red line indicates the time period when 10 µM clozapine-N-oxide (CNO) solution was added to the extracellular solution. (C) Action potential frequency of dSPNs and iSPNs elicited by light-evoked terminal glutamate release from ChR2-containing corticostriatal axons in the pDMS 1 min before (black), immediately before (grey) and 1 min after (red) the application of CNO in the extracellular solution. Each data point represents individually recorded cells. (D) Schematic depicting the viral injections to target dSPNs; retro-Cre was injected bilaterally into the SNr and DIO-hM4D or DIO-mCherry was injected bilaterally into the pDMS. (E) Confocal image (scale bar, 1000 µm) from one rat showing retro-Cre expression in the SNr (left) and a representative confocal image (scale bar, 100 µm) showing DIO-hM4D expression in pDMS dSPNs (right). (F) Same as D but for targeting iSPNs; retro-Cre was injected into the GPe. (G) Same as E but showing retro-Cre expression in the GPe and DIO-hM4D expression in pDMS iSPNs. (H) Summary of experimental groups; blue arrows indicate intact direct pathway function, red arrows indicate intact indirect pathway function, and unfilled arrows indicate inhibited pathway. (I) Summary of the experimental design; R1 and R2 indicate left and right lever responses; Oc, O1, and O2 indicate distinct food outcomes; CNO and VEH indicate injections of clozapine-N-oxide or vehicle, respectively. (J) Mean (± SEM) lever presses per minute averaged across each day of instrumental training for each group. (K) Mean lever presses per minute on the devalued and valued lever for each rat in each group, averaged across two choice tests under extinction. For all data, bars represent group means. *p<0.05.
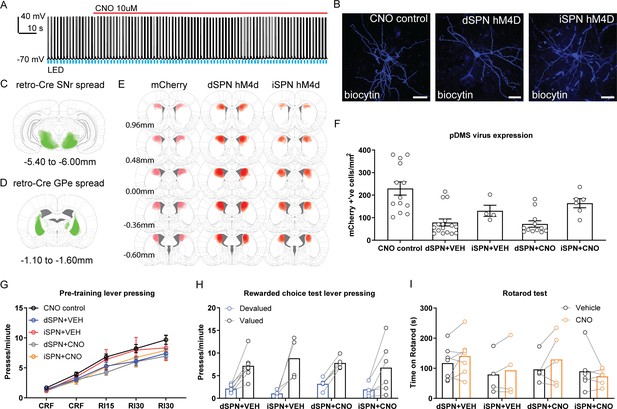
Verification of hM4D virus and supplemental behavior for the DREADDs suppression experiment.
(A) Example trace from one recorded direct spiny projection neuron (dSPN) expressing DIO-mCherry. Upward deflection black lines represent action potentials, which were elicited by the pulsing of a 473 nm wavelength LED light (blue) onto corticostriatal terminals (see Materials and methods). The red line indicates the time period when 10 µM clozapine-N-oxide (CNO) solution was added to the extracellular solution. (B) Example biocytin-labelled SPNs from Group CNO control (left), Group dSPN hM4D (middle), and Group indirect spiny projection neuron (iSPN) hM4D (right) from recordings (scale bars, 30 µm). (C) The extent of retro-Cre spread in the SNr, imaged at the widest point of expression for all included animals. (D) Same as C but for retro-Cre expression in the GPe. (E) The extent of virus spread for DIO-mCherry (left), DIO-hM4D in dSPNs (middle), and DIO-hM4D in iSPNs (right) for all included animals mapped at five different anteroposterior coordinates. (F) Total number of mCherry positive cells/mm2 in the pDMS for each rat in each experimental group. Bars represent group means ± SEM. (G) Mean (± SEM) lever presses per minute averaged across each day of instrumental pre-training for each group. (H) Mean lever presses per minute on the devalued and valued lever for each rat in each group, averaged across 2 days of rewarded choice testing. (I) Mean amount of time spent on the rotarod, averaged across two trials for each rat in each group following either vehicle or CNO injections. For all data, bars represent group means.
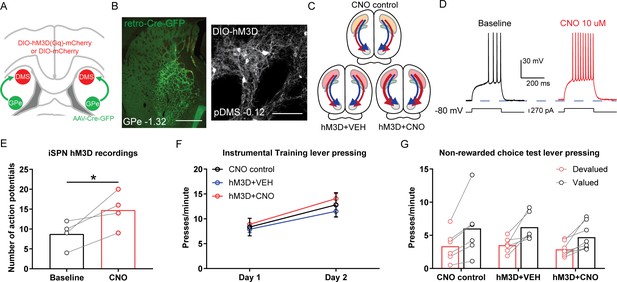
Chemogenetic stimulation of indirect spiny projection neurons (iSPNs) in the posterior dorsomedial striatum (pDMS) leaves goal-directed learning intact.
(A) Schematic depicting the viral injections, retro-Cre was injected bilaterally into the globus pallidus (GPe) and DIO-hM3D or DIO-mCherry was injected bilaterally into the pDMS. (B) Confocal image (scale bar, 1000 µm) from one rat showing retro-Cre expression in the GPe (left) and a confocal image (scale bar, 100 µm) from one rat showing DIO-hM3D expression on pDMS iSPNs. (C) Summary of experimental groups; blue arrows represent direct pathway, red arrows represent indirect pathway, and thicker arrows indicate increased activity in stimulated pathway. (D) An example trace from one iSPN from injection of a depolarizing current step (at resting membrane potential) before and after bath application of clozapine-N-oxide (CNO). (E) Number of action potentials evoked from an identical size current step injection before and during CNO application in hM3D-expressing iSPNs. The chosen step size in each neuron was when current injection first elicited action potentials, before drug. Individual data points represent each recorded cell. (F) Mean (± SEM) lever presses per minute averaged across each day of instrumental training for each group. (G) Mean lever presses per minute on the devalued and valued lever for each rat in each group, averaged across 2 days of non-rewarded choice tests. For all data, bars represent group means. *p<0.05.
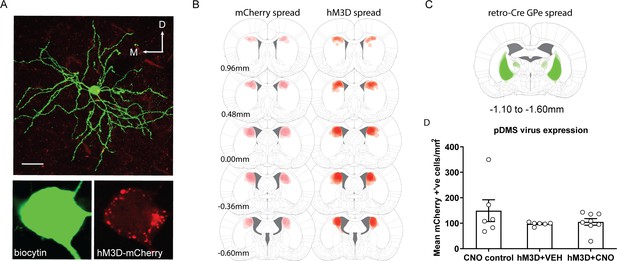
Verification of the hM3D virus for the DREADDs stimulation experiment.
(A) An example biocytin-labelled indirect spiny projection neuron expressing hM3D from recording (scale bar, 30 µm). (B) The extent of virus spread for DIO-mCherry (left) and DIO-hM3D (right), for all included animals, mapped at five different anteroposterior coordinates. (C) Schematic showing the extent of retro-Cre spread in the globus pallidus, imaged at the widest point of expression for all included animals. (D) Total number of mCherry positive cells/mm2 in the pDMS for each rat in each experimental group. Bars represent group means ± SEM.
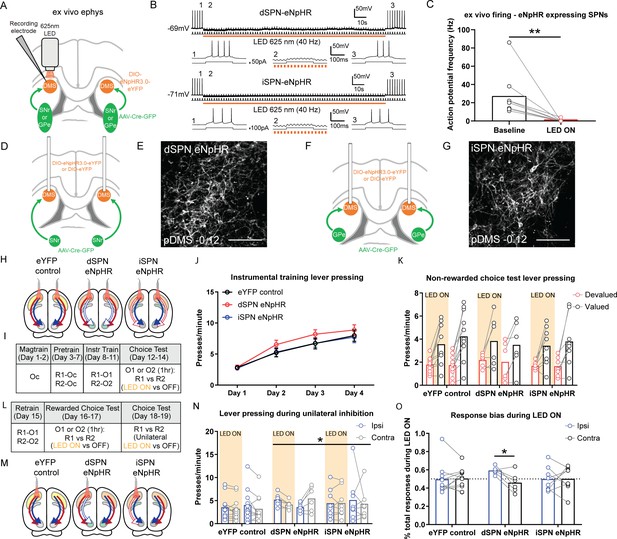
Bilateral optogenetic inhibition of direct spiny projection neurons (dSPNs) or indirect spiny projection neurons (iSPNs) leaves the expression of goal-directed learning intact but unilateral inhibition of dSPNs biases performance.
(A) Schematic depicting the design for ex vivo recordings; retro-Cre was injected into the substantia nigra pars reticulata (SNr) or globus pallidus (GPe) and DIO-eNpHR into the posterior dorsomedial striatum (pDMS); direction of arrows indicates retrograde transport of the virus. (B) Example trace from one dSPN (top) and iSPN (bottom) expressing eNpHR recorded under current-clamp with an injection of +50 or +100 pA current (200 ms, 0.5 Hz). Orange bar denotes time period when 625 nm LED was applied to the neuron (40 Hz, 150 s). Bottom three panels of each SPN are expanded traces from the above at time points before (1), during (2), and after (3) LED illumination. (C) Action potential frequency of eNpHR-labelled SPNs (four dSPNs and five iSPNs) at baseline (10 s period before LED) versus LED (10 s period from beginning of LED). (D) Surgery design for targeting dSPNs; retro-Cre was injected bilaterally into the SNr and DIO-eNpHR or DIO-eYFP was injected bilaterally into the pDMS. Cannulae were inserted bilaterally into the pDMS. (E) Confocal image (scale bar, 100 µm) showing DIO-eNpHR expression in pDMS dSPNs. (F and G) Same as D and E but for pDMS iSPNs; retro-Cre was injected into the GPe. (H) Summary of experimental groups; blue arrows indicate intact direct pathway, red arrows indicate intact indirect pathway, and unfilled arrows indicate inhibited pathway. (I) Summary of the experimental design; R1 and R2 indicate left and right lever responses; Oc, O1, and O2 indicate distinct food outcomes; LED ON versus OFF indicates training or test days when LED light was delivered. (J) Mean (± SEM) lever presses per minute averaged across each day of instrumental training for each group. (K) Mean presses per minute on the devalued and valued lever for each rat in each group, averaged across two tests, during LED ON (orange shaded) and LED OFF (non-shaded) periods. (L) Continuation of I. (M) Same as H but showing unilateral LED manipulation. (N) Mean presses per minute on the lever ipsilateral and contralateral to unilateral inhibition for each rat in each group during periods of LED ON (orange shaded) and LED OFF (non-shaded), averaged across two tests with inhibition in each hemisphere. (O) Mean proportion of responding on the ipsilateral and contralateral lever during the LED ON period, as a proportion of the total responding on each respective lever, for each animal in each group, averaged across two tests. For all data, bars represent group means. *p<0.05, **p<0.01.
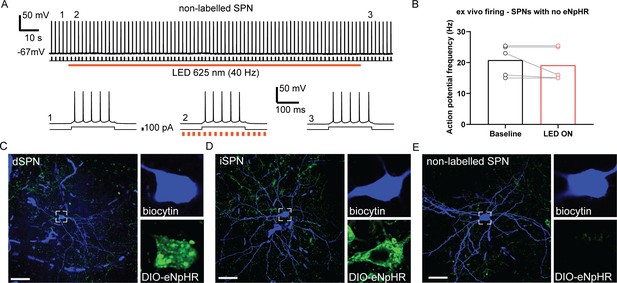
Verification of cellular expression and non-labelled SPN activity for the optogenetic inhibition experiment.
(A) A representative raw trace of a non-halorhodopsin-positive spiny projection neuron (SPN) recorded under current-clamp with an injection of +100 pA current (duration 200 ms pulse, triggered at 0.5 Hz) to evoke action potentials. Orange bar directly below denotes time period when LED 625 nm was applied to the neuron pulsing at 40 Hz for 150 s. Bottom three panels are expanded traces from the above at time points before (1), during (2), and after (3) LED illumination. The SPN was resting at −67 mV. (B) Action potential frequency of non-halorhodopsin labelled SPNs (n=5) at baseline (10 s time period before LED) versus LED illumination (10 s time period from the beginning of LED). (C) Biocytin (blue)-labelled dSPN with halorhodopsin (green) expression from recording in Figure 4B (top trace) (scale bar, 30 µm). (D) Biocytin (blue)-labelled iSPN with halorhodopsin (green) expression from recording in Figure 4B (bottom trace) (scale bar, 30 µm). (E) Biocytin-labelled SPN without halorhodopsin expression from cell recording in A.
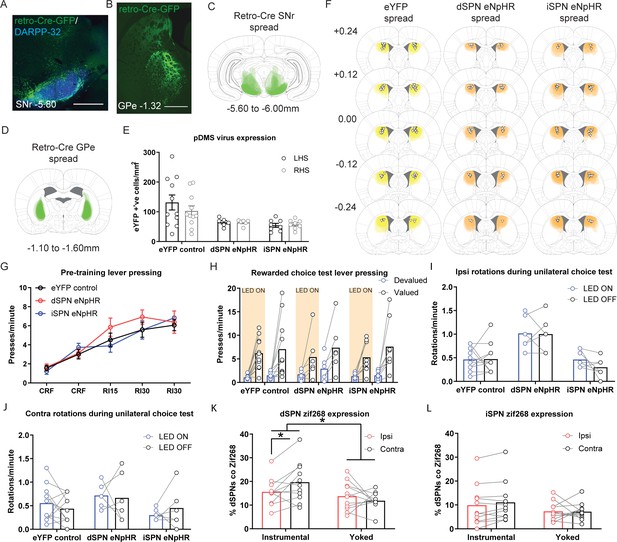
Verification of the eNpHR virus and supplemental behavioral data related to the optogenetic inhibition experiment.
(A) Confocal image (scale bar, 1000 µm) from one rat showing retro-Cre expression in the substantia nigra pars reticulata (SNr). (B) Same as A but image shows retro-Cre expression in the globus pallidus (GPe). (C) The extent of retro-Cre spread in the SNr, imaged at the widest point of expression for all included animals. (D) Same as C but schematic shows the extent of retro-Cre spread in the GPe. (E) Total number of eYFP positive cells/mm2 in left and right posterior dorsomedial striatum (pDMS) for each rat in each experimental group. Bars represent group means ± SEM. (F) Extent of DIO-eYFP (left), DIO-eNpHR in direct spiny projection neurons (dSPNs; middle) and DIO-eNpHR in indirect spiny projection neurons (iSPNs; right) for all included animals in each group, imaged at five different AP locations encompassing where virus expression is at its widest. Triangles represent locations of cannula placements for each rat. (G) Mean (± SEM) lever presses per minute averaged across each day of instrumental pre-training for each group. (H) Mean lever presses per minute on the devalued and valued lever for each rat in each group during LED ON (orange shaded) and LED OFF (non-shaded) periods, averaged across 2 days of rewarded choice tests. (I) Mean number of ipsilateral rotations (relative to hemisphere of LED light delivery) per minute for each rat in each group, averaged across two tests during LED ON and LED OFF periods. (J) Same as I but for contralateral rotations (relative to hemisphere of LED light delivery). (K) Data from SPN activity marker experiment (Figure 1) showing the percentage of labelled dSPNs that were co-labelled with Zif268 for each rat in Group Instrumental and Group Yoked in the hemisphere ipsilateral to lever position and contralateral to lever position. Data presented are means of four pDMS sections per rat. (L) Same as K but for iSPNs. For all data, bars represent group means *p<0.05.
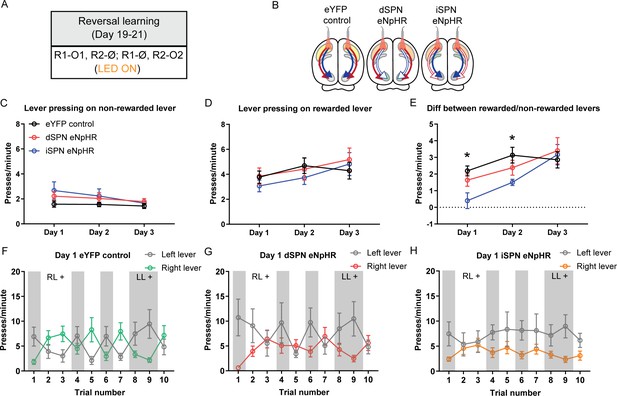
Optogenetic inhibition of indirect spiny projection neurons, but not direct spiny projection neurons, in the posterior dorsomedial striatum impairs response flexibility.
(A) Summary of the experimental design; R1 and R2 indicate left and right lever responses; O1 and O2 indicate distinct food outcomes; Ø indicates non-rewarded responses; LED light was delivered during all lever presentations. (B) Summary of experimental groups; blue arrows indicate intact direct pathway function, red arrows indicate intact indirect pathway function, and unfilled arrows indicate inhibited pathway. (C) Mean (± SEM) presses per minute on the non-rewarded lever across 3 days of reversal training in all groups. (D) Mean (± SEM) presses per minute on the rewarded lever across 3 days of reversal training in all groups. (E) Mean (± SEM) difference in press rate (expressed as presses per minute) between the rewarded and non-rewarded levers for each group. (F–H) Mean (± SEM) lever presses per minute on the left lever and right lever for rats in each group on Day 1 of training, averaged across each 2.5 min trial – grey-shaded regions indicate trials in which the left lever was rewarded (LL+) and non-shaded regions indicate trials in which the right lever was rewarded (RL+). *p<0.05.
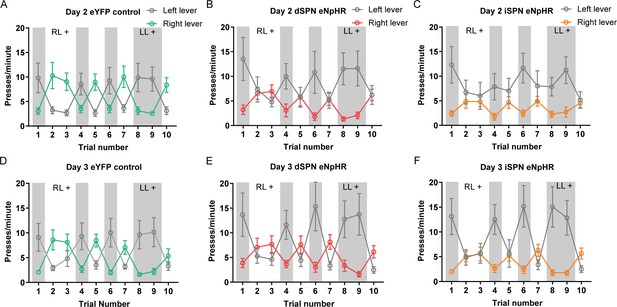
Days 2 and 3 of reversal training in the optogenetic inhibition experiment.
(A–C) Mean (± SEM) lever presses per minute on the left lever and right lever for rats in each group on Day 2 of reversal training, averaged across each 2.5 min trial – grey-shaded regions indicate trials in which the left lever was rewarded (LL+) and non-shaded regions indicate trials in which the right lever was rewarded (RL+). (D–F) Same as previous but for Day 3 of training.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Rattus norvegicus, Long-Evans) | Long-Evans Rat | Randwick Rat Breeding Facility, Randwick, NSW, Australia | NA | Wild-type rats |
| Strain, strain background (Rattus norvegicus, Long-Evans) | Long-Evans Rat | Animal Resources Centre, Perth, WA, Australia | NA | Wild-type rats |
| Antibody | Anti-goat Alexa Fluor 647 (donkey polyclonal) | Invitrogen | Cat# A-21447; RRID:AB_2535864 | (1:1000) |
| Antibody | Anti-mouse Alexa Fluor 488 (donkey polyclonal) | Invitrogen | Cat# A-21202; RRID:AB_141607 | (1:1000) |
| Antibody | Anti-mouse Alexa Fluor 546 (donkey polyclonal) | Invitrogen | Cat# A10036; RRID:AB_2534012 | (1:1000) |
| Antibody | Anti-rabbit Alexa Fluor 488 (donkey polyclonal) | Invitrogen | Cat# A-21206; RRID:AB_2535792 | (1:1000) |
| Antibody | Anti-rabbit Alexa Fluor 546 (donkey polyclonal) | Invitrogen | Cat# A10040; RRID:AB_2534016 | (1:1000) |
| Antibody | Anti-cholera-toxin B subunit (goat polyclonal) | LIST Biological Laboratories Inc | Cat# 703; RRID:AB_10013220 | (1:2000) |
| Antibody | Anti-DARPP-32 (mouse monoclonal) | BD Biosciences | Cat# 611520; RRID:AB_398980 | (1:1000) |
| Antibody | Anti-EGR1 (rabbit polyclonal) | Santa Cruz Biotechnology | Cat# C-19; RRID:AB_2231020 | (1:300) |
| Antibody | Anti-GFP (rabbit polyclonal) | Invitrogen | Cat# A-11122; RRID:AB_221569 | (1:1000) |
| Antibody | Streptavidin, Alexa Fluor 488 | Invitrogen | Cat# S11223; RRID:AB_2315383 | (1:500) |
| Recombinant DNA reagent | AAV-Cre: rAAV5.CMV.HI.eGFP-CRE.WPRE.SV40 | AddGene | Cat# 105545 | |
| Recombinant DNA reagent | ChR2: AAV5-CaMKIIa-hChR2(H134R)-eYFP | AddGene | Cat# 26969 | |
| Recombinant DNA reagent | Cre-eNpHR: AAV5-Ef1a-DIO-eNpHR3.0-eYFP | AddGene | Cat# 26966 | |
| Recombinant DNA reagent | Cre-eYFP: AAV5-Ef1a-DIO-eYFP | AddGene | Cat# 27056 | |
| Recombinant DNA reagent | Cre-hM3D: rAAV5/hSyn-DIO-hM3D-mCherry | AddGene | Cat# 44361 | |
| Recombinant DNA reagent | Cre-hM4D: rAAV5/hSyn-DIO-hM4D(Gi)-mCherry | AddGene | Cat# 44362 | |
| Recombinant DNA reagent | Cre-hM4D: rAAV5/hSyn-DIO-hM4D(Gi)-mCherry | UNC Vector Core | SCR_002448 | |
| Recombinant DNA reagent | Cre-mCherry: rAAV5/hSyn-DIO-mCherry | AddGene | Cat# 50459 | |
| Chemical compound, drug | Clozapine-N-oxide | RTI International | NA, Batch ID: 13626–76 | |
| Software, algorithm | GraphPad Prism | GraphPad Software Inc | RRID:SCR_002798 | |
| Software, algorithm | Image J | NIH | RRID:SCR_003070 | |
| Software, algorithm | Med-PC program | Med Associates | RRID:SCR_012156 | |
| Software, algorithm | PSY statistical program | School of Psychology, UNSW | NA | |
| Other | AMCA Avidin D | Vector Laboratories | Cat# A-2008 | |
| Other | Biocytin | Sigma-Aldrich | Cat# B1758 | |
| Other | Cholera-toxin B subunit | List Biological Laboratories | Cat# 104; RRID:AB_2313636 | |
| Other | Fluorogold | Fluorochrome | NA |
| Experiment # | N (total rats) | J (number of groups) | df |
|---|---|---|---|
| SPN tracing and immunofluorescence | 22 | 2 (inst vs yoked) | 20 |
| M4 DREADDs | 51 | 5 (CNO control, dSPN+VEH, iSPN+VEH, dSPN+CNO, iSPN+CNO) | 46 (subset of rats analysed separately N=22, J=4 (no mCherry), df=18) |
| M3 DREADDs | 20 | 3 (mCherry+CNO, hM4D+VEH, hM4D+CNO) | 17 |
| Optogenetic inhibition | 25 | 3 (eYFP control, dSPN eNpHR, iSPN eNpHR) | 22 |
| Experiment | Training/Test stage | Group | Stats |
|---|---|---|---|
| DREADDs suppression | Instrumental training (press rate; male vs female) | CNO control | F(1,11)=2.43, p=0.147 |
| dSPN+VEH | F(1,14)=0.27, p=0.616 | ||
| iSPN + VEH | F(1,2)=4.613, p=0.165 | ||
| dSPN+CNO | F(1,10)=0.15, p=0.710 | ||
| iSPN+CNO | F(1,4)=11.55, p=0.027 | ||
| Non-rewarded choice test (magnitude of devaluation; male vs female) | CNO control | F(1,11)=5.425, p=0.040 | |
| dSPN+VEH | F(1,14)=1.41, p=0.254 | ||
| iSPN + VEH | F(1,2)=1.02, p=0.419 | ||
| dSPN+CNO | F(1,10)=0.17, p=0.685 | ||
| iSPN+CNO | F(1,4)=3.97, p=0.117 | ||
| DREADDs stimulation | Instrumental training (press rate; male vs female) | mCherry+CNO | F(1,4)=0.41, p=0.556 |
| hM3D+VEH | F(1,4)=0.07, p=0.806 | ||
| hM3D+CNO | F(1,6)=0.60, p=0.815 | ||
| Non-rewarded choice test (magnitude of devaluation; male vs female) | mCherry+CNO | F(1,4)=2.44, p=0.913 | |
| hM3D+VEH | F(1,4)=0.30, p=0.612 | ||
| hM3D+CNO | F(1,6)=0.64, p=0.454 | ||
| Optogenetic inhibition | Instrumental training (press rate; male vs female) | eYFP control | F(1,9)=0.00, p=0.975 |
| dSPN eNpHR | F(1,4)=4.98, p=0.090 | ||
| iSPN eNpHR | F(1,6)=0.43, p=0.534 | ||
| Non-rewarded choice test (sex x LED interaction on magnitude of devaluation) | eYFP control | F(1,9)=0.03, p=864 | |
| dSPN eNpHR | F(1,4)=0.00, p=1.00 | ||
| iSPN eNpHR | F(1,6)=0.13, p=0.734 | ||
| Choice test with unilateral stimulation (gender x LED effect on lever bias) | eYFP control | F(1,9)=0.197, p=0.668 | |
| dSPN eNpHR | F(1,4)=0.067, p=0.809 | ||
| iSPN eNpHR | F(1,6)=0.170, p=0.694 | ||
| Reversal (difference between rewarded and non-rewarded; male vs female) | eYFP control | F(1,9)=0.01, p=0.931 | |
| dSPN eNpHR | F(1,4)=0.28, p=0.628 | ||
| iSPN eNpHR | F(1,6)=0.63, p=0.458 |





