Regenerative neurogenic response from glia requires insulin-driven neuron-glia communication
Figures
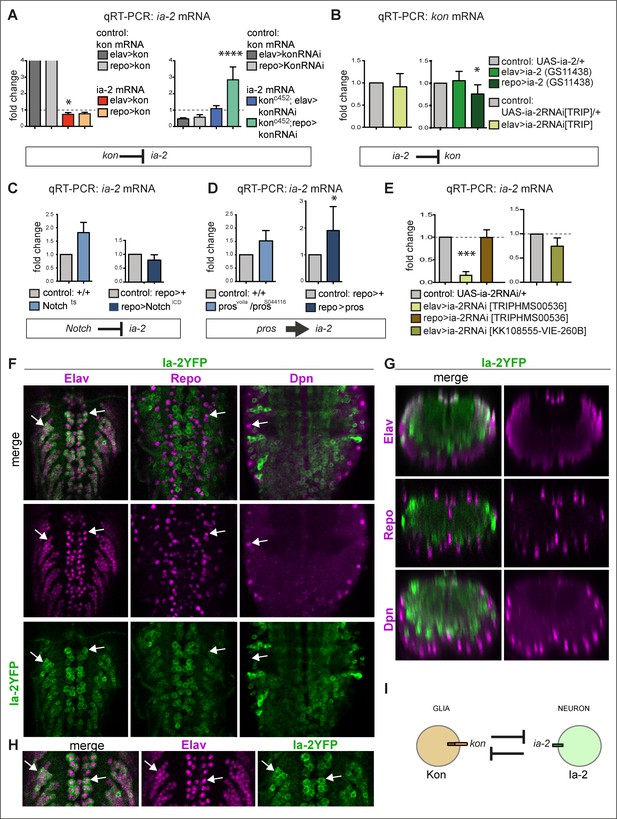
ia-2 interacts genetically with kon, Notch, and pros.
(A) Quantitative real-time PCR (qRT-PCR) showing that gain of kon function reduced ia-2 mRNA levels by 25% (one-way ANOVA p=0.045), whereas loss of kon function in glia caused practically a threefold increase in ia-2 mRNA levels (genotype: konc452/UASkonRNAi; repoGAL4/+; one-way ANOVA p<0.0001). Post hoc Dunnett’s test multiple comparisons to control. N = 4 replicates. (B) qRT-PCR showing that overexpression of ia-2 in glia downregulated kon mRNA levels. Left: Unpaired Student's t-test with Welch correction p=0.457. Right: one-way ANOVA p<0.045, post hoc Dunnett’s test multiple comparisons to control. N = 4–6 replicates. (C) Ia-2 is functionally related to Notch: qRT-PCR showing that ia-2 mRNA levels increased in Nts mutant larvae at the restrictive temperature of 25°C. Unpaired Student's t-test with Welch correction. Left: p=0.4123; Right: p=0.2182. N = 3 replicates. (D) ia-2 is functionally related to pros: qRT-PCR showing that overexpression of pros in glia increased ia-2 mRNA levels by twofold. Unpaired Student's t-test with Welch correction. Left: p=0.1368; Right: p=0.0428. N = 3 replicates. (E) qRT-PCR showing that UAS-ia-2 RNAi[TRIPHMS00536] knock-down in neurons (with elavGAL4) lowered ia-2 mRNA levels to 20%, whereas in glia it has no effect, meaning that ia-2 is expressed in neurons. A second UAS-ia-2RNAi[KK108555-VIE-260B] line lowered mRNA levels by 25%. One-way ANOVA p=0.0004, post hoc multiple comparisons to control Dunnett’s test. N = 3 replicates. (F–H) Fusion protein Ia-2YFP revealed expression exclusively in neurons, as all Ia-2YFP+ cells were also Elav+, but Repo− and Dpn−. Genotype: ia-2[CPTI100013]. N = 4–16 larval ventral nerve cords (VNCs). (I) Illustration showing that kon and ia-2 functions are restricted to glia and neurons, respectively, and they mutually exclude each other. (G) Transverse views; (F and H) horizontal views; (H) higher magnification views. With more than two sample types, asterisks indicate multiple comparison post hoc tests to controls: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. For full genotypes and further statistical analysis details, see Supplementary file 1.
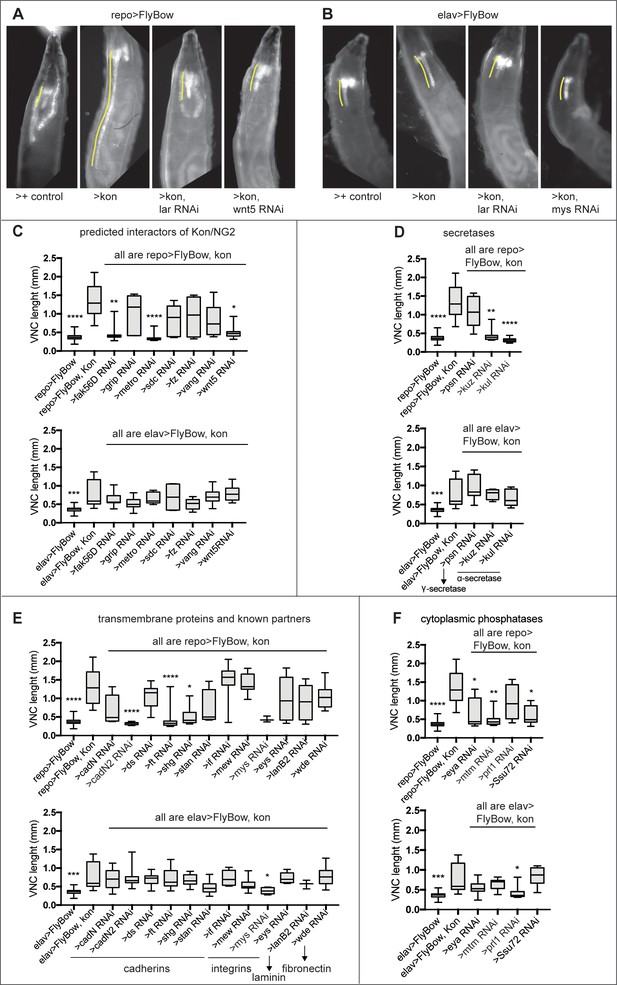
Modifier genetic screens identify genes interacting with kon.
(A and B) Overexpression of kon in glia with repoGAL4 (repo>UASFlyBow, UASkon-full-length) caused a phenotype of very long ventral nerve cord (A), and in neurons with elavGAL4 too, although to a lesser extent (B). These phenotypes were quantified by using the reporter UASFlyBow, and the ventral nerve cord (VNC) measured using ImageJ tools. RNAi knock-down of candidate genes could rescue these phenotypes; some examples are shown here. (C–F) The kon gain of function (GOF) phenotype resulting from overexpressing kon-full-length in either neurons or glia could be rescued by RNAi knock-down of: (C) predicted interactors of Kon or NG2, most prominently in glia; Kruskal–Wallis ANOVA p<0.0001, post hoc Dunn test to >FlyBow, kon controls. N = 4–24 VNCs. (D) α- and γ-secretases that cleave NG2 and Notch, from glia; Kruskal–Wallis ANOVA p<0.0001, post hoc Dunn test to >FlyBow, kon controls. N = 4–24 VNCs. (E) Known Kon partners, for example, integrins, and other transmembrane proteins from neurons; Kruskal–Wallis ANOVA p<0.0001, post hoc Dunn test to >FlyBow, kon controls. N = 3–24 VNCs. (F) cytoplasmic phosphatases, from either glia or neurons; Kruskal–Wallis ANOVA p<0.0001, post hoc Dunn test to >FlyBow, kon controls. N = 7–24 VNCs. VNC length indicated in yellow in (A and B). Asterisks indicate multiple comparison post hoc tests to controls: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Data shown in box-plots. For full genotypes and further statistical analysis details, see Supplementary file 1.

Modifier candidate genetic screens identify genes encoding transmembrane phosphatases and insulin signalling factors as interacting with kon.
(A and B) Overexpression of kon in glia causes a very long ventral nerve cord (VNC) (A), and in neurons too, but to a lesser extent (B). RNAi knock-down of candidate genes could rescue these gain of function phenotypes; some examples are given. (C and D) Quantification of normalised VNC length shows rescue prominently by knock-down of most transmembrane phosphatases, the Notch-related Akap200, and genes functionally related to the insulin signalling pathway (Akt, lar and ia-2), most prominently lar. Normalised measurements are given as a ratio of the VNC over total larval length. Kruskal–Wallis ANOVA p<0.0001, post hoc Dunn’s test comparison to controls repo>kon or elav>kon. (C) N = 2–28; (D) N = 2–31. Data shown in box-plots. Asterisks indicate multiple comparison post hoc tests to controls: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. For full genotypes and further statistical analysis details, see Supplementary file 1.
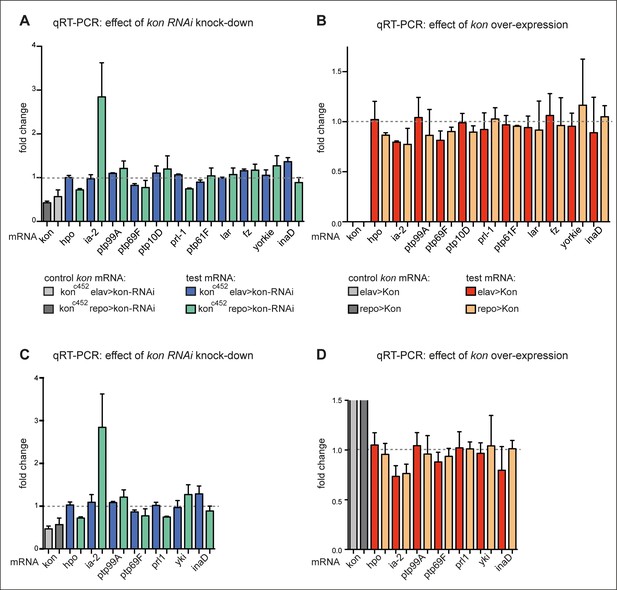
Loss and gain of kon function prominently affected ia-2 expression.
(A and B) Exploratory quantitative real-time PCR (qRT-PCR), N = 2 replicates each: (A) showing the change in mRNA levels for candidate genes upon kon RNAi targeted to either neurons (with elavGAL4) or glia (with repoGAL4). ia-2 mRNA levels increased at least threefold when kon was knocked-down in glia; (B) showing the effect of kon gain of function. kon overexpression in either neurons or glia decreased ia-2 mRNA levels. The first two columns have been left cut out as they are controls with the increase in kon mRNA with kon overexpression, which are very high compared to the rest. (C and D) Further replicates were carried out for a selected group of genes, and they validate that kon prominently regulates ia-2 expression. N = 4 replicates each. (D) The first columns represent the very high increase in kon mRNA with kon overexpression, and they have been cut as they go well beyond this scale compared to the rest. For full genotypes and further statistical analysis details, see Supplementary file 1.
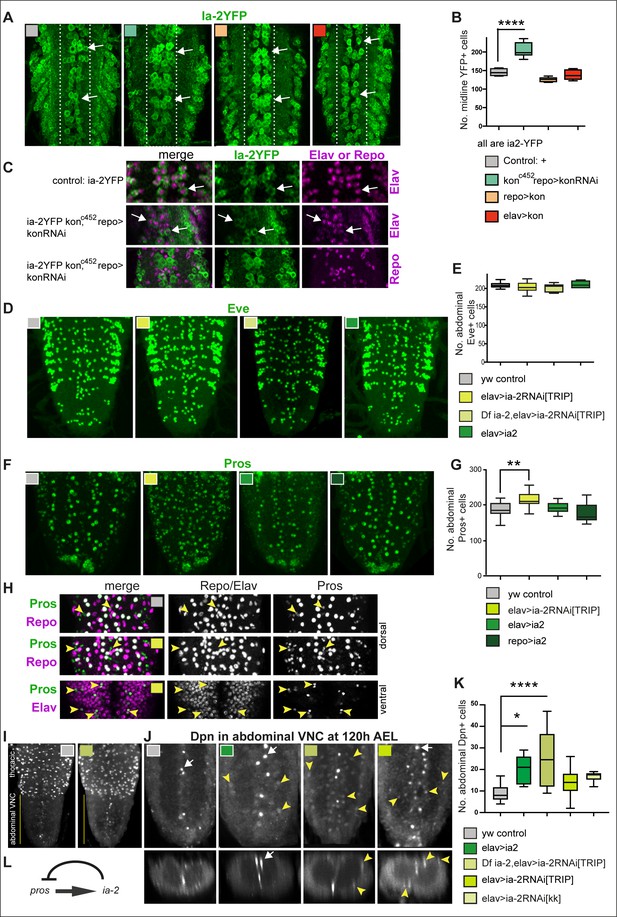
ia-2 influences neural cell fate non-autonomously.
(A and B) Loss of kon function in glia (konc452/UASkonRNAi; repoGAL4/+) increased the number of Ia-2YFP+ cells along the midline. One-way ANOVA p<0.0001, post hoc Tukey’s test. N = 5–8 ventral nerve cords (VNCs). (C) The ectopic Ia-2YFP+ cells in kon loss of function were Elav+ and not Repo+. N = 5–7 VNCs. (D and E) Neither loss nor gain of ia-2 function affected the number of Eve+ neurons. One-way ANOVA p=0. 2374. N = 7–12 VNCs. (F and G) Loss of ia-2 function (elavGAL4>UASia-2RNAi[TRIPHSM00536]) increased Pros+ cell number, and supernumerary cells were small. Kruskal–Wallis ANOVA p=0.0003, post hoc Dunnett’s test. N = 8–26 VNCs. (H) Small Pros+ cells in ia-2 knock-down (genotype as in F and G) did not have the glial marker Repo, but could have the neuronal marker Elav. (I–K) Dpn+ cells visualised at 120 hr AEL, after developmental neuroblasts have disappeared. (I) Dpn signal in thorax was strong as normal, and in abdomen lower Dpn was found ectopically in ia-2 loss or gain of function. (J and K) Both loss and gain of ia-2 function increased the number of abdominal Dpn+ cells, which also were in ectopic locations. (L) Genetic inference: ia-2 negatively regulates pros, most likely non-autonomously. All images are horizontal views, except for I (bottom row) which are transverse views. One-way ANOVA p=0.0002, post hoc Dunnett. N = 7–15. Data shown in box-plot s: box represents 50% values around with median, and whiskers 25% top and bottom values. Asterisks indicate multiple comparison post hoc tests to a fixed control: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. For further statistical analysis details, see Supplementary file 1.
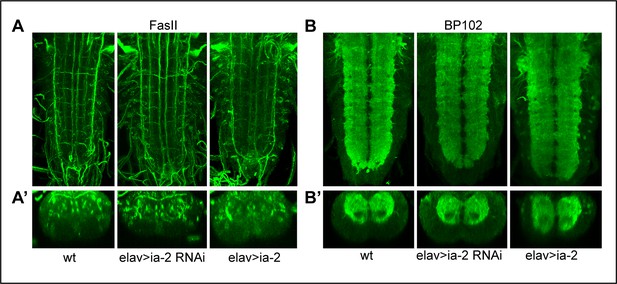
Alterations in ia-2 levels cause no obvious neuronal phenotypes.
(A) Neurons and their axonal fascicles are visualised with anti-FasII. N = 7–11 ventral nerve cords (VNCs). (B) Neurons and their dendrites are visualised with anti-BP102. N = 9–10 VNCs. No abnormal phenotypes were observed. (A and B) Horizontal views; (A’ and B’) transverse views.
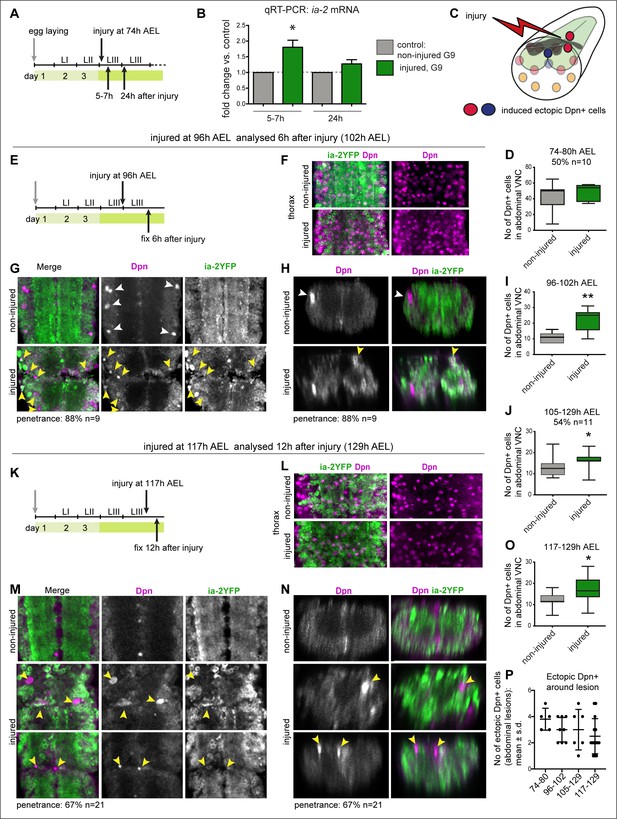
Injury induced ia-2 expression and ectopic Dpn+ cells.
(A, E, and K) Time course of crush-injury experiments in the larval abdominal ventral nerve cord (VNC), indicating the age of the larvae (after egg laying, AEL) when crush was applied (top arrows), followed by various recovery periods, and when they were dissected and fixed (bottom arrows). (C) Diagram showing that crush injury induced ectopic Dpn+ cells. (A, B, and D) Crush injury in the larval abdominal VNC at 74 hr AEL: (B) increased the levels of ia-2 mRNA at 5–7 hr post-injury, which recovered homeostatically by 24 hr, detected by qRT-PCR. N = 3 biological replicates. (D) Induced ectopic abdominal Dpn+ cells by 5–7 hr post-injury (74–80 hr AEL) in half of injured VNCs (penetrance 50%, N = 10), and increased (albeit not significantly) the total number of abdominal Dpn+ cells inthose samples. Mann–Whitney U-test, p=0.24. (F and L) Thoracic Ia-2YFP and Dpn signal were normal in thoracic neuroblasts, in samples shown in (G and H) and (M and N), respectively. (E–I) Injury at 96 hr AEL caused ectopic abdominal Dpn+ cells by 6 hr post-injury (arrowheads). Most Dpn+ cells were Ia-2YFP−. At this stage, some developmental neuroblasts could still remain (white arrowheads), but ectopic abdominal Dpn+ cells were dorsal (yellow arrowheads, H). Most injured VNCs had ectopic abdominal Dpn+ cells (88% penetrance N=9). (I) Unpaired Student's t-test p=0.0063. (J) Injury at 105 hr AEL and fixation at 129 hr AEL, when no developmental neuroblasts remain, induced a significant increase in abdominal Dpn+ cells (54.5% penetrance N = 11). Mann–Whitney U-test p=0.0375. (K–O) Injury at 117 hr AEL caused ectopic abdominal Dpn+ cells by 12 hr post-injury (129 hr AEL). . Ectopic abdominal Dpn+ cells were found in ectopic dorsal positions (yellow arrowheads, N). This stage is devoid of developmental neural stem cells. Over two thirds of injured VNCs had ectopic Dpn+ cells (67% penetrance N=21). (O) Student's t-test p=0.0302. (I,J,O) All abdominal Dpn+ cells were counted in all injured samples. (P) Temporal profile of number of ectopic Dpn+ cells surrounding the lesions, number in X-axis indicate time points of injury and fixation. (F, G, L, and M) Horizontal views, (H and N) transverse views. (D, I, J, O ) Graphs show box-plots . (P) Shows dot plots, with mean and error bars (±s.d.) indicated. *p<0.05, **p<0.01. For full genotypes and further statistical analysis details, see Supplementary file 1.
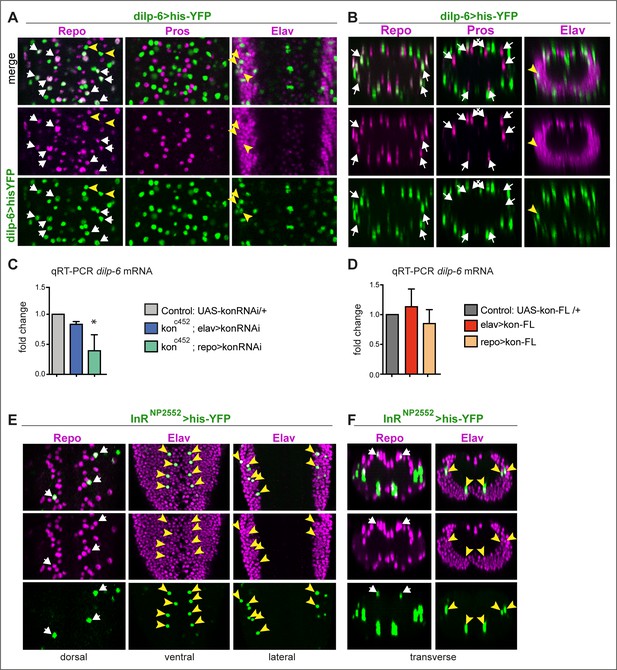
dilp-6 is expressed in neurons and cortex glia and received by neuropile glia.
(A and B) Dilp-6GAL4>UAShisYFP cells are mostly Repo+ Pros− glia that do not surround the neuropile (white arrows), and from position appear to be cortex and surface glia. No YFP+ cells have Pros. Some cells are Repo− Pros− Elav+ (yellow arrowheads) meaning they are neurons. (C and D) qRT-PCRs showing that: (C) kon knock-down in glia (konc452/UASkonRNAi; repoGAL4/+) downregulates dilp-6 mRNA levels; (D) overexpression on kon does not cause a significant effect. N = 3 replicates for both. (C and D) One-way ANOVA, only differences in (C) for dilp-6 mRNA significant p=0.0362, *p<0.05. (E and F) inR expression visualised with reporter InRNP2552GAL4> UAShistoneYFP is expressed stochastically in some dorsal Repo+ neurople glia (white arrows), and other glia, and in some Elav+ neurons (yellow arrowheads). (A and E) Horizontal views of the abdominal ventral nerve cord (VNC); (B and F) transverse views. For full genotypes and further statistical analysis details, see Supplementary file 1.
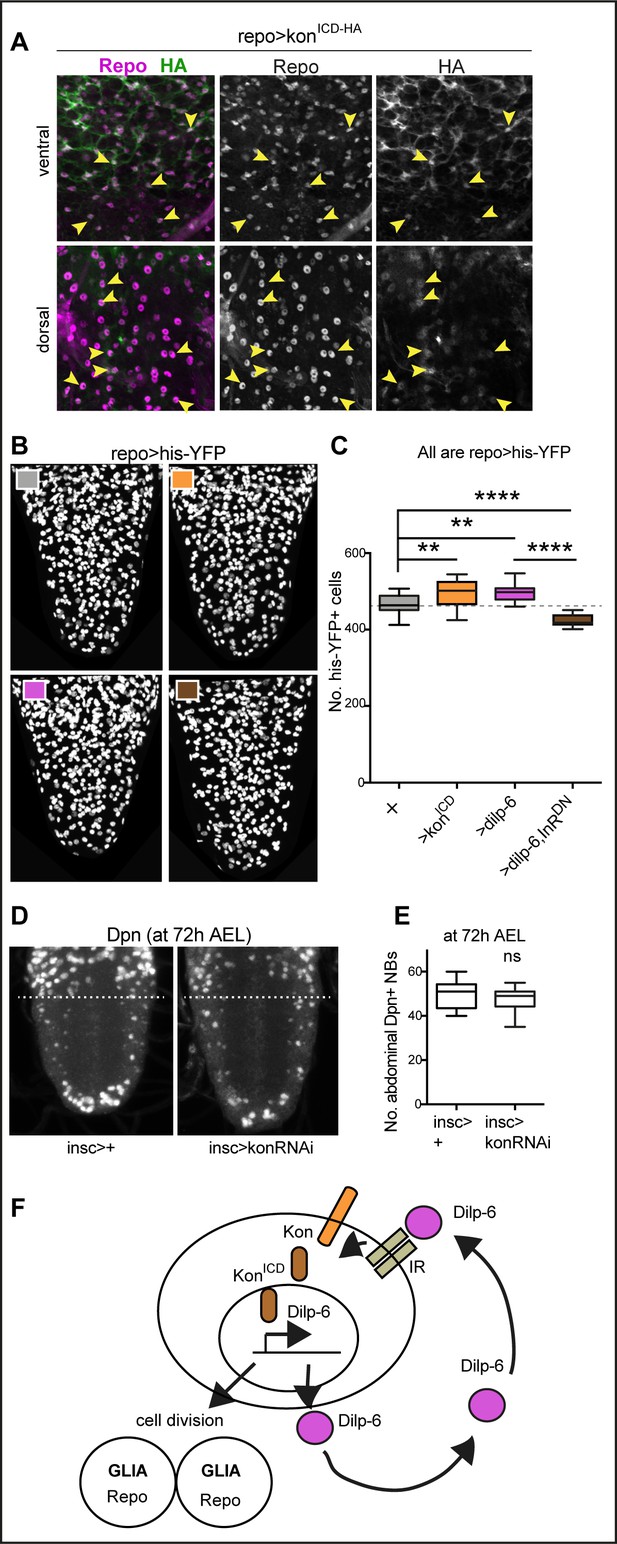
Ia-2, Kon, and Dilp-6 are linked though a neuron-glia communication loop.
(A) Overexpressed HA-tagged KonICD in glia (repoGAL4>UASkonICD::HA) visualised with anti-HA antibodies in third instar wandering larvae, localises to both glial cytoplasms and nuclei (arrows). (B and C) Overexpression of the intracellular domain of kon (konICD) or dilp-6 increased glial cell number, visualised with repoGAL4>UAShistone-YFP, and quantified automatically with DeadEasy in (C). Overexpression of a dominant negative form of the insulin receptor rescues the increase in cell number caused by Dilp-6 (repo>hisYFP, dilp-6, InRDN), meaning that autocrine InR signalling regulates glial proliferation. Box-plots. One-way ANOVA p<0.0001, post hoc Tukey’s test multiple comparisons between all samples. N = 15–28 ventral nerve cords (VNCs). (D and E) Third star larvae at 72 hr AEL to visualise abdominal developmental neuroblasts: kon-RNAi knock-down in neural stem cells with insGAL4 does not affect Dpn+ cell number. Box-plots. Unpaired Student's t-test, p=0.3111. N = 10 VNCs. (F) Illustration summarising that a positive feedback autocrine loop involving Dilp-6, InR, and Kon promotes both glial proliferation and Dilp-6 production. All images are horizontal views. Asterisks refer to multiple comparison post hoc tests, all samples vs. all: **p<0.01, ****p<0.0001. All graphs show box-plots. For full genotypes and further statistical analysis details, see Supplementary file 1.
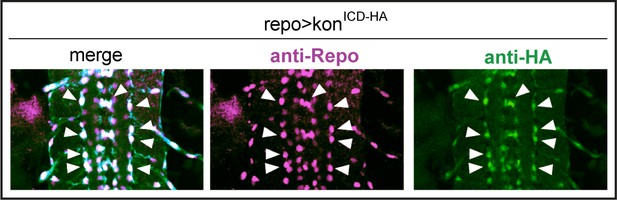
Over-expressed HA-tagged konICD localised to nuclei in embryos.
Overexpressed HA-tagged KonICD in glia (repoGAL4>UASkonICd::HA) visualised with anti-HA antibodies in stage 16 embryos localises to glial nuclei stained with the pan-glial marker anti-Repo (arrows).
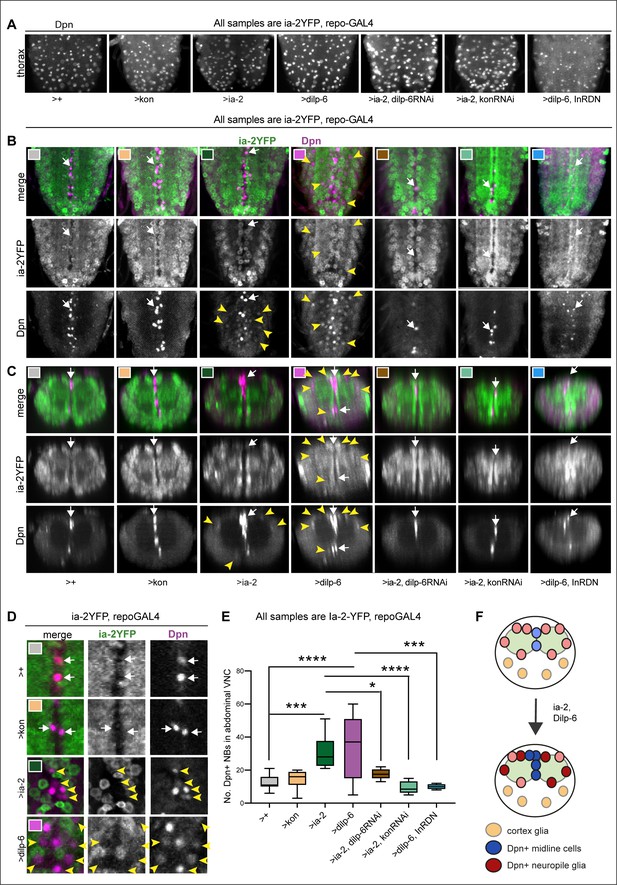
Ia-2 and Dilp-6 induce ectopic neural stem cells from InR signalling in glia.
All samples were analysed at 120 hr AEL, after disappearance of abdominal developmental neuroblasts. (A) Dpn signal in thorax was normally strong and clear, except with the overexpression of both dilp-6 and InRDN in glia, which reduced Dpn levels and NB size. (B and C) Overexpression of ia-2 and dilp-6, but not kon-full-length, induced Dpn+ cells in the abdominal ventral nerve cord (VNC), at the midline and in lateral positions. Ectopic abdominal Dpn was at lower levels than normal thoracic signal in NBs. (D) Ectopic Dpn+ cells did not express Ia-2YFP (arrowheads). (E) Overexpression of ia-2 or dilp-6 increased abdominal Dpn+ cell number. Quantification of all abdominal VNC Dpn+ cells, and genetic epistasis analysis showing that: the increase in Dpn+ cell number caused by ia-2 overexpression was rescued by dilp-6 RNAi and kon-RNAi knock-down in glia, meaning that ia-2 requires Dilp-6 and glial Kon to induce Dpn; and preventing insulin signalling with InRDN in glia rescued the increase in Dpn+ cell number caused by dilp-6 overexpression, meaning that Dilp-6 induced Dpn via InR signalling in glia. One-way ANOVA p<0.0001, post hoc Tukey’s test multiple comparisons all samples vs. all. N = 5–13 VNCs. (F) Illustration showing that Ia-2 and Dilp-6 can induce Dpn via InR signalling in glial cells. (A and B) Horizontal views; (C) transverse views; (D) higher magnification. Graphs show quantifications in box-plots. Asterisks refer to multiple comparison post hoc tests: *p<0.05, ***p<0.0001, ****p<0.0001. For full genotypes and further statistical analysis details, see Supplementary file 1.
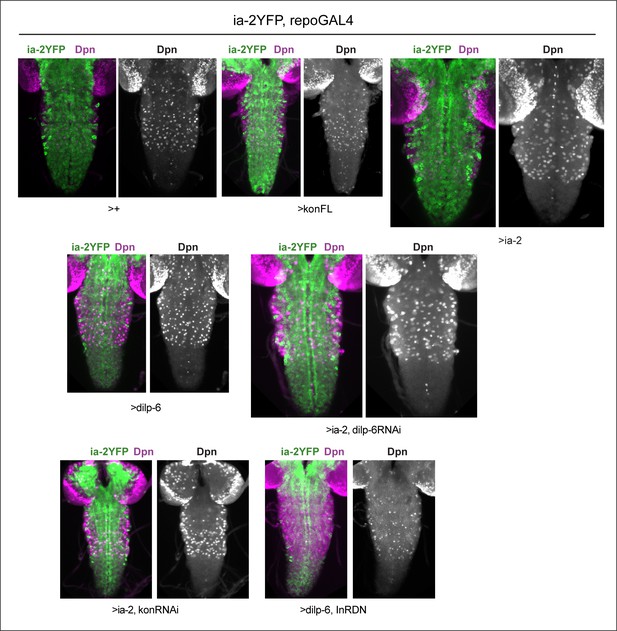
Dpn in thoracic neuroblasts of specimens shown in Figure 6.
Virtually full ventral nerve cord (VNC) projections showing that Dpn signal was robust in normal thoracic NBs and variations are genotype-specific. All samples carry Ia-2YFP (shown in merged images) and are stained with anti-Dpn, shown also in single channel images.
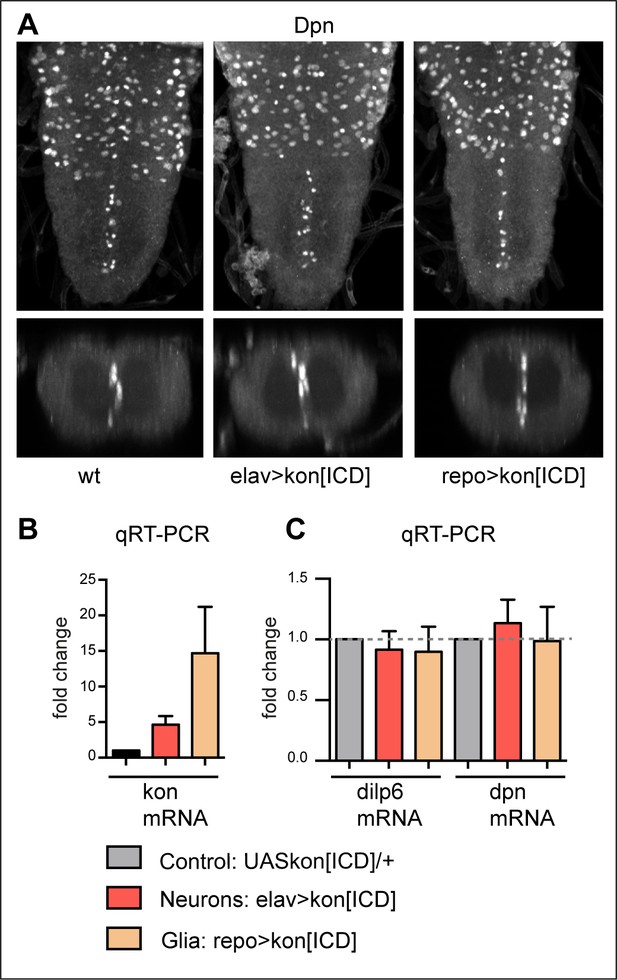
KonICD does not induce dilp-6 nor dpn expression.
(A) Overexpression of konICD in glia with repoGAL4 did not induce ectopic abdominal Dpn+ cells. N = 8. (B) Overexpressed konICD in glia with repoGAL4 was verified with qRT-PCR and resulted in virtually a 20-fold increase in konICD mRNA levels. (C) qRT-PCR data showing that overexpression of konICD in glia with repoGAL4, or in neurons with elavGAL4, did not significantly alter mRNA levels of dilp-6 nor dpn, n = 4 biological replicates.
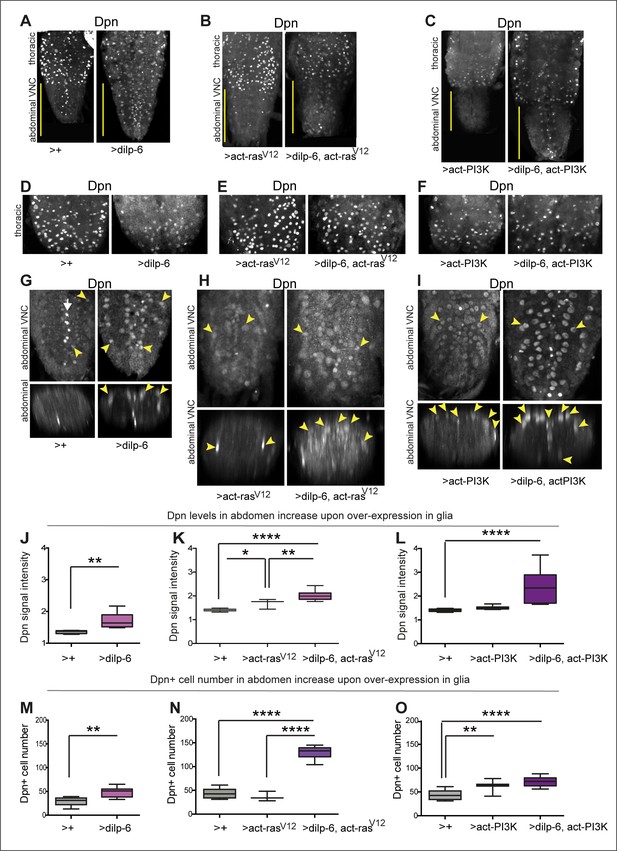
Activation of Ras or PI3K downstream of insulin signalling in glia induces ectopic Dpn+ cells.
(A–C) Full ventral nerve cord (VNC) projections of VNCs showing normal thoracic levels of Dpn, and genotype-specific modulations, using anti-Dpn from Wang Lab. (D–F) Projections of thoracic sections, showing Dpn signal in normal NBs. (G, J, and M) Overexpression of dilp-6 with repoGAL4 induced ectopic abdominal Dpn (G). Some ectopic Dpn+ cells were also observed at low levels in control samples. (J) Dilp-6 increased ectopic abdominal Dpn levels, unpaired Student's t-test; and (M) abdominal Dpn+ cell number, Mann–Whitney U-test. (H, I, K, L, N, and O) tubGAL80ts repoGAL4 was used to overexpress dilp-6 together with activated rasV12 or PI3K in glia to prevent embryonic lethality (48 hr 18°C, followed by 30°C until dissection ). (H, K, and N) Overexpression of dilp-6 with activated rasV12 induced ectopic Dpn in possibly all glia (H). Ectopic Dpn+ cells were large and neuropile integrity was disrupted. (K) Ectopic abdominal Dpn levels increased. One-way ANOVA, p<0.0001, Tukey’s multiple comparison test. (N) Abdominal Dpn+ cell number increased threefold, indicating widespread Dpn+ cell proliferation. One-way ANOVA, p<0.0001, Tukey’s multiple comparison test. (I, L, and O) Overexpression of dilp-6 with activated PI3K induced ectopic abdominal Dpn in neuropile glia only (astrocytes and midline glia). Cells and VNC were large. (L) Ectopic abdominal Dpn levels increased. One-way ANOVA, p<0.0001, Tukey’s multiple comparison test. (O) Abdominal Dpn+ cell number increased, One-Way ANOVA p<0.0001, Tukey’s multiple comparison test. Graphs show quantifications in box-plots. Asterisks refer to multiple comparison tests: *p<0.05, **p<0.01, ****p<0.0001. N=3-10 VNCs. For full genotypes, sample sizes and further statistical analysis details, see Supplementary file 1.
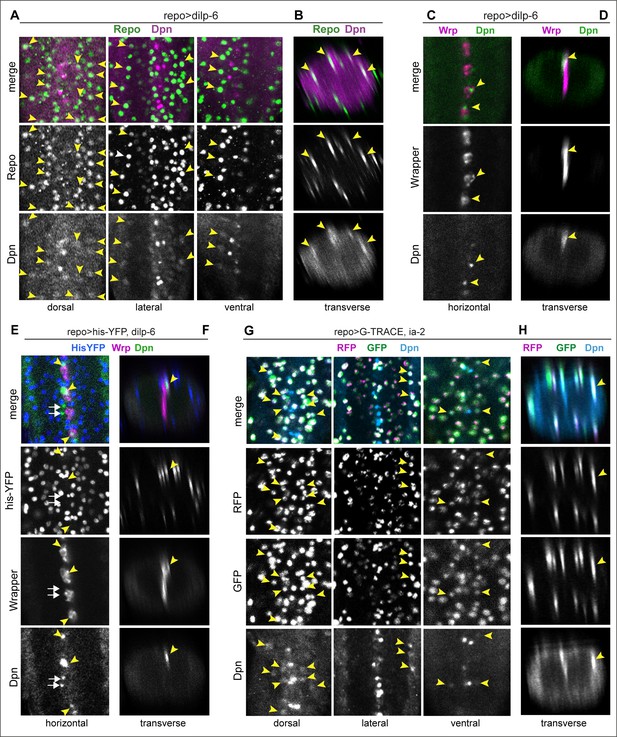
Ia-2 and Dilp-6 induced ectopic neural stem cells originate from glia.
All samples were analysed at 120 hr AEL, after disappearance of abdominal developmental neuroblasts. (A and B) Overexpression of dilp-6 from glia (repoGAL4>UAS-dilp-6) induced Dpn expression in Repo+ neuropile glial cells (arrowheads). N = 10 ventral nerve cords (VNCs). (C and D) Overexpressed dilp-6 also induced Dpn in Wrp+ midline glia (arrowheads). N = 6 VNCs. (E and F) When dilp-6 was overexpressed, and all glia except midline glia were visualised with nuclear repoGAL4>Histone-YFP and midline glia with anti-Wrp, Dpn+ YFP−Wrp− cells were found, which therefore were not glia (white arrows; yellow arrowheads point to Dpn+Wrp+ cells). N = 6 VNCs. (G and H) G-TRACE expression in glia with repoGAL4 revealed with GFP cells that were originally glia or originated from a glial cell lineage, even if they switched off the glial repo promoter, and with RFP newly generated glial cells. Dpn colocalised in neuropile glia with both GFP and RFP, meaning that Dpn+ cells originated from glia, and at that point in time these cells still retained active the glial repo promoter. N = 8 VNCs. (A, C, E, and G) Horizontal and (B, D, F, and H) transverse views. For full genotypes and sample sizes, see Supplementary file 1.
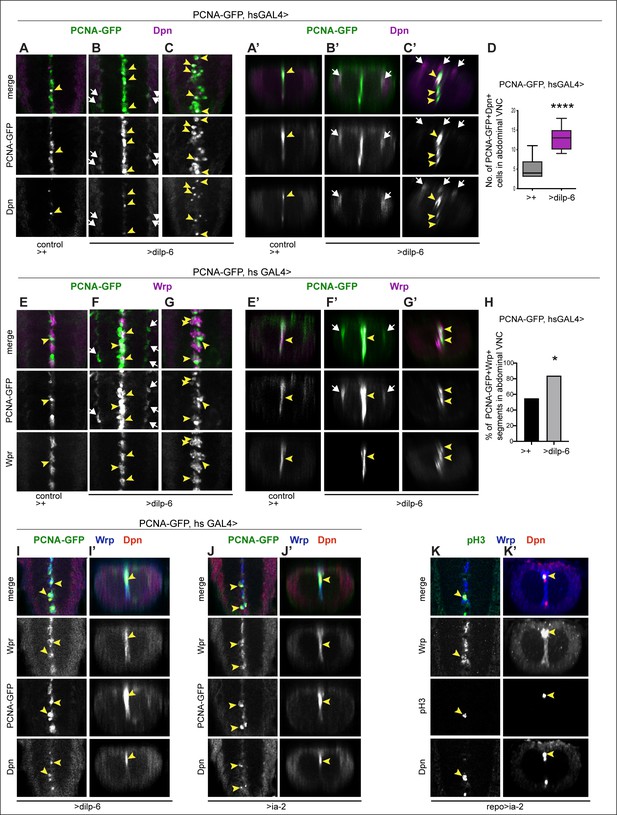
Ia-2 and Dilp-6 induced ectopic neural stem cells can divide.
All samples were analysed at 120 hr AEL, after disappearance of abdominal developmental neuroblasts. (A–C’ and E–G’) Cell proliferation was visualised with the S-phase marker PCNA-GFP, quantification in (D and H). dilp-6 expression was induced in all cells with heat-shock-GAL4, raising the temperature to 37°C for 30 min at the end of the third instar larval stage at 110.5 hr AEL, and then larvae were kept at 25°C for 9 hr, visualising Dpn+ and PCNA-GFP at 120 hr AEL. (A–C’) Overexpression of dilp-6 resulted in Dpn+ PCNA-GFP+ cells laterally around the neuropile (B and B’ white arrows) and along the midline (C and C’ yellow arrowheads), showing that these ectopic Dpn+ cells were in S-phase. Quantification box-plots in (D), Student's t-test. There were also some Dpn+ cells that were not dividing (white arrows in C’). (E–G’) Overexpression of dilp-6 resulted in PCNA-GFP+ Wrp+ midline glia (yellow arrowheads) that therefore were dividing. In (G) there is a notable increase in the number of Wrp+ cells. In (E and E’) lateral PCNA-GFP+Wrp−Dpn+ cells around the neuropile (white arrows) most likely correspond to neuropile glia. (H) Quantification showing phenotypic penetrance: percentage of segmentally repeated Wrp+ cell clusters that contain PCNAGFP+ cells. Fisher’s exact test p=0.0276. (I–J’) Overexpression of either dilp-6 or ia-2 with hsGAL4 upregulated the S-phase marker PCNA-GFP in Wrp+ Dpn+ midline cells, meaning these ectopic Dpn+ cells were dividing. Penetrance: >dilp-6 25% N = 4; >ia-2: 18% N = 11 ventral nerve cords (VNCs). (K and K’) Overexpression of ia-2 in glia with repoGAL4 induced non-autonomously proliferation of ectopic Wrp+ Dpn+ cells, visualised with the mitotic marker pH3. Penetrance: 60% N = 10 VNCs.
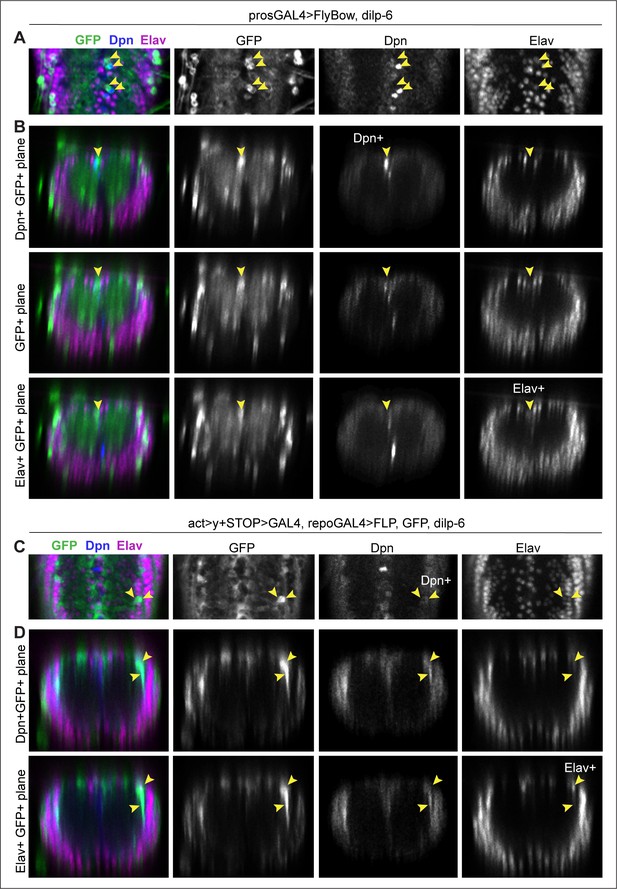
Neurons were detected from glial-derived neural stem cells.
All samples were analysed at 120 hr AEL, after disappearance of abdominal developmental neuroblasts. (A and B) prosGAL4>FlyBow can reveal expression of neural stem cells, ganglion mother cells, neurons, and glia. Overexpression of dilp-6 with prosGAL4 resulted in clusters of 3 GFP+ cells along the midline that comprised one GFP+Dpn+ neural stem cell (top row in B), a GF+Dpn−Elav− progeny cell (middle row, B) and one GFP+Elav+ progeny neuron (bottom row, B). (C and D) Progeny cells of a glial cell-lineage were visualised with GFP, expressed originally under the control of the glial repo promoter, then switched using Flipase, to the permanent actin promoter activated only in glial cells (act>y+STOP>UAS-GFP/UAS-FLP; repoGAL4/UAS-Dilp-6). Overexpression of dilp-6 resulted in clusters of two to three GFP+ cells that comprised a GFP+Dpn+ neural stem cell and two progeny GFP+Elav+ neuronal progeny cells. (A and C) Horizontal and (B and D) transverse views. For full genotypes, sample sizes, and statistical details, see Supplementary file 1.
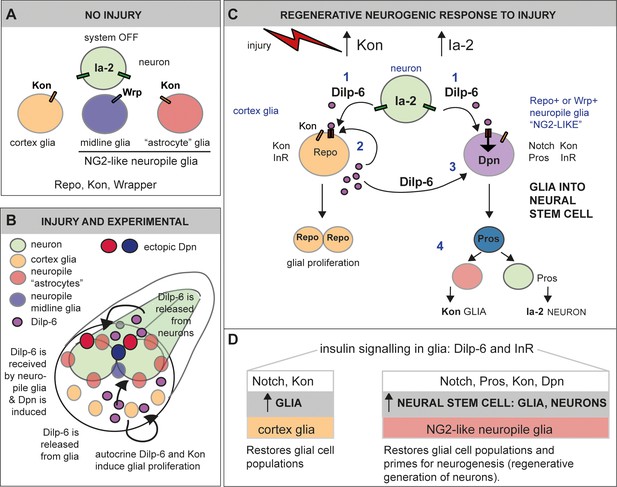
Ia-2 and Dilp-6 drive a regenerative neurogenic response to central nervous system (CNS) injury.
(A) In the abdominal larval ventral nerve cord (VNC), neurons have Ia-2, glia have Kon, and Ia-2 and Kon are mutually exclusive; non-midline glia have the transcription factor Repo and midline glia the membrane protein Wrapper. In the normal, uninjured abdominal VNC, InR is in glial cells and some neurons; Ia-2 expression is constantly present in neurons; kon is switched off, and there are no neural stem cells (neuroblasts). (B) Diagram showing that Dilp-6 can be secreted from neurons, amplified and secreted by cortex glia, and received by all glial types. Dilp-6 production and secretion depend on Kon and Ia-2, which increase in injury. (C) Injury to the abdominal VNC provokes a dramatic surge in Ia-2 and Kon. This drives the initial secretion of Dilp-6 from neurons (1). Secreted Dilp-6 binds InR in glia, and InR signalling may facilitate cleavage and activation of Kon. KonICD activates glial proliferation (2). In an autocrine Kon and InR dependent manner, Dilp-6 sets off a positive feedback loop that amplifies Dilp-6 production from cortex glia (2). Once secreted, Dilp-6 and InR signalling cause the upregulation of Dpn+ in neuropile glia – including Notch+ Pros+ lateral (astrocytes) and Wrp+ midline glia (3). Neuropile glia can stochastically switch on Dpn. Glial-derived Dpn+ neural stem cells can divide and generate new neurons – although to a rather limited extent (4). After cell division, Kon may determine whether daughter cells become glia, to the exclusion of Ia-2. (D) Insulin signalling involving Ia-2, Dilp-6, and InR can increase cell number of various glial cell types – including cortex glia, neuropile astrocytes and midline glia – induce neural stem cells, and potentially generate new neurons. The neurogenic potential of glia may depend on the availability of Notch and Pros and the downregulation of Kon. Together, these genes can potentially induce neurogenesis and gliogenesis, matching cell populations for regeneration.
Additional files
-
Supplementary file 1
Genotypes, sample sizes, and statistical analysis details.
This table contains full genotypes for all experiments, sample sizes used, and statistical analysis details including normality tests, tests applied, and multiple comparison correction tests.
- https://cdn.elifesciences.org/articles/58756/elife-58756-supp1-v2.xls
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58756/elife-58756-transrepform-v2.docx





