Gap junctions deliver malonyl-CoA from soma to germline to support embryogenesis in Caenorhabditis elegans
Figures
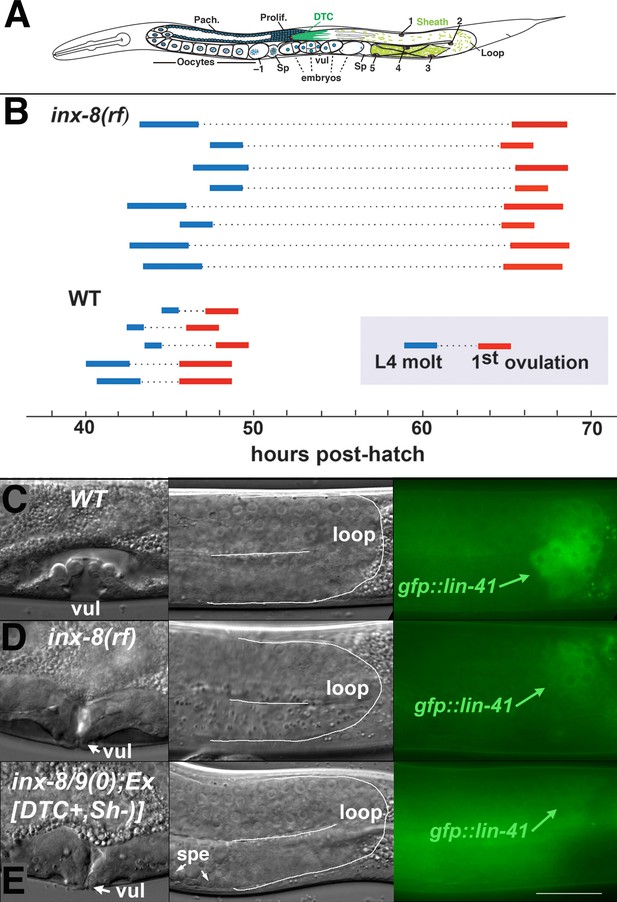
Ovulation and oocyte development are delayed in inx-8(rf).
(A) Diagram of adult C. elegans gonad. Posterior arm shows the positions of the somatic distal tip cell (DTC) and five pairs of sheath cells (nuclei are numbered). Anterior arm shows the underlying germline, approximate regions of germ cell proliferation (Prolif.), meiotic pachytene stage (Pach.), and developing oocytes. Embryos typically undergo some cell division in the uterus before being laid. Sp, spermatheca; vul, vulva. (B) Compared to the wild type (N2), ovulation in inx-8(tn1513 tn1555rf) inx-9(ok1502null) mutants (a.k.a. inx-8[rf]) is delayed ~18 hr. Bars representing 1–3 hr windows indicate approximate time of the fourth larval stage (L4 to adult) molt, followed by time of appearance of the first fertilized embryo ovulated into the uterus. (C–E) Onset of gfp::lin-41 expression as a marker of oocyte development is delayed when sheath–germline gap junction coupling is perturbed. Panels show DIC and fluorescent images from individual worms of vulval development (left), loop region of gonad arm (middle), and expression of gfp::lin-41 (right). (C) Wild-type hermaphrodite expressing gfp::lin-41 by the approximate L4.7 sub-stage of vulval development (vul). (D) inx-8(rf) hermaphrodite does not express gfp::lin-41 until after vulval development is complete but before the L4-to-adult molt. (E) Null inx-8(tn1474) inx-9(ok1502) hermaphrodite, rescued for inx-8::gfp expression in the DTC with Ex[lag-2p::inx-8::gfp]—a.k.a. inx-8/9 (0); Ex[inx-8(DTC+, Sh–)]—shows a similar delayed onset of gfp::lin-41 expression as inx-8(rf). spe, sperm. Bar, 20 μm.
-
Figure 1—source data 1
inx-8(rf) exhibits reduced brood size and interacts with FA synthesis pathway genes.
- https://cdn.elifesciences.org/articles/58619/elife-58619-fig1-data1-v2.docx
-
Figure 1—source data 2
Brood counts for Figure 1.
- https://cdn.elifesciences.org/articles/58619/elife-58619-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Reduced soma-germline gap junction coupling delays gametogenesis.
- https://cdn.elifesciences.org/articles/58619/elife-58619-fig1-data3-v2.docx
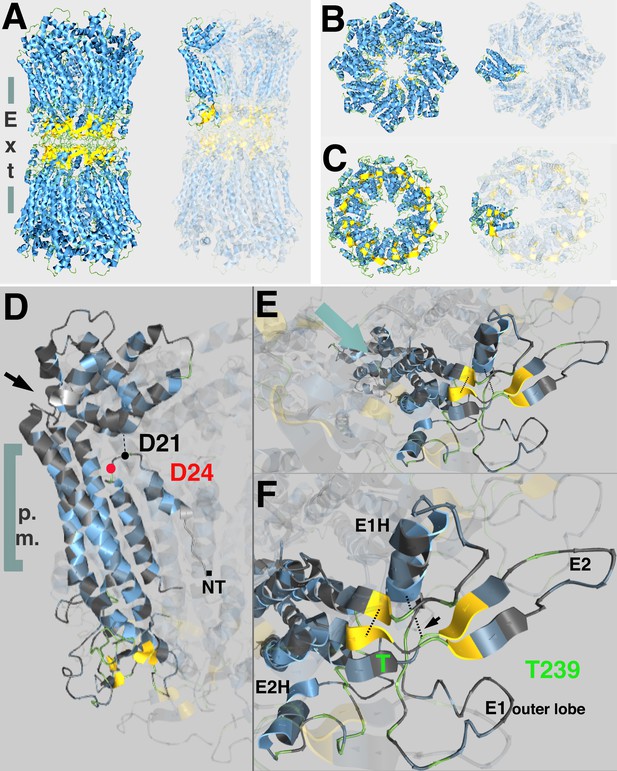
Predicted topological sites of amino acids mutated in inx-8(tn1513 tn1555) inx-9(0).
(A) Side view of atomic structure of a C. elegans homomeric, homotypic INX-6 gap junction full channel, determined by Oshima et al., 2016 (PDB I.D. 5H1R, rcsb.org; Berman et al., 2000), viewed at aquaria.ws (O'Donoghue et al., 2015). Vertical gray bars indicate approximate location of the plasma membranes of apposed cells; ‘Ext’ indicates the extracellular region where octameric hemichannels associate via extracellular loops to form an enclosed channel in the region classically identified by TEM as the ‘gap’ between closely aligned plasma membranes. Beta-strands (yellow) in the extracellular loops contribute to walling off the pore. Single innexin subunit is highlighted in the representation on the right. (B,C) Model for INX-8 hemichannel based on homology alignment (25% identity) to INX-6 (PDB 5H1Q) as viewed from the cytoplasm (B) or extracellular region (C). Single subunits highlighted in representations on right. In wild-type, this hemichannel is comprised of both INX-8 and INX-9 subunits (84% identity), and associates with germline hemichannels comprised of INX-14+INX-21 or INX-14+INX-22. (D) Position of INX-8 D24N, suppressor mutation of T239I. NT indicates approximate region of N-terminus. A kinked region (black arrow) allows the cytoplasmic structure of the subunits (cytoplasmic loops and C-termini) to form a dome at the pore entry. Amino termini line the pore as a funnel; a loop in the amino terminus allows for an interaction with residues in the dome (Oshima et al., 2016). D24 (red) lies near the D21 residue predicted by homology with INX-6 (D25) to interact with the cytoplasmic dome. (E) View of INX-8 subunit from extracellular face. Aqua arrow indicates approximate angle of channel, which lies in a plane above the highlighted subunit. (F) INX-8 T239 is predicted to lie adjacent to beta-strands (yellow) contributing to extracellular loop formation integral to hemichannel-hemichannel association. Disulfide bonds forming between cysteines that are invariant in the innexin family are indicated by dotted lines (one marked with arrow). E1H, helix formed in the first extracellular loop that contributes to constriction of the pore in the extracellular region. E2H, helix formed in the second extracellular loop that lies external to the channel. E1 and E2 are loops formed in the respective extracellular regions. Oshima et al., 2016 for details.
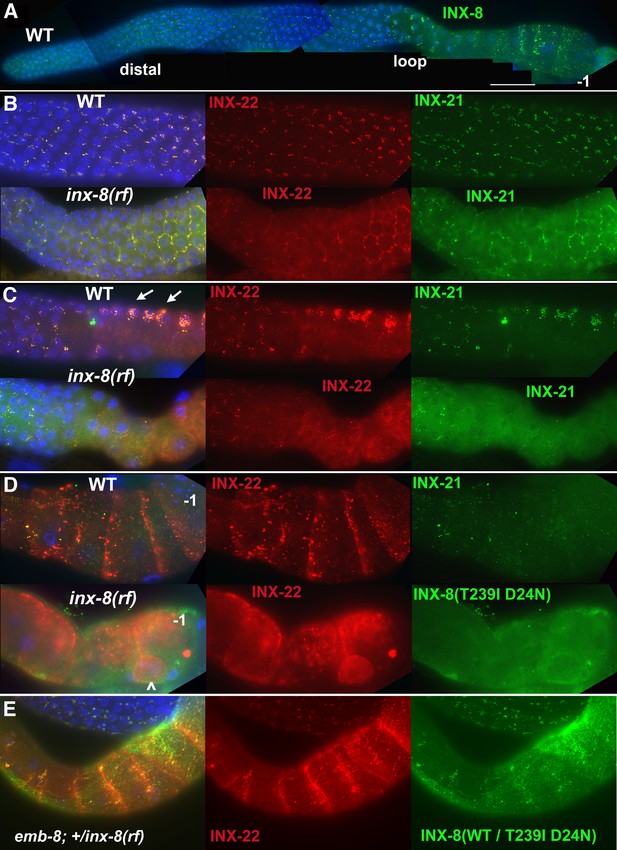
INX-8(T239I D24N) compromises gap junction formation between soma and germline.
(A) Somatic INX-8 in wild-type (WT) gonads localizes to puncta representing soma-germline gap junctions where DTC or sheath cells overlie germ cells. (B) In WT distal gonad arms, gap junction puncta aggregate in small clusters associated with each germ cell; in inx-8(rf) distal arms, gap junction puncta localize more apically, where sheath dips down between germ cells, outlining the cell (honeycomb pattern). Localization of INX-21 and INX-22 define the two different classes of germline gap junction hemichannels, both of which include INX-14 subunits. (C) At the loop region, WT gap junction puncta form large aggregates (arrows) which are absent in inx-8(rf). (D) In WT proximal arms, gap junction formation with INX-22-containing hemichannels is extensive; in inx-8(rf), INX-22 and INX-8(T239I D24N) are strongly expressed but show little evidence of gap junction formation. ‘–1’ indicates the most proximal oocyte; inx-8(rf) hermaphrodites frequently show undersized oocytes at this position as well (<). (E) Heterozygous +/inx-8(rf) shows little effect on gap junction localization in an emb-8(hc69ts) background at 22°C. Antibodies specific to INX-8, INX-21, or INX-22 were used as indicated. DAPI-stained DNA in blue. Bar, 50 μm.
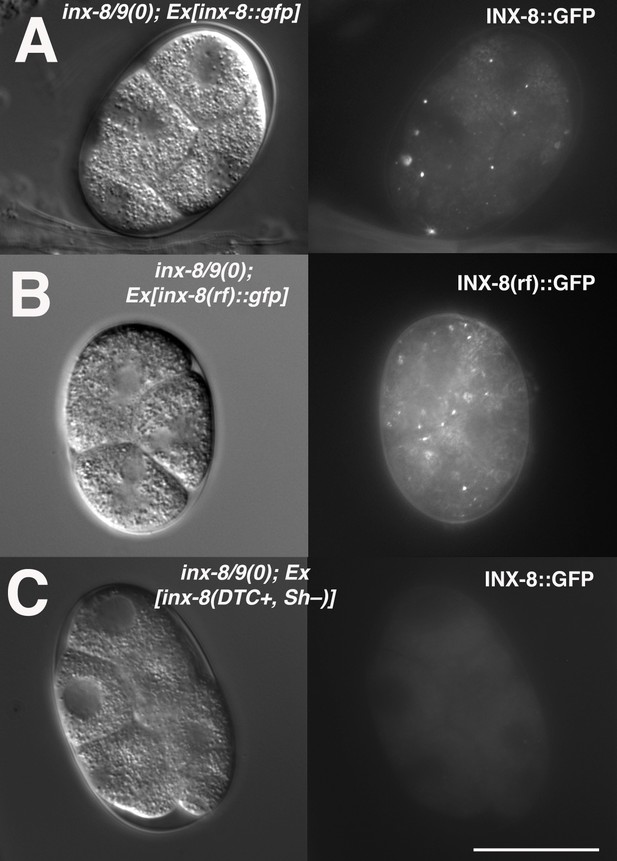
INX-8(rf)::GFP can be endocytosed by oocytes.
(A) inx-8(+)::gfp expressed in the somatic gonad sheath is endocytosed during ovulation and appears as distinct fluorescent puncta in early embryos. (B) inx-8(rf)::gfp expressed on an extrachromosomal array in inx-8(0) inx-9(0) animals is endocytosed by ovulating oocytes. (C) inx-8::gfp expressed by the DTC in inx-8/9 (0); Ex[inx-8(DTC+,Sh-)] is not observed in early embryos. Bar, 20 μm.
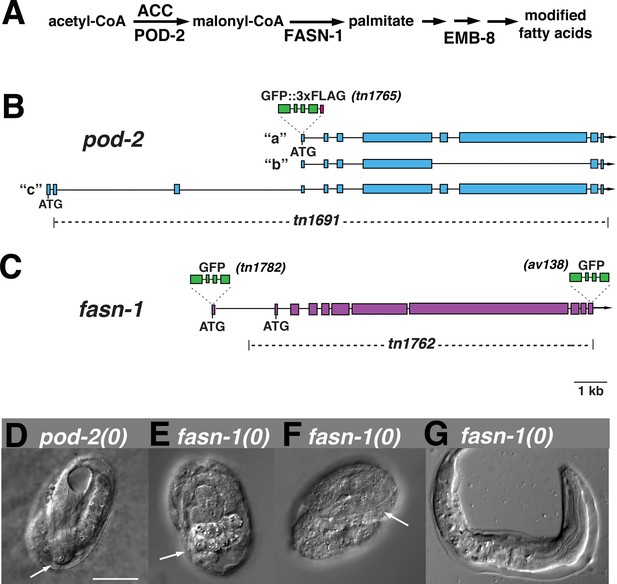
Genome editing of pod-2 and fasn-1.
(A) Predicted assignment of FAS gene products in a simplified FAS pathway. Targeted deletions and GFP insertions are shown for pod-2 (B) and fasn-1 (C). Predicted isoforms for POD-2 are indicated. (D–G) Terminal phenotypes of (D) pod-2(0) and (E–G) fasn-1(0) mutant embryos from heterozygous +/(0) parents. Arrows in (D) and (F) indicate differentiated anterior pharynx; arrow in (E) indicates differentiated intestinal cells that formed gut granules. Bar, 20 μm.
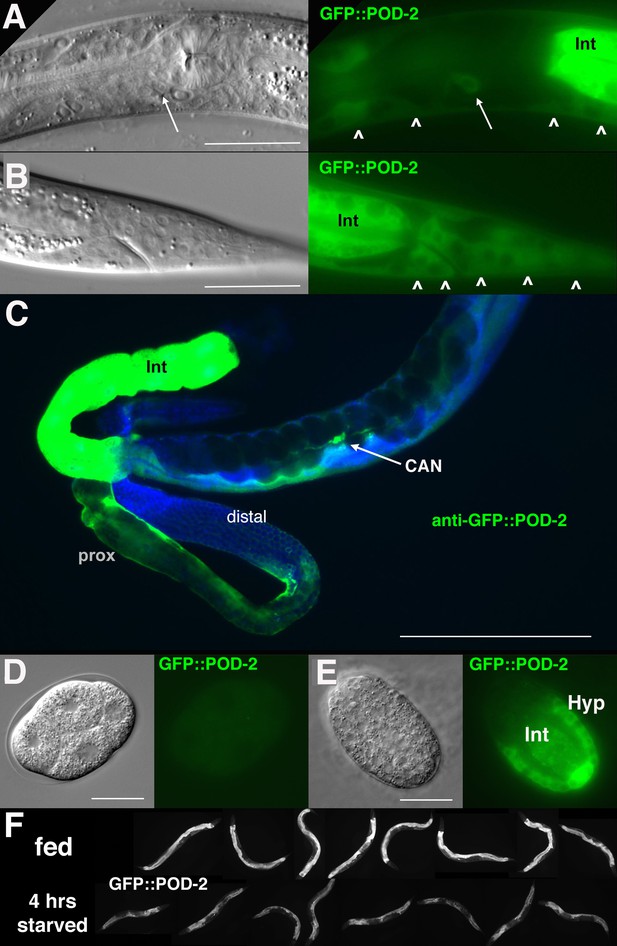
GFP::POD-2 expression.
(A) Expression in the head includes hypodermis (carets) and excretory duct cell (arrow). Int, intestine. (B) Primary expression in the tail includes hypodermis (carets) and intestine (Int). (C) Dissected animal stained with anti-GFP antibody shows abundant intestinal expression (Int) and strong expression in proximal sheath of exposed gonad arm (prox) that diminishes in the distal arm. No germline expression is seen in the distal arm where germ cells are revealed due to the absence of sheath coverage. Arrow indicates CAN neuron, one of the few neurons essential for viability in C. elegans. DAPI-staining of DNA in blue. (D, E) Earliest expression in embryos is evident in developing hypodermis and intestine. (A, B, D, E) DIC images in left panels, live GFP fluorescence in right panels. Bars, 20 μm, except (C) 200 μm. (F) GFP::POD-2 is responsive to starvation. Lower row, animals briefly washed and transferred to bacteria-free NGM plates for 4 hr; upper row, animals similarly-treated but fed. Fluorescent images captured from live animals at identical exposures.
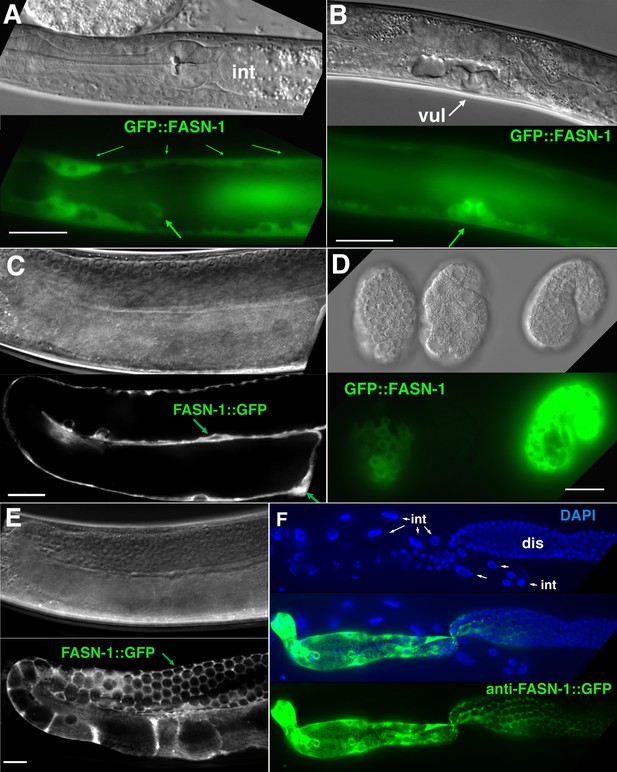
Expression of N-terminal GFP::FASN-1 or C-terminal FASN-1::GFP CRISPR-Cas9 constructs.
(A) Expression is prominent throughout the hypodermis (small arrows) and excretory duct cell (larger arrow), but not intestine (int). (B) Expression in developing vulva (arrow). (C) Gonadal sheath expression, with absence of apparent germline expression. (D) Earliest expression is seen in the hypodermis of developing embryos, and hypodermal expression continues throughout development. (E) Expression is apparent in all somatic sheath cells (Sh1–5). (F) Anti-GFP antibody staining of dissected gonad and intestine expressing FASN-1::GFP. DAPI staining shows large nuclei (int) of intestine crossing underneath the gonad. Strong expression is detected in the proximal gonad, less so in distal gonad (dis), but no intestinal expression is apparent. (A–E) DIC images in upper panels, live GFP fluorescence in lower panels. Bars, 20 μm.
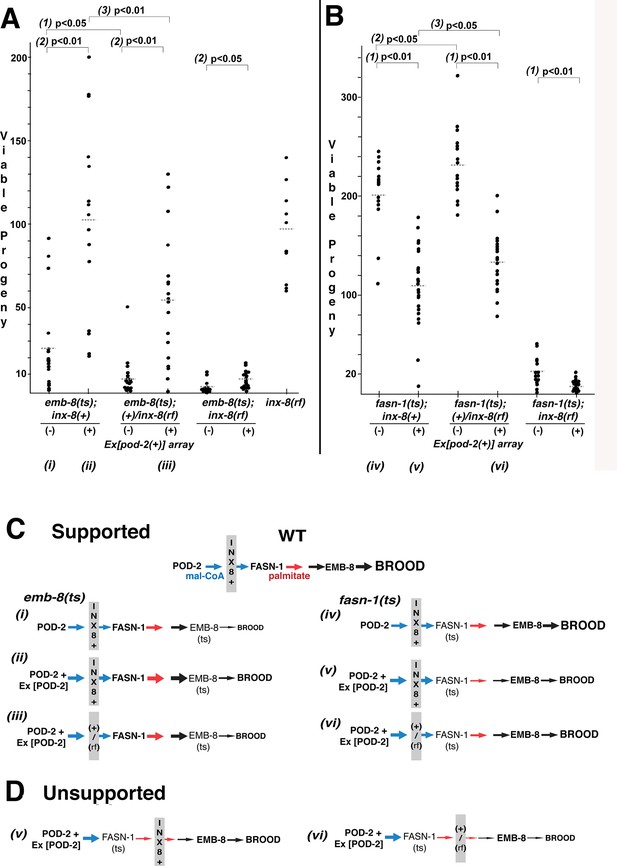
Wild-type INX-8 facilitates effects of pod-2(+) overexpression on conditional mutations downstream in the fatty acid synthesis pathway.
Brood size was measured as the number of viable progeny produced. The balancer mIs11 provided inx-8(+) for all strains listed. (A) Embryonic lethality in emb-8(ts) mutants at 22°C is more severe in heterozygous +/inx-8(rf) than inx-8(+) backgrounds (1; p=0.013)). Additional pod-2(+) expression furnished by Ex[pod-2(+)] in emb-8(ts) significantly rescues embryonic lethality (2; p=1.9 E-05, inx-8[+]; p=1.9 E-05, +/inx-8[rf]; p=0.022, inx-8[rf]). Rescue of emb-8(ts) by Ex[pod-2(+)] is also dependent on inx-8(+) copy number (3; p=5.5 E-03). (B) Expression of Ex[pod-2(+)] in fasn-1(ts) mutants at 20°C reduces brood size (1; p=2.2 E-08, inx-8[+]; p=1.5 E-10, +/inx-8[rf]; p=7.0 E-04, inx-8[rf]). fasn-1(ts); inx-8(+) homozygotes show greater embryonic lethality than fasn-1(ts); (+)/inx-8(rf) heterozygotes in the absence (2; p=0.026) or presence (3; p=0.026) of Ex[pod-2(+)]. Averages indicated with dotted lines. Probability was calculated using Student’s two-tailed t-test. (C) Interpretation of genetic interaction consistent with the results from (A) and (B) in a pathway representation. Relative size of arrows corresponds to expected relative amount of product. Sheath–germline gap junctions composed of INX-8(+) or a mixture of INX-8(+) and INX-8(rf) are positioned as facilitating transfer of malonyl-CoA from soma to germline. Roman numerals correspond to genotypes indicated in (A) and (B). (D) Placement of INX-8 gap junctions downstream of fasn-1 predicts brood size from genotype (vi) would be less than from (v), which is not consistent with the results observed in (B). Figure 6—source data 1 Viable progeny counts for panels (A, B).
-
Figure 6—source data 1
Brood counts for Figure 6.
- https://cdn.elifesciences.org/articles/58619/elife-58619-fig6-data1-v2.xlsx
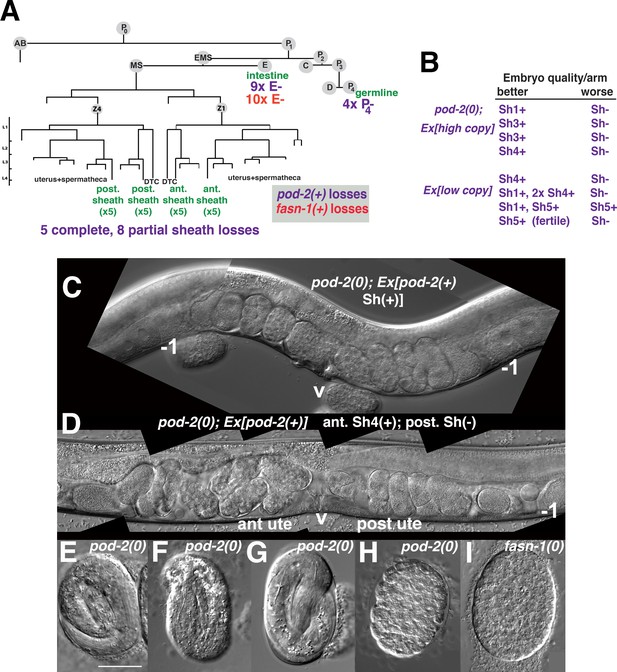
Genetic mosaic analyses of pod-2 reveal a mal-CoA requirement in somatic gonad sheath.
(A) Abbreviated cell lineage of C. elegans. Mosaics characterized correspond to loss of sur-5::gfp expression in the germline (P4), intestine (E), or somatic gonad. Five pod-2(0); Ex[pod-2(+); sur-5::gfp] animals lost the array from all somatic gonad sheath cells and produced no viable embryos. (B) Eight animals showed differences in array retention in each gonad arm with phenotypic consequences. Arrays were generated by microinjecting cosmid W09B6 [pod-2(+)] at 4 ng or 20 ng/μl (low/high copy). (C) Uterus of non-mosaic pod-2(0); Ex[pod-2(+)—high copy] hermaphrodite; developmental progression of embryos from each gonad arm is comparable. (D) Mosaic pod-2(0); Ex[pod-2(+)—high copy] with array lost from posterior gonad arm sheath but retained in a single Sh4 cell of anterior arm. Anterior arm embryos have more distinct eggshells and develop to later stages. (E–H) Embryonic development depends on somatic levels of mal-CoA. Shown are terminal-stage pod-2(0) embryos derived from parents of designated genotypes. (E) Pretzel-stage embryo from +/pod-2(0). (F) Threefold stage embryo from pod-2(0); Ex[pod-2(+)—low copy], with array maintained in soma but lost in the germline (P4–). (G) Pretzel-stage embryo from P4(–) mosaic pod-2(0); Ex[pod-2(+)—high copy]. (H) Embryo from mosaic pod-2(0); Ex[pod-2(+)—low copy] with uncharacterized somatic loss of array, but array retained in the germline (P4+). This parent produced exclusively dead embryos. (I) Typical terminal undifferentiated fasn-1(0) Pod embryo from fasn-1(0); Ex[fasn-1(+)]. ant ute, anterior uterus; post ute, posterior uterus; v, vulva; –1, most proximal oocyte. Bar, 20 μm.
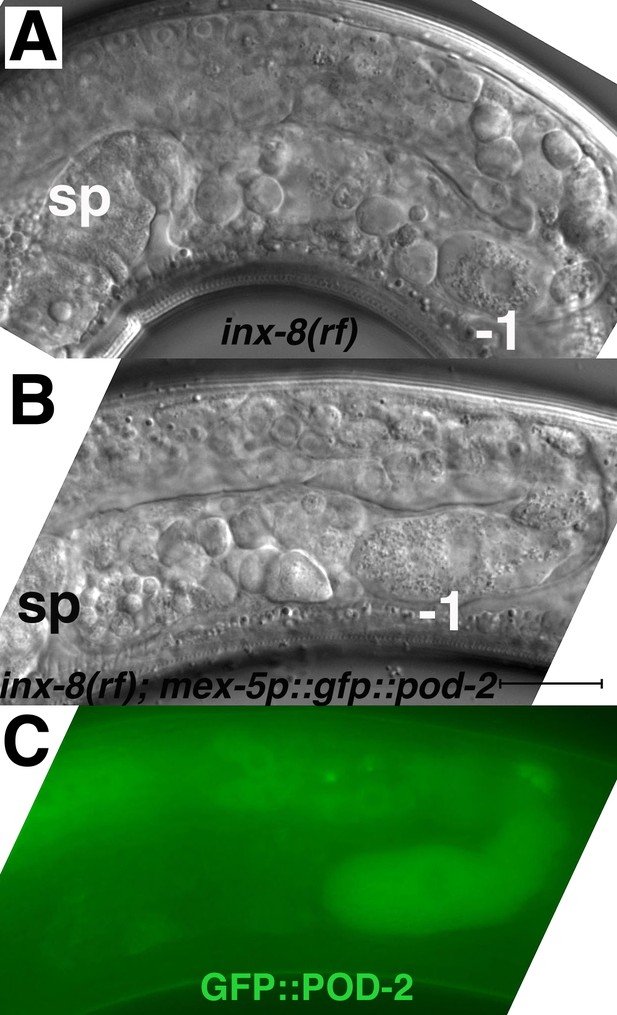
Expression of gfp::pod-2 in the germline fails to rescue inx-8(rf) phenotypes.
(A) Young (first-day) adult inx-8(rf) gonad arm. (B, C) Young adult inx-8(rf) gonad arm expressing mex-5p::gfp::pod-2::tbb-2–3’UTR. The pod-2 isoform in this construct corresponds to gfp::pod-2 isoform ‘a’ in Figure 3B. This isoform encompasses sequence homology to all the enzymatic sites in other ACC multi-functional enzymes. sp, spermatheca; –1, most proximal oocyte; bar, 20 μm.
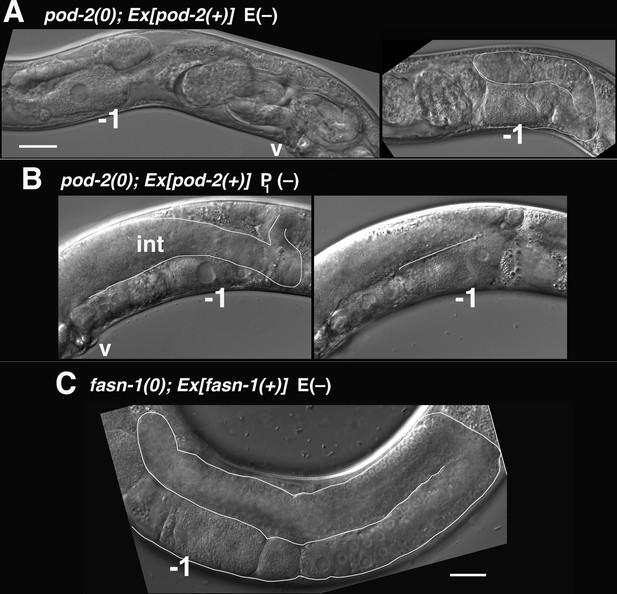
Mosaic loss from the intestine of pod-2, but not fasn-1, leads to an ARD-like gonad arm.
(A) E(–) genetic mosaic pod-2(0); Ex[pod-2(+); sur-5::gfp] gonad arms (from same animal). Gonad arm on right partially outlined. (B) Two focal planes of a presumptive P1(–) pod-2(0); Ex[pod-2(+); sur-5::gfp] mosaic with constipated intestine. Upper plane (left) with partially outlined intestine; lower plane (right) shows underlying distal gonad arm. int, intestine; p, pachytene region; v, vulva. (C) E(–) genetic mosaic fasn-1(0); Ex[fasn-1(+); sur-5::gfp] gonad arm. –1, most proximal oocyte. Bars, 20 μm.
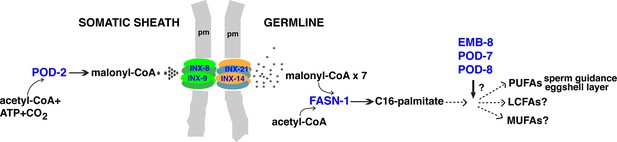
Model for malonyl-CoA transfer from soma to germline.
Enzymes (blue) implicated in germline FAS. Malonyl-CoA transits from somatic sheath to germline through gap junction channels comprised of INX-8/9 (soma) and INX-14/INX-21 (germline) hemichannels. Germline FASN-1 uses malonyl-CoA to synthesize palmitate, which may be further extended and modified to produce other long-chain fatty acids. EMB-8 (NADPH-dependent CYP450 reductase) and POD-7/8 (CYP450s) are necessary to synthesize the lipid-rich (inner) layer of the eggshell and support polarity establishment (Rappleye et al., 2003; Benenati et al., 2009); the composition of lipid layer fatty acids is unknown. de novo PUFA synthesis is required in the germline for embryonic viability and is implicated in sperm guidance (Kubagawa et al., 2006; Watts et al., 2018). The identities of other long-chain fatty acids that may be synthesized in the germline are unknown. PUFAs, polyunsaturated fatty acids; LFAs, long-chain fatty acids; MUFA, monounsaturated fatty acids; pm, plasma membrane.
Tables
Brood sizes suggest FA synthesis influences embryo output.
| Genotype | Larvae | Dead embryos | Total (embryo output) |
|---|---|---|---|
| Wild type (N2) | 291 ± 29 (n=33) | 2 ± 2 | 293 ± 29 |
| inx-8(rf)* | 108 ± 40 (n=90) | 11 ± 6 (n=30) | 119 ± 42 |
| inx-8/9(0); Ex[inx-8(DTC+,Sh–)] | 2 ± 2 (n=32) | 19 ± 13 | 20 ± 14 |
| pod-2(0); Ex[pod-2(+)] high copy (20 ng/μl) | 164 ± 50 (n=31) | 118 ± 41 | 282 ± 59 |
| pod-2(0); Ex[pod-2(+)] low copy (4 ng/μl) | 91 ± 24 (n=41) | 156 ± 37 | 247 ± 50 |
| fasn-1(0); Ex[fasn-1(+)] | 0.05 (n=80) | 276 ± 18 (n=16) | 276 ± 18 |
| fasn-1(0); inx-8(rf); Ex[fasn-1(+)] | 17 ± 15 (n=47) | 43 ± 27 | 60 ± 37 |
-
*Full genotype inx-8(tn1513 tn1555) inx-9(ok1502null).
-
Table 1—source data 1
Brood counts for genotypes shown in Table 1.
- https://cdn.elifesciences.org/articles/58619/elife-58619-table1-data1-v2.xlsx
Additional files
-
Supplementary file 1
C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/58619/elife-58619-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58619/elife-58619-transrepform-v2.docx





