SWELL1 regulates skeletal muscle cell size, intracellular signaling, adiposity and glucose metabolism
Figures
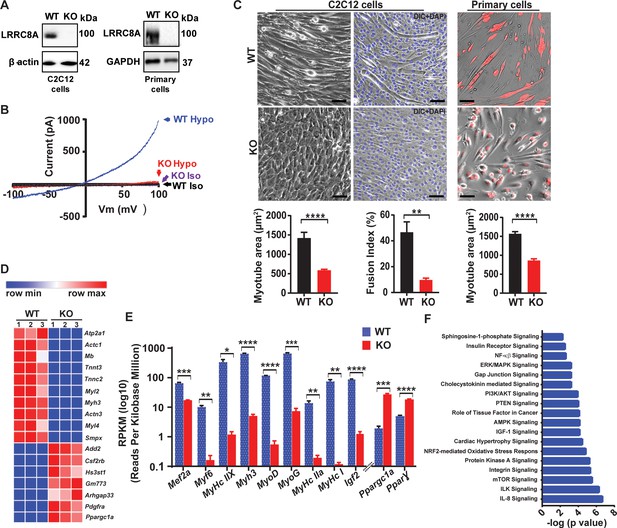
Skeletal muscle LRRC8A is required for myotube formation and regulates multiple myogenic signaling pathways.
(A) Western blots from WT and Lrrc8a KO C2C12 (KO) (left) and primary myotubes (right). (B) Current-voltage curves from WT and Lrrc8a KO C2C12 myoblast measured during a voltage-ramp from −100 to +100 mV +/- isotonic and hypotonic (210 mOsm) solution. (C) Bright field merged with fluorescence images of differentiated WT and Lrrc8a KO C2C12 myotubes (left, middle) and skeletal muscle primary cells (right). DAPI stains nuclei blue (middle). Red is mCherry reporter fluorescence from adenoviral transduction. Scale bar: 100 µm. Mean myotube surface area measured from WT (n = 21) and Lrrc8a KO (n = 21) C2C12 myotubes (left), and WT (n = 22) and Lrrc8a KO (n = 15) primary skeletal myotubes (right). Fusion index (%multinucleated cells) measured from WT (n = 5 fields) and Lrrc8a KO (n = 5 fields) C2C12 (shown below the representative image). (D) Heatmap of top 17 differentially expressed genes in WT versus Lrrc8a KO C2C12 myotubes derived from RNA sequencing. (E) Reads Per Kilobase Million for select myogenic differentiation genes (n = 3, each). (F) IPA canonical pathway analysis of genes significantly regulated in Lrrc8a KO C2C12 myotubes in comparison to WT. n = 3 for each group. For analysis with IPA, FPKM cutoffs of 1.5, fold change of ≥1.5, and false discovery rate <0.05 were utilized for significantly differentially regulated genes. Statistical significance between the indicated values were calculated using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. n = 3, independent experiments.
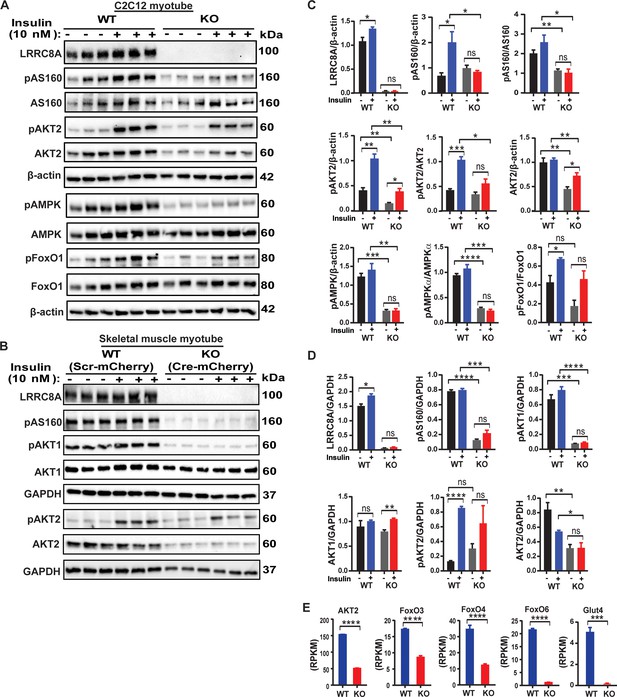
LRRC8A regulates multiple insulin dependent signaling pathways in skeletal myotubes.
(A) Western blots of LRRC8A, pAKT2, AKT2, pAS160, AS160, pAMPK, AMPK, pFoxO1, FoxO1 and β-actin in WT and Lrrc8a KO C2C12 myotubes upon insulin-stimulation (10 nM). (B) Western blots of LRRC8A, pAS160, pAKT1, AKT1, pAKT2, AKT2 and GAPDH in WT (Ad-CMV-mCherry) and Lrrc8a KO (Ad-CMV-Cre-mCherry) primary skeletal muscle myotubes following insulin-stimulation (10 nM). (C and D) Densitometric quantification of proteins depicted on western blots normalized to corresponding β-actin and GAPDH respectively. (E) Gene expression analysis of insulin signaling associated genes AKT2, FOXO3, FOXO4, FOXO6 and GLUT4 in WT and Lrrc8a KO C2C12 myotubes. Statistical significance between the indicated values were calculated using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. n = 3, independent experiments.
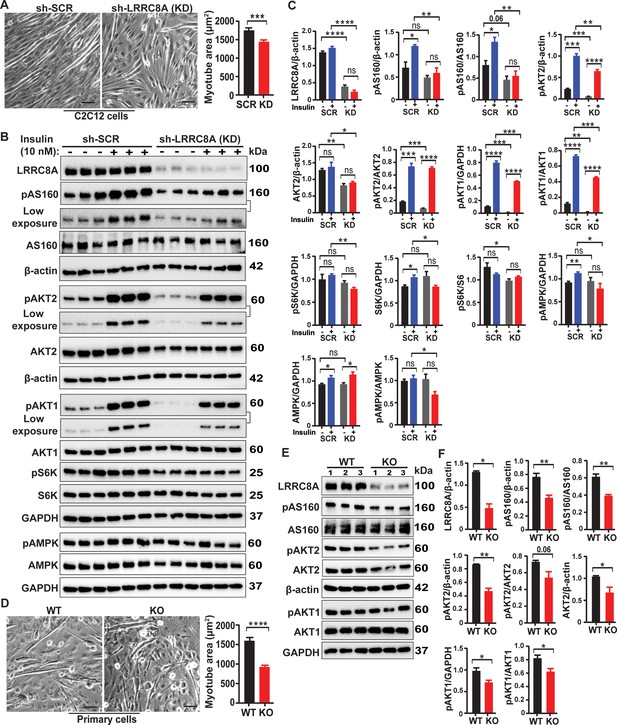
LRRC8A regulates myotube size and insulin dependent signaling pathways in differentiated skeletal myotubes.
(A) C2C12 myoblasts were differentiated for 6 days, transduced with either Ad5-shSCR-mCherry (sh-SCR) or Ad5-shLRRC8A-mCherry (KD), and then analyzed after 3 days. Bright field images of differentiated sh-SCR and shLRRC8A (KD) treated C2C12 myotubes. Mean myotube surface area measured from C2C12-sh-SCR (n = 78) and C2C12-shLRRC8A (n = 95) myotubes (right). Scale bar: 100 µm. (B) Western blots of LRRC8A, pAS160, AS160, pAKT2, AKT2, pAKT1, AKT1, pS6K, S6K, pAMPK, AMPK, β-actin and GAPDH in WT C2C12-sh-SCR and LRRC8A KD myotubes upon insulin-stimulation (10 nM). (C) Densitometric quantification of proteins depicted on western blots normalized to corresponding β-actin and GAPDH, respectively. For pAS160, pAKT2 and pAKT1 lower exposure of western blots were used for quantification. (D) Primary skeletal muscle cells differentiated 3 days and subsequently transduced with Ad-CMV-mCherry (WT) and Ad-CMV-Cre-mCherry (Lrrc8a KO). Bright field images of differentiated WT and Lrrc8a KO primary skeletal muscle myotubes. Mean myotube surface area measured from WT (n = 73) and Lrrc8a KO (n = 83) primary skeletal muscle myotubes (right). Scale bar: 100 µm. (E) Western blots of LRRC8A, pAS160, AS160, pAKT2, AKT2, pAKT1, AKT1, β-actin and GAPDH in WT and Lrrc8a KO primary skeletal muscle myotubes measured under basal culture conditions. (F) Densitometric quantification of proteins depicted on western blots normalized to corresponding β-actin and GAPDH respectively. Statistical significance between the indicated values were calculated using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. n = 3, independent experiments.
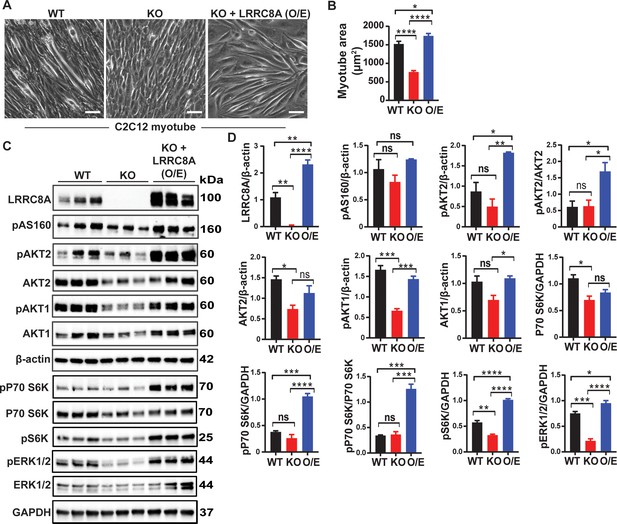
LRRC8A over-expression in Lrrc8a KO C2C12 myotubes rescues myogenic differentiation and augments intracellular signaling.
(A) Bright-field image of differentiated WT, Lrrc8a KO (KO) and Lrrc8a KO + LRRC8A O/E C2C12 myotubes. (A) Quantification of mean myotube surface areas in WT (n = 35), Lrrc8a KO C2C12 (n = 26) and Lrrc8a KO + LRRC8A O/E C2C12 (n = 45) cells. Scale bar: 100 µm. (C) Western blots of LRRC8A, AKT2, pAKT2, pAS160, pAKT1, AKT1, pP70S6K, P70S6K, pS6K, pERK1/2, ERK1/2, β-actin and GAPDH from WT, Lrrc8a KO and Lrrc8a KO + LRRC8A O/E C2C12 myotubes. (D) Densitometric quantification of proteins depicted on western blots normalized to β-actin and GAPDH, respectively. Statistical significance between the indicated group were calculated with one-way ANOVA, Tukey's multiple comparisons test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. n = 3, independent experiments.
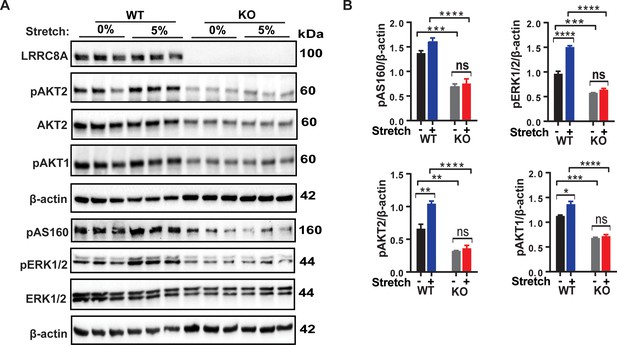
LRRC8A is required for intact stretch-induced PI3K-pAKT2-pAS160-MAPK signaling in C2C12 myotubes.
(A) Western blot of LRRC8A, AKT2, pAKT2, pAKT1, pAS160, pERK1/2, ERK1/2 and β-actin in WT and Lrrc8a KO myotube in response to 15 min of 0% and 5% static stretch. (B) Densitometric quantification of each signaling protein relative to β-actin. Statistical significance between the indicated group calculated with one-way Anova, Tukey's multiple comparisons test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. n = 3, independent experiments.
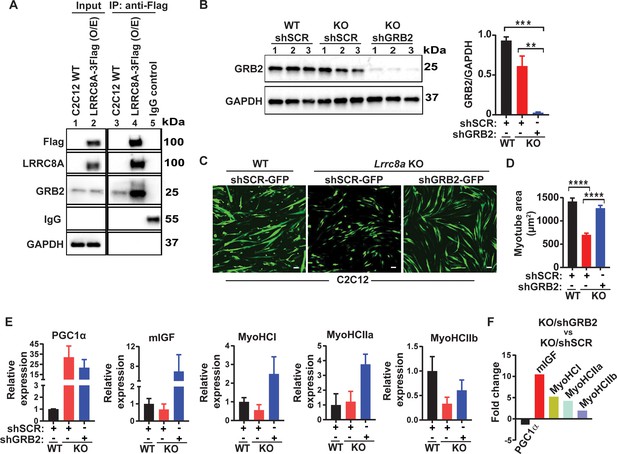
LRRC8A interacts with GRB2 in C2C12 myotubes and regulates myogenic differentiation.
(A) LRRC8A-3xFlag over expressed in C2C12 cells followed by immunoprecipitation (IP) with Flag antibody. Western blot of Flag, LRRC8A, GRB2 and GAPDH. IgG serves as a negative control. (B) Western blot of GRB2 to validate GRB2 knock down efficiency in Lrrc8a KO/GRB2 knock-down (Ad-shGRB2-GFP) compared to WT C2C12 (Ad-shSCR-GFP) and Lrrc8a KO (Ad-shSCR-GFP). Densitometric quantification of GRB2 knock-down relative to GAPDH (right). (C) Fluorescence image of WT C2C12/shSCR-GFP, Lrrc8a KO/shSCR-GFP and Lrrc8a KO/shGRB2-GFP myotubes. Scale bar: 100 µm. (D) Quantification of mean myotube area of WT C2C12/shSCR-GFP (n = 25), Lrrc8a KO/shSCR-GFP (n = 28) and Lrrc8a KO/shGRB2-GFP (n = 24). (E) Relative mRNA expression of selected myogenic differentiation genes in Lrrc8a KO/shSCR and Lrrc8a KO/shGRB2 compared to WT C2C12/shSCR (n = 3 each), and of Lrrc8a KO/shGRB2 compared to Lrrc8a KO/shSCR (F), fold change of mRNA’s in KO shGRB2 relative to KO cells with preserved GRB2 expression. Statistical significance between the indicated group were calculated with one-way ANOVA, Tukey's multiple comparisons test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. n = 3, independent experiments.
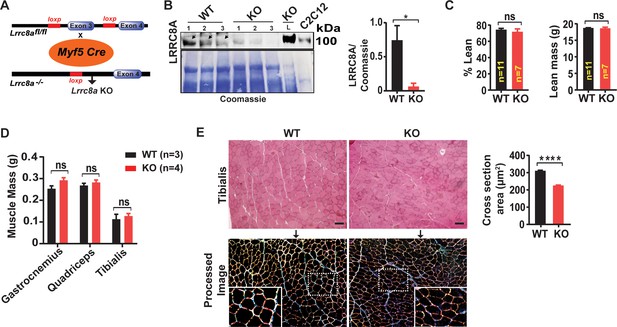
Skeletal muscle-targeted Lrrc8a KO mice develop smaller myofibers but normal muscle mass.
(A) Schematic representation of Cre-mediated recombination of loxP sites flanking Exon three using muscle-specific Myf5-Cre mice to generate skeletal muscle targeted Lrrc8a KO mice. (B) Western blot of gastrocnemius muscle protein isolated from of WT and Myf5Cre/Lrrc8afl/fl (KO) mice. Liver sample from KO and C2C12 cell lysates used as a positive control for LRRC8A. Coomassie gel, below, serves as loading control for skeletal muscle protein. Densitometric quantification for LRRC8A deletion in skeletal muscle of KO mice (n = 3) compared to WT (n = 3; Lrrc8afl/fl) (right). (C) NMR measurement of lean mass (%) and absolute fat mass of WT (n = 11) and KO (n = 7) mice. (D) Absolute muscle mass of muscle groups freshly isolated from WT (n = 3) and KO (n = 4). (E) Hematoxylin and eosin staining of tibialis muscle of WT and KO mice fed on regular chow diet for 28 weeks (above). Scale bar: 100 µm. Below, ImageJ converted image highlights distinct surface boundaries of myotubes. Inset, enlarged image shows smaller fiber size in KO muscle tissue. Quantification of average cross-sectional area of muscle fiber of WT (n = 300) and KO (n = 300) mice from 10 to 12 different view field images (right). Statistical significance between the indicated values were calculated using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001.
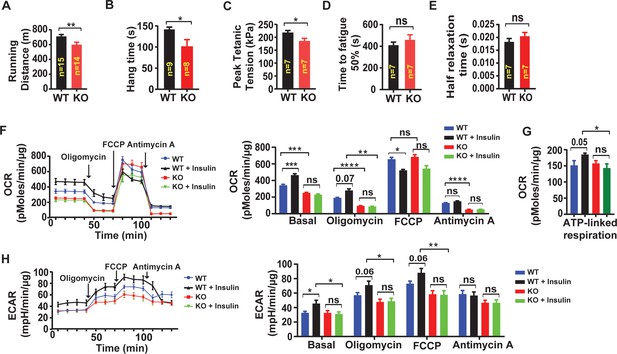
Skeletal muscle-targeted LRRC8A deletion impairs muscle endurance, force generation and insulin-stimulated oxygen consumption.
(A) Exercise treadmill tolerance test for KO mice (n = 14) compared to WT littermates (n = 15). (B) Hang times on inversion testing of KO (n = 8) and WT (n = 9) mice. (C–E) Ex vivo isometric peak tetanic tension (C), time to fatigue (D) and half relaxation time (E) of isolated soleus muscle from KO (n = 7) compared to WT (n = 7) mice. (F) Oxygen consumption rate (OCR) in WT and Lrrc8a KO primary myotubes +/- insulin stimulation (10 nM) (n = 6 independent experiments) and quantification of basal OCR, OCR post-Oligomycin, OCR post-FCCP and OCR post-Antimycin A. (G) ATP‐linked respiration obtained by subtracting the OCR after oligomycin from baseline cellular OCR. (H) Extracellular acidification rate (ECAR) in WT and Lrrc8a KO primary myotubes +/- insulin stimulation (10 nM) (n = 6 independent experiments) and quantification of basal OCR, OCR-post Oligomycin, OCR-post FCCP and OCR post-Antimycin A. Statistical significance between the indicated values were calculated using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001.
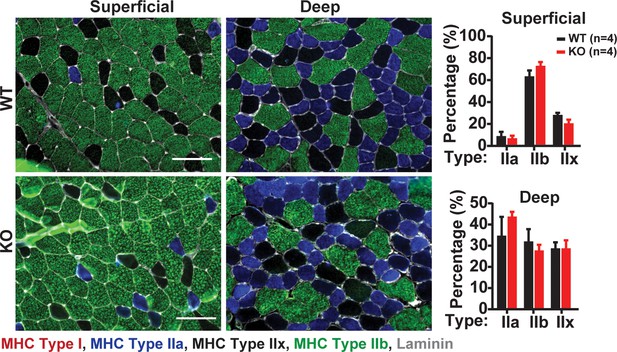
Quantification of skeletal muscle fiber type in WT and Skm KO TA muscle.
TA muscles of WT and Skm KO were sectioned and immunostained with myosin heavy chains (MHC) isoforms Type I (red), Type IIa (blue), Type IIx (black), Type IIb (green) and Laminin (Gray). Scale bar: 100 µm. Mean percentage of fibers expressing MHC Type I, Type IIa, Type IIx, Type IIb and Laminin relative to total number of fibers in a TA muscle sections of WT (n = 4) and Skm KO (n = 4) mice (right).
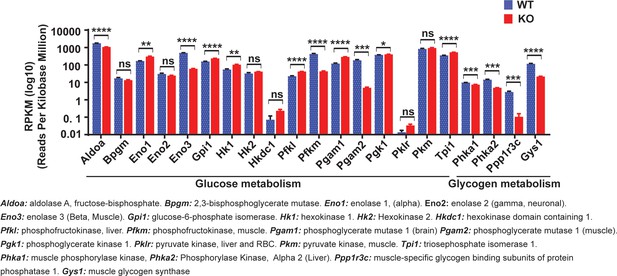
Lrrc8a ablation in C2C12 myotubes downregulates glucose and glycogen-associated genes.
Differentially expressed glucose and glycogen metabolism-associated gene after RNA-seq analysis of C2C12 WT and Lrrc8a KO myotube (n = 3, each). Statistical significance between the indicated values were calculated using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001.
Exercise Treadmill Testing of WT and Myf5-Cre/ SWELL1flflmice.
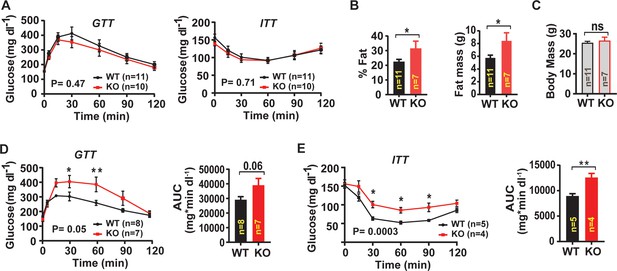
Skeletal muscle-targeted Lrrc8a ablation increases adiposity and induces glucose intolerance with overnutrition.
(A) Glucose and insulin tolerance tests of mice raised on chow diet of WT (n = 11) and KO (n = 10) mice. (B) NMR measurement of fat mass (%) and absolute fat mass of WT (n = 11) and KO (n = 7) mice. (C) Body mass of WT (n = 11) and KO (n = 7) mice on regular chow diet. (D) Glucose tolerance test of WT (n = 8) and KO (n = 7) mice fed HFD for 16 weeks after 14 weeks of age. Corresponding area under the curve (AUC) for glucose tolerance for WT and KO mice. (E) Insulin tolerance tests of WT (n = 5) and KO (n = 4) mice fed HFD for 18 weeks after 14 weeks of age. Corresponding area under the curve (AUC) for insulin tolerance for WT and KO mice. Statistical significance test between the indicated group B, C, D and E (AUC) were calculated by using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. Two-way ANOVA was used for A, D and E (p-value in bottom corner of graph). Error bars represent mean ± s.e.m. *, p<0.05, **, p<0.01, ***, p<0.001.
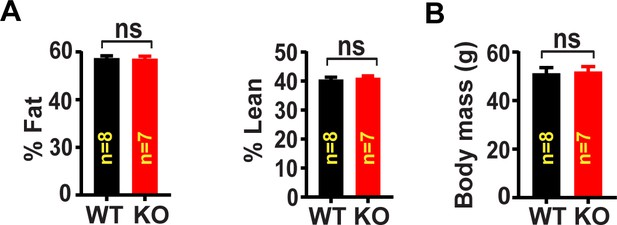
Body composition of skeletal muscle specific Lrrc8a KO mice raised on a high-fat-diet (HFD).
(A) NMR measurement of fat mass (%) and lean mass (%) of WT (n = 8) and KO (n = 7) mice raised on HFD (16 weeks) after 14 weeks of age. (B) Body mass of WT (n = 8) and KO (n = 7) mice.
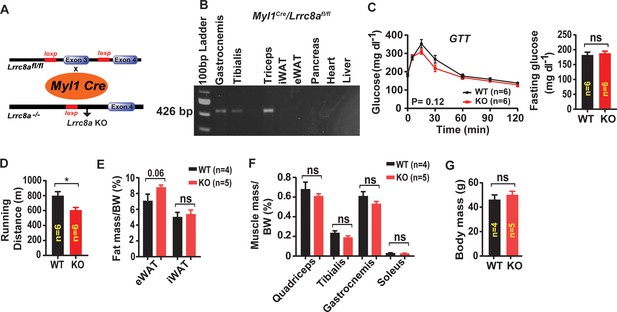
Skeletal muscle-targeted Lrrc8a ablation impairs muscle endurance and induces adiposity.
(A) Schematic representation of Cre-mediated recombination of loxP sites flanking Exon three using muscle-specific Myl1-Cre mice to generate skeletal muscle-targeted Lrrc8a KO mice (Myl1Cre/Lrrc8a fl/fl; KO) (B) PCR band of LRRC8A recombination in Myl1Cre/Lrrc8a fl/fl mice from isolated tissues. (C) Glucose tolerance test of WT (n = 6) and KO (n = 6) mice raised on chow food diet for 14 weeks. Fasting glucose level for WT and KO mice (right). (D) Exercise treadmill tolerance test for KO (n = 6) compared to WT (n = 6) littermates. (E) Epididymal (eWAT) and inguinal (iWAT) fat mass normalized to body mass (BM) isolated from KO (n = 5) and WT (n = 4) mice. (F) Skeletal muscle mass normalized to body mass (BM) isolated from KO (n = 5) and WT (n = 4) mice. (G) Body mass of KO (n = 5) and WT (n = 4) mice raised on regular chow diet. Statistical significance between the indicated values were calculated using a two-tailed Student’s t-test. Error bars represent mean ± s.e.m. *, p<0.05.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Lrrc8a (Lrrc8afl/fl) | This paper | Sah lab | SWELL1 is a regulator of adipocyte size, insulin signaling and glucose homeostasis (Zhang et al., 2017) |
| Strain, strain background (Mus musculus) | Myf5Cre | Jackson lab | JAX# 007893, RRID:IMSR_JAX:007893 | |
| Strain, strain background (Mus musculus) | Myl1Cre | Jackson lab | JAX# 24713, RRID:IMSR_JAX:024713 | |
| Cell line (Mus musculus) | C2C12 | ATCC | CRL-1772, RRID:CVCL_0188 | |
| Biological sample (Mus musculus) | Skeletal muscle primary cell | Lrrc8afl/fl | Freshly isolated from Lrrc8afl/flmice | |
| Antibody | Anti-β-actin (Rabbit monoclonal) | Cell signalling | Cat#8457 s, RRID:AB_10950489 | WB (1:2000) |
| Antibody | Anti-p-AKT1 (Rabbit monoclonal) | Cell signalling | Cat#9018 s, RRID:AB_2629283 | WB (1:1000) |
| Antibody | Anti- Akt1 (Rabbit monoclonal) | Cell signalling | Cat#2938 s, RRID:AB_915788 | WB (1:1000) |
| Antibody | Anti- pAKT2 (Rabbit monoclonal) | Cell signalling | Cat#8599 s, RRID:AB_2630347 | WB (1:1000) |
| Antibody | Anti- Akt2 (Rabbit monoclonal) | Cell signalling | Cat#3063 s, RRID:AB_2225186 | WB (1:1000) |
| Antibody | Anti- pAS160 (Rabbit polyclonal) | Cell signalling | Cat#4288 s, RRID:AB_10545274 | WB (1:1000) |
| Antibody | Anti- AS160 (Rabbit monoclonal) | Cell signalling | Cat#2670 s, RRID:AB_2199375 | WB (1:1000) |
| Antibody | Anti- AMPKα (Rabbit monoclonal) | Cell signalling | Cat#5831 s, RRID:AB_10622186 | WB (1:1000) |
| Antibody | Anti-pAMPKα (Rabbit monoclonal) | Cell signalling | Cat#2535 s, RRID:AB_2106495 | WB (1:1000) |
| Antibody | Anti-FoxO1 (Rabbit monoclonal) | Cell signalling | Cat#2880 s, RRID:AB_2106495 | WB (1:1000) |
| Antibody | Anti-pFoxO1 (Rabbit polyclonal) | Cell signalling | Cat#9464 s, RRID:AB_329842 | WB (1:1000) |
| Antibody | Anti- p70 S6 Kinase (Rabbit polyclonal) | Cell signalling | Cat#9202 s, RRID:AB_331676 | WB (1:1000) |
| Antibody | Anti- p-p70 S6 Kinase (Rabbit polyclonal) | Cell signalling | Cat#9205 s, RRID:AB_330944 | WB (1:1000) |
| Antibody | Anti-pS6 Ribosomal (Rabbit monoclonal) | Cell signalling | Cat#5364 s, RRID:AB_10694233 | WB (1:2000) |
| Antibody | Anti-S6 Ribosomal (Mouse monoclonal) | Cell signalling | Cat#2317 s, RRID:AB_2238583 | WB (1:1000) |
| Antibody | Anti- GAPDH (Rabbit monoclonal) | Cell signalling | Cat#5174 s, RRID:AB_1062202 | WB (1:2000) |
| Antibody | Anti-pErk1/2 (Rabbit polyclonal) | Cell signalling | Cat#9101 s, RRID:AB_331772 | WB (1:1000) |
| Antibody | Anti-Erk1/2 (Rabbit polyclonal) | Cell signalling | Cat#9102 s, RRID:AB_330744 | WB (1:1000) |
| Antibody | Anti- Grb2 (Mouse monoclonal) | BD | Cat#610111 s, RRID:AB_397517 | WB (1:1000) |
| Antibody | Anti-flag (Mouse monoclonal) | Sigma-Aldrich | Cat#F3165, RRID:AB_259529 | WB (1:2000) |
| Antibody | Anti-LRRC8A (Rabbit polyclonal) | Pacific antibodies | Custom made | Epitope: QRTKSRIEQGIVDRSE, WB (1:1000), SWELL1 is a glucose sensor regulating β-cell excitability and systemic glycaemia (Kang et al., 2018) |
| Antibody | Anti-BA‐F8 (Mouse monoclonal) | Developmental Studies Hybridoma Bank, Iowa City | Cat#AB_10572253, RRID:AB_10572253 | IF (1:100) |
| Antibody | Anti-SC‐71 (Mouse monoclonal) | Developmental Studies Hybridoma Bank, Iowa City | Cat#AB_2147165, RRID:AB_2147165 | IF (1:100) |
| Antibody | Anti-BF‐F3 (Mouse monoclonal) | Developmental Studies Hybridoma Bank, Iowa City | Cat#AB_2266724, RRID:AB_2266724 | IF (1:100) |
| Antibody | Anti-laminin (Rabbit polyclonal) | Abcam | Cat# ab11575, RRID:AB_298179 | IF (1:100) |
| Antibody | Anti-IgG (Normal mouse IgG) | Santa Cruz | Cat# sc-2027, RRID:AB_737197 | WB (1:1000) |
| Antibody | Anti-rabbit-HRP | BioRad | Cat# 170–6515, RRID:AB_11125142 | WB (1:10000) |
| Antibody | Anti-mouse-HRP | BioRad | Cat# 170–5047, RRID:AB_11125753 | WB (1:10000) |
| Recombinant DNA reagent | (Ad5-CMV-mCherry) | University of Iowa viral vector core facility | Ad3518 | |
| Recombinant DNA reagent | Ad5-CMV-Cre-mCherry | University of Iowa viral vector core facility | Ad3494 | |
| Recombinant DNA reagent | Ad5-CAG-LoxP-stop-LoxP-3XFlag- LRRC8A | Vector biolabs | 20180313T#1 | |
| Recombinant DNA reagent | Ad5-U6-m-shGRB2-GFP | Vector biolabs | shADV-260737 | |
| Recombinant DNA reagent | Ad5-U6-shSCR-GFP | Vector biolabs | 1122N | |
| Recombinant DNA reagent | Ad5-CMVmCherry-U6-hLRRC8A-shRNA | Vector biolabs | AD3535 | |
| Recombinant DNA reagent | Ad5-U6-scramble-mCherry | Vector biolabs | 3086 | |
| Commercial assay or kit | RNA isolation (PureLinkRNA mini kit) | Invitrogen | 12183018A | |
| Chemical compound, drug | Polybrene (Hexadimethrine bromide) | Sigma Aldrich | H9268 | (4 µg/ml) |
| Software, algorithm | GraphPad Prism8 | La Jolla California USA, www.graphpad.com’ RRID:SCR_002798 | ||
| Software, algorithm | Fiji, ImageJ | Schindelin et al., 2012 (PMID:22743772) | RRID:SCR_002285 | |
| Other | DAPI stain | Invitrogen | D1306 | (1 µg/ml) |
Additional files
-
Supplementary file 1
RNA sequencing data.
- https://cdn.elifesciences.org/articles/58941/elife-58941-supp1-v3.xlsx
-
Supplementary file 2
IPA canonical pathway analysis.
- https://cdn.elifesciences.org/articles/58941/elife-58941-supp2-v3.xlsx
-
Supplementary file 3
Genotypes fromMyf5-Cre x SWELL1flflbreeding.
- https://cdn.elifesciences.org/articles/58941/elife-58941-supp3-v3.docx
-
Supplementary file 4
Primers used for qRT-PCR of muscle differentiation gene.
- https://cdn.elifesciences.org/articles/58941/elife-58941-supp4-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58941/elife-58941-transrepform-v3.docx





