Cytomegalovirus restricts ICOSL expression on antigen-presenting cells disabling T cell co-stimulation and contributing to immune evasion
Figures
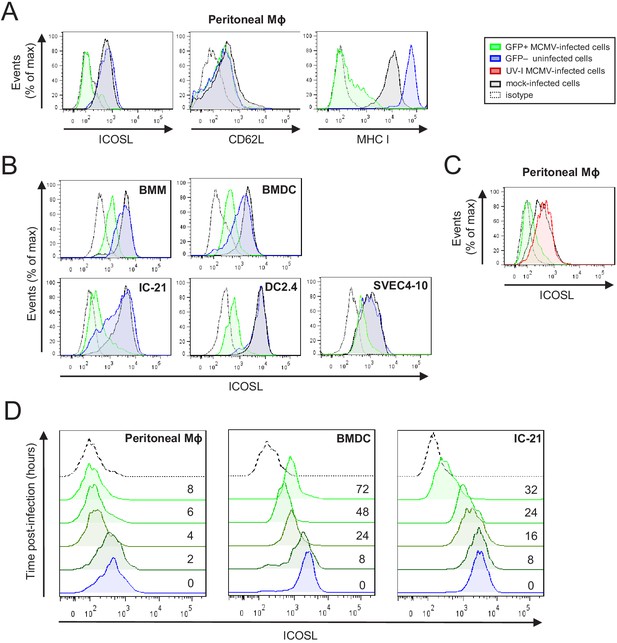
Downregulation of ICOSL upon MCMV infection of antigen-presenting cells (APCs).
(A) Peritoneal macrophages (MΦ) were mock-infected or infected for 72 hr with MCMV-GFP at an moi of 5 and analyzed by flow cytometry for surface expression of ICOSL, MHC I, or CD62L using specific mAbs against each of these molecules. Gray histograms represent the expression on mock-infected cells, green histograms represent the expression on MCMV-infected (GFP+) cells, and blue histograms represent the expression on uninfected (GFP-) cells from the same culture. (B) Bone-marrow-derived macrophages (BMM), bone-marrow-derived dendritic cells (BMDC), IC-21, DC2.4, and SVEC4-10 cells were infected with MCMV-GFP at different moi (10, 20, 20, and 5, respectively) to obtain an infection of around 50% of the cell culture, and analyzed by flow cytometry for surface expression of ICOSL. Gray, green, and blue histograms, as indicated in A. (C) Same as in A, except that peritoneal macrophages were infected with MCMV-GFP at an moi of 10, or treated for 72 hr with the same amount of MCMV-GFP UV-inactivated. Gray and green as indicated in A, and red histograms represent the expression on MCMV-GFP UV-inactivated infected cells. (D) Peritoneal macrophages, BMDCs, and IC-21 cells were mock-infected (time 0) or infected with MCMV-GFP at an moi of 10, 40, and 20, respectively, and analyzed by flow cytometry for surface expression of ICOSL at the different time points after infection indicated. In A, B, C, and D, the isotype for each antibody was used as a negative control (dotted lines). Data are representative of at least two independent experiments.
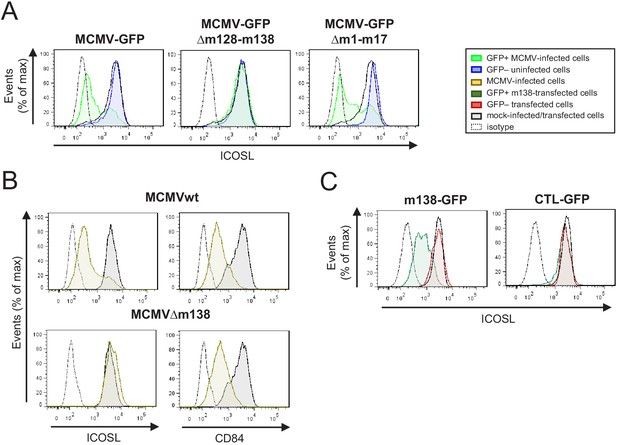
Identification of the MCMV gene involved in the decreased cell surface expression of ICOSL.
(A) IC-21 cells were mock-infected or infected for 72 hr with MCMV-GFP, MCMV-GFPΔm128-m138, or MCMV-GFPΔm1-m17 at an moi of 10 and examined by flow cytometry for surface expression of ICOSL. Gray histograms represent the expression of mock-infected cells, green histograms represent the expression on MCMV-infected (GFP+) cells, and blue histograms represent the expression on uninfected (GFP-) cells from the same culture. (B) IC-21 cells were mock-infected or infected with MCMVwt or MCMVΔm138 for 72 hr at an moi of 20 and analyzed by flow cytometry for surface expression of ICOSL and CD84. Gray histograms as indicated in A, and brown histograms represent the expression on MCMVwt and MCMV∆m138 IC-21-infected cells. (C) IC-21 cells were mock-transfected or transfected with the m138-GFP construct or with the control GFP empty vector and stained with the anti-ICOSL mAb or an isotype control (dotted lines). Dark green and red histograms represent cells from transfected cultures expressing or not expressing GFP, respectively, and gray histograms represent surface expression of ICOSL on untransfected cells. In A, B, and C, the isotype for each antibody was used as a negative control (dotted lines). Data are representative of at least two independent experiments.
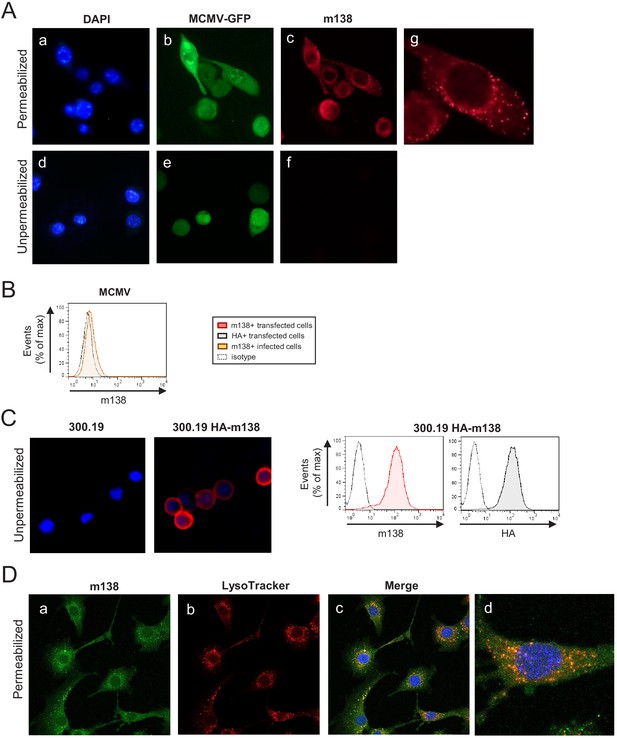
Localization of m138 in MCMV-infected cells.
(A) NIH3T3 cells were infected with MCMV-GFP at an moi of 5 for 24 hr, fixed with 4% formaldehyde (panels d-f) or fixed and permeabilized with 0.05% Triton (panels a-c and g), and stained with the anti-m138 mAb followed by an anti-mouse IgG-A555. Nuclei were stained with the DAPI reagent. The cells were examined under a fluorescence microscope. Magnification, ×20 (a-f); an enlarged individual cell from panel c is shown in panel g. (B) MCMVwt-infected NIH3T3 cells were examined by flow cytometry for surface expression of m138 with the anti-m138 mAb (red histogram). Dotted line represents the isotype control. (C) 300.19 cells untransfected or stably transfected with the HA-m138 construct were fixed with 4% formaldehyde and stained with the anti-m138 mAb and DAPI and examined under a fluorescence microscope as indicated in A. Magnification, ×20. Overlaid images are shown. Transfected cells from the same cultures were analyzed by flow cytometry (right panels) with the anti-m138 mAb (red histogram), an anti-HA mAb (gray histogram) or isotype controls (dotted lines). (D) NIH3T3 cells were infected with MCMV at an moi of 5 for 24 hr, treated with LysoTracker DND99, fixed with 4% formaldehyde and permeabilized with 0.02% Saponin, and stained with the anti-m138 mAb followed by an anti-mouse IgG-A488. Nuclei were stained with the DAPI reagent. The cells were examined under a confocal microscope. Representative fields are shown in panels a, b, and c, with an enlargement of an individual and more exposed cell from panel c, in panel d. Magnification, ×63.
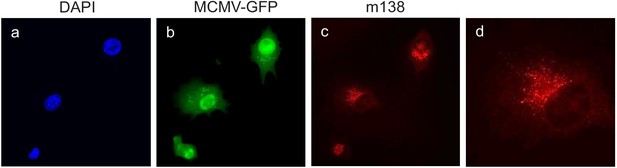
Localization of m138 in MCMV-infected DC2.4 cells.
D2.4 cells were infected with MCMV-GFP at an moi of 5 for 24 hr, fixed with 4% formaldehyde, permeabilized with 0.05% Triton, and stained with the anti-m138 mAb followed by an anti-mouse IgG-A555. Nuclei were stained with the DAPI reagent. The cells were examined under a fluorescence microscope. Magnification, ×20 (a-c); an enlarged individual cell from panel c is shown in panel d.
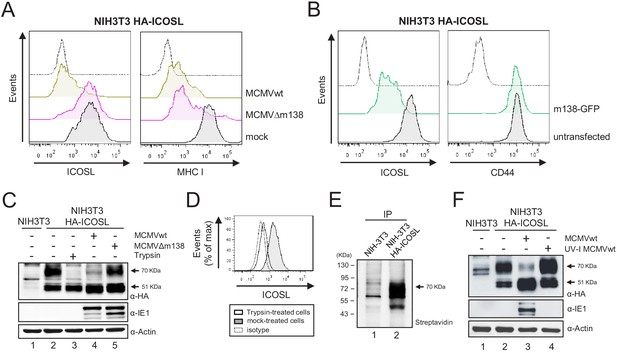
m138 leads to a reduction of the expression of ICOSL on the cell surface of infected cells but not of its overall intracellular levels.
(A) NIH3T3 cells stably transfected with HA-ICOSL were mock-infected (gray histograms) or infected with MCMVwt or MCMV∆m138 (brown or pink histograms, respectively) and analyzed by flow cytometry for surface expression of ICOSL and MHC I. (B) NIH3T3 HA-ICOSL cells non-transfected (gray histograms) or transfected with the m138-GFP construct (green histograms) were analyzed by flow cytometry for surface expression of ICOSL and CD44. In A and B, the isotype for each antibody was used as a negative control (dotted lines). (C) NIH3T3 HA-ICOSL cells were mock-infected (lane 2), treated with trypsin (lane 3), or infected with MCMVwt (lane 4) or MCMV∆m138 (lane 5). Untransfected NIH3T3 cells were used as a control (lane 1). Samples were analyzed by western blot using an anti-HA mAb, followed by anti-rabbit IgG-HRP. An anti-IE1 and an anti-actin mAbs followed by anti-mouse IgG-HRP, were used as controls of MCMV infection and loading, respectively. (D) Flow cytometry analysis of NIH3T3 HA-ICOSL treated with trypsin (open histogram) or mock-treated (gray histogram) and stained with an anti-HA mAb or an isotype control (dotted line), followed by anti-mouse IgG PE. (E) NIH3T3 cells (lane 1) or NIH3T3 HA-ICOSL (lane 2) were surface labeled with biotin, immunoprecipitated with an anti-HA mAb, and analyzed by western blot analysis using streptavidin-HRP conjugate. (F) NIH3T3 HA-ICOSL cells were mock-infected (lane 2), or infected with MCMVwt (lane 3) or MCMVwt UV-inactivated (lane 4). Samples were lysed and processed as indicated in C. A lysate of untransfected NIH3T3 cells (lane 1) was also subjected to western blot and employed as a control. All infections were performed at an moi of 5 for 24 hr. Data are representative of at least two independent experiments. In C, E, and F, the size of the bands corresponding to ICOSL are indicated on the right margin in kilodaltons (kDa). In E, the sizes of the molecular weight markers are shown on the left side.
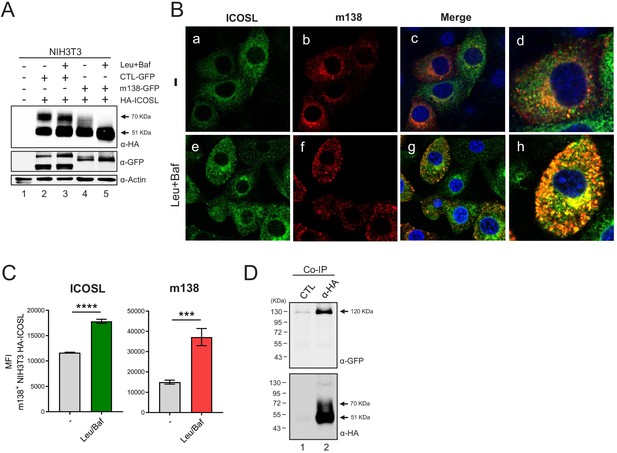
m138 impedes ICOSL maturation redirecting it to lysosomal degradation.
(A) Western blot analysis of NIH3T3 cells untransfected (lane 1) or transiently co-transfected with either the HA-ICOSL and the control-GFP (CTL-GFP) constructs (lanes 2 and 3), or the HA-ICOSL and the m138-GFP constructs (lanes 4 and 5) and, when indicated, treated with 250 μM of leupeptin and 20 nM of bafilomycin A1 (lanes 3 and 5). The expression of ICOSL and actin was assessed as described in Figure 4C, and for the expression of the m138-GFP and CTL-GFP proteins a polyclonal antibody anti-GFP followed by anti-rabbit IgG-HRP were used. (B) NIH3T3 HA-ICOSL were infected with MCMVwt at an moi of 5 for 24 hr in the absence (-; panels a, b, c and d) or presence of leupeptin and bafilomycin (panels e, f, g and h). Cells were fixed, permeabilized, and stained with anti-m138 and anti-ICOSL mAbs followed by an anti-mouse IgG-A555 and anti-rat IgG-A488, respectively. Nuclei were stained with the DAPI reagent and samples were examined under a confocal fluorescence microscope. Shown are representative cells from cultures stained for m138 (panels a, e), ICOSL (b, f), and overlaid images (c, d, g, and h). Enlarged and more exposed individual cells from panels c and g are presented on panels d and h, respectively. Magnification, ×63. (C) NIH3T3 HA-ICOSL infected and treated with the lysosomal inhibitors as in B were fixed, permeabilized with the Foxp3 staining buffer set, and analyzed by flow cytometry with anti-m138 and anti-ICOSL mAbs followed by an anti-mouse IgG-A555 and anti-rat IgG-A488, respectively. Conditions were analyzed in triplicates and median fluorescence intensity (MFI) values +/- SD were extracted from pre-gated m138 positive cells. (D) NIH3T3 cells were co-transfected with the m138-GFP and HA-ICOSL constructs. HA-ICOSL was immunoprecipitated from 0.5% Triton-treated lysates using an anti-HA mAb or an anti-hFc as a control. Recovered immunoprecipitates were subjected to SDS-PAGE and western blot with antibodies against GFP and HA, as described in A and Figure 4C. Data are representative of at least two independent experiments. In A and D, the size of the two bands corresponding to ICOSL, and in D, the band corresponding to m138-GFP, are indicated on the right margin in kilodaltons (kDa). In D, the sizes of the molecular weight markers are shown on the left side.
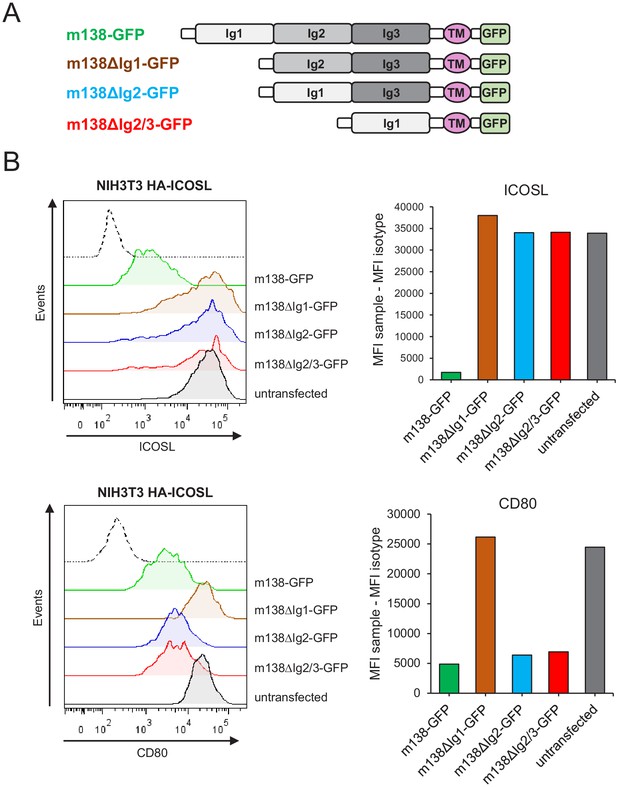
Identification of the m138 domains involved in the downmodulation of ICOSL and CD80.
(A) Schematic representation (not drawn to scale) of m138-GFP protein mutants. (B) NIH3T3 ICOSL-HA cells were untransfected or transfected with m138-GFP, or m138-GFP mutant constructs and analyzed by flow cytometry for ICOSL and CD80 surface levels. Panels on the right represent the MFI of each sample minus the MFI of the control isotypes. Gray histograms represent the expression of untransfected cells and colored histograms represent the expression of cells transfected with m138-GFP (green), m138∆Ig1-GFP (brown), m138∆Ig2-GFP (blue), or m138∆Ig2/3-GFP (red). The isotype for each antibody was used as a negative control (dotted lines). A representative experiment out of two performed is shown.
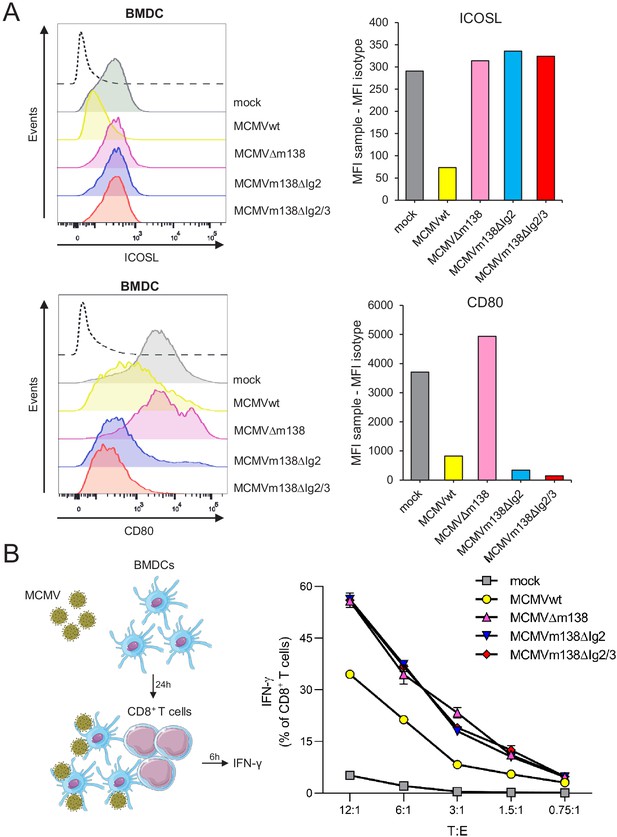
m138 is capable to restrict CD8+ T-cell activation by downregulating ICOSL on the surface of antigen-presenting cells (APCs).
(A) C57BL/6-derived BMDCs were mock-infected or infected for 24 hr with MCMVwt or MCMVm138 mutants at an moi of 3 and analyzed by flow cytometry for surface expression of ICOSL and CD80 using specific mAbs against each of these molecules. Gray histograms represent the expression of mock-infected cells and colored histograms represent the expression of cells infected (positive for the MCMV m04 protein) with MCMVwt (yellow), MCMVΔm138 (pink), MCMVm138ΔIg2 (blue), or MCMVm138ΔIg2/3 (red). The isotype for each antibody was used as a negative control (black line histograms). Panels on the right represent the MFI of each sample minus the MFI of the control isotypes. Results are representative of two independent experiments (B) On the left, schematic representation of the in vitro antigen-presentation assay. BMDCs infected as indicated in A with MCMVwt (yellow), MCMVΔm138 (pink), MCMVm138ΔIg2 (blue), or MCMVm138ΔIg2/3 (red), or left uninfected (gray) were further co-cultured with naïve CD8+ T cells from Maxi mice. After 6 hr, IFN-γ production by CD8+ T cells was determined by flow cytometry. Results are representative of three independent experiments. Results are expressed as the mean +/-SEM of the percentage values obtained for samples in each group.
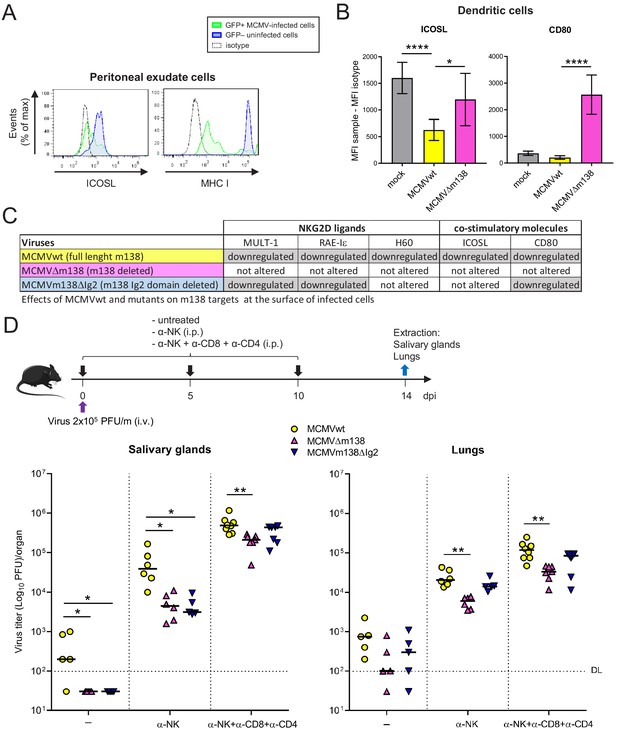
Via targeting ICOSL, m138 reduces T-cell mediated control of MCMV infection in vivo.
(A) BALB/c mice were intraperitoneally (i.p.) inoculated with 2 × 106 PFU MCMV-GFP. Two days post-infection mice were sacrificed and peritoneal exudate cells were extracted and analyzed by flow cytometry for surface expression of ICOSL and MHCI, using specific mAbs against each of these molecules. Blue histograms represent the expression of uninfected (GFP-) cells and green histograms represent the expression of MCMV-infected (GFP+) cells from the same mouse. The isotype for each antibody was used as a negative control (dotted lines). The results obtained from a representative infected mouse out of two are shown. (B) BALB/c mice (n = 4/group) were mock-infected or i.p. infected with 1 × 106 PFU of MCMVwt or MCMVΔm138. At 6 hpi, peritoneal exudate cells were extracted and surface expression of ICOSL and CD80 assessed by flow cytometry on the surface of DCs (CD11+ MHC II+ CD3-CD19- NKp46- cells), gating on m04-positive cells when derived from MCMVwt- or MCMVΔm138-infected mice. Results are expressed as the mean +/-SD of the MFI values obtained for samples from two independent experiments. (C) Table displaying the immunomodulatory effects of the different MCMVs on the cellular targets of m138. (D) On the top, a schematic representation of the MCMVΔm138 and MCMVm138ΔIg2 in vivo infection assay is shown. C57BL/6 mice (n = 5–7/group) with or without NK, or NK, CD4+ and CD8+ T-cell depletion, as indicated, were intravenously (i.v.) inoculated with 2 × 105 PFU/mouse of MCMVwt (yellow circles), MCMVΔm138 (pink triangles) or MCMVm138ΔIg2 (blue triangles). At day 14 post-infection mice were sacrificed and viral titers in salivary glands and lungs of individual mice were determined by standard plaque assays. Horizontal bars indicate the median values. The Kruskal-Wallis test was used to assess statistical differences between experimental groups. *p<0.05, **p<0.01. A representative experiment out of two performed is shown.
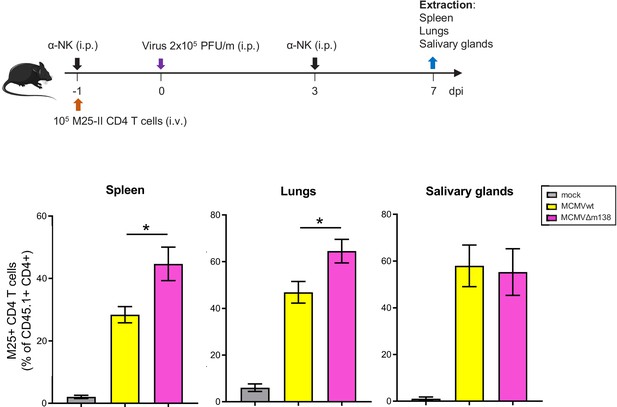
m138 contributes to immune evasion of CD4+ T cells in spleens and lungs of MCMV-infected mice.
On the top, a schematic representation of the adoptive transfer in vivo assay is shown. 1 × 105 M25-specific transgenic CD4+ T cells were transferred (i.v.) into C57BL/6 SCID mice (n = 4–5/group), which were depleted of NK cells as indicated, and either mock-infected or i.p. infected with 2 × 105 PFU/mouse of MCMVwt or MCMVΔm138. Seven dpi, spleens, lungs, and salivary glands were harvested, and analyzed by flow cytometry using anti-CD45.1 and anti-CD4-specific mAbs. The frequency of CD45.1+ CD4+ cells for each organ is indicated. Results are expressed as the mean +/-SEM of the percentage values obtained for samples in each group.
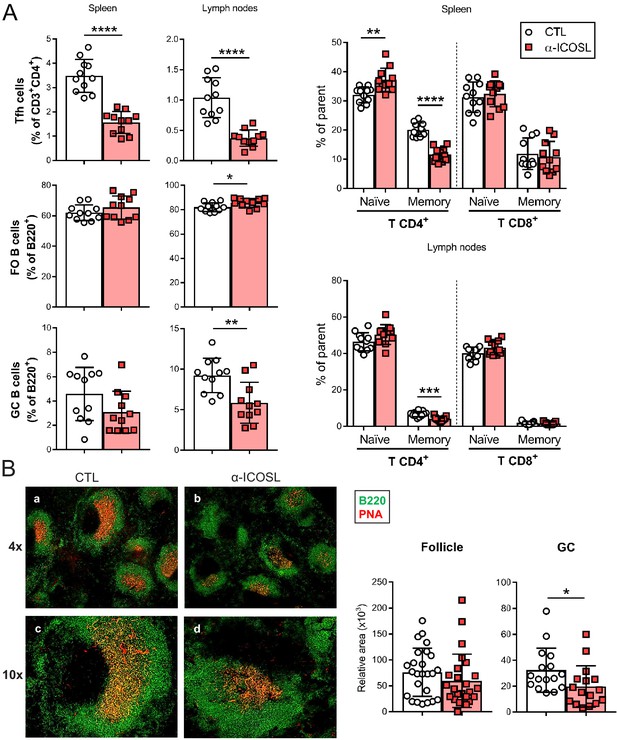
Blockade of ICOSL reduces the frequency of particular lymphocyte subsets in the spleen and lymph nodes of MCMV-infected mice.
(A) BALB/c mice (n = 11–12/group) were i.p. inoculated with 2 × 106 PFU of MCMVΔm138 and treated with (α-ICOSL) or without (CTL) 100 mg of anti-ICOSL mAb. At day 14 post-infection mice were sacrificed, and spleens and lymph nodes were isolated and disaggregated. Cell suspensions were analyzed by flow cytometry using anti-mouse CD3, CD4, CD8, CD44, CD62L, CXCR5, and PD1 for T-cell phenotype. For B cell phenotype B220, CD21, CD23, CD95, and GL7 mAbs were used. Percentages of Tfh cells (CD3+CD4+CXCR5hiPD1hi), germinal center (GC) B cells (CD95hiGL7hiB220+), follicular (FO) B cells (CD21+CD23+B220+), and CD3+CD4+ or CD3+CD8+ naive (CD44lowCD62Lhi) and memory (CD44hiCD62Llow) T cells from these two organs are shown. (B) Representative immunofluorescence of spleens from mice infected and treated with (panels b and d) or without (panels a and c) the anti-ICOSL mAb, as indicated in A. Two color colocalization with B220 in green and peanut agglutinin (PNA) in red are shown at ×4 and ×10 magnifications. Panels on the right show a graphic representation of follicle and GC relative areas from tissue sections of a representative mouse of each experimental group. In graphs shown in A and B, results are expressed as mean +/-SD. Mice treated with anti-ICOSL mAb are represented as red squares and mice not receiving the mAb are represented as open circles. Two-tailed unpaired t-tests were used to assess statistical differences between experimental groups. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Data are pooled from two independent experiments.
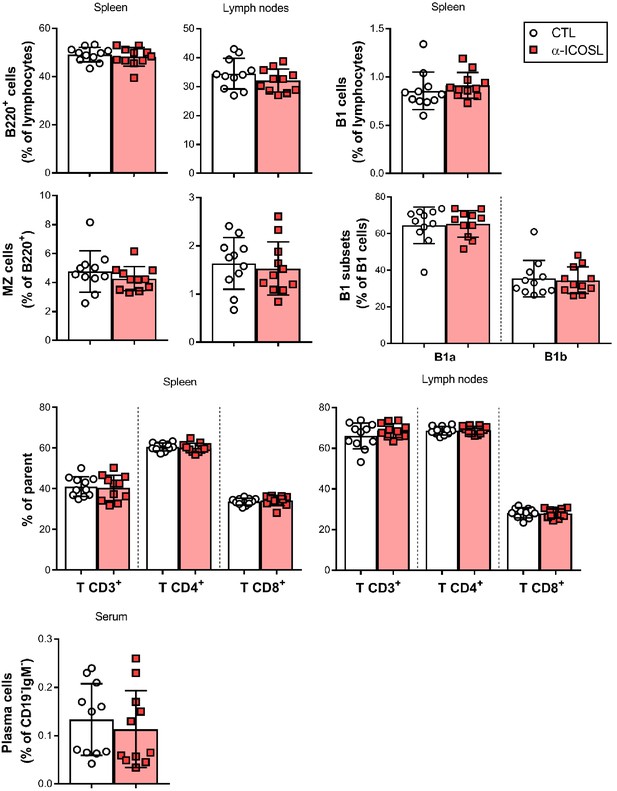
Effect of ICOSL blockade on the frequency of several lymphocyte subsets in the spleen and lymph nodes of MCMV-infected mice.
Cell suspensions obtained from spleens and lymph nodes of BALB/c mice infected and treated as indicated in Figure 8A were analyzed by flow cytometry using anti-mouse CD3, CD4, CD8, CD5, B220, CD23, CD21, CD138, and CD19 mAbs, and anti-mouse IgM polyclonal Ab. Percentages of total B cells (B220+), marginal zone (MZ) B cells (CD21+CD23-B220+), B1 B cells (CD19hiB220int), B1a (CD19hiB220intCD5+), and B1b (CD19hiB220intCD5-) subsets, CD3+CD4+ or CD3+CD8+ T cells, and plasma cells (CD138hiB220lowCD19-IgM-) are shown. Results are expressed as mean +/- SD. Mice treated with anti-ICOSL mAb are represented as red squares and mice untreated are represented as open circles. Two-tailed unpaired t-test was used to assess statistical differences between experimental groups.
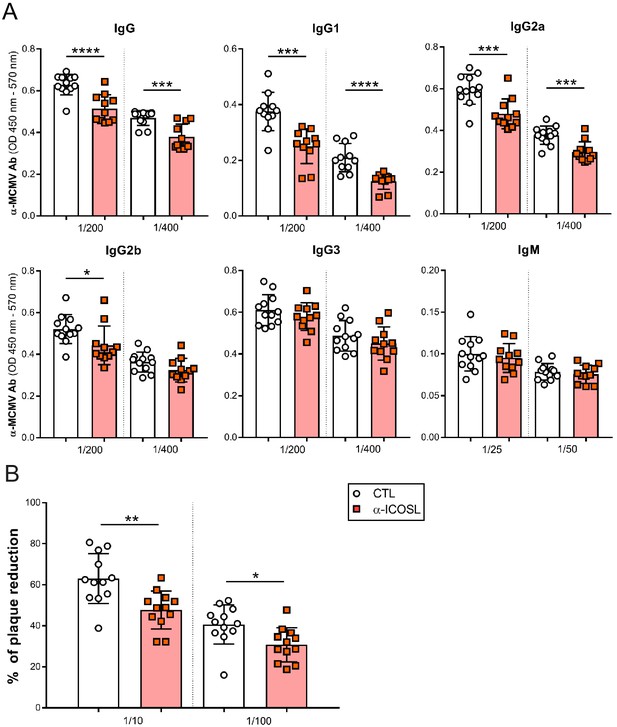
ICOSL blockade limits the generation of MCMV specific antibodies.
(A) Total levels of IgG, IgG1, IgG2a, IgG2b, IgG3 (at dilutions 1:200 and 1:400), and IgM (at dilutions 1:25 and 1:50) were determined by ELISA from sera collected at day 14 of the same infected BALB/c mice indicated in Figure 8A. (B) Sera (at dilutions 1:10 and 1:100) as in A were tested in a viral neutralization assay. Results are presented as the percentage of plaque reduction determined by the ratio of the number of plaques counted in the sample wells relative to the ratio of plaques in wells containing the serum of an uninfected mouse used as a negative control. In A and B, results are expressed as mean +/- SD. Mice treated with anti-ICOSL mAb are represented as red squares and mice that did not receive the mAb are represented as open circles. Two-tailed unpaired t-tests were used to assess statistical differences between experimental groups. *p<0.05, **p<0.01, ***p<0.001. Data are pooled from two independent experiments.
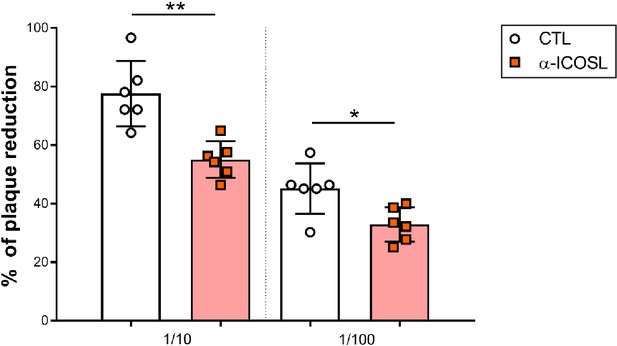
Effect of ICOSL blockade on MCMV neutralization in the presence of complement.
Same as in Figure 9B, except that the MCMV neutralization assay was performed in the presence of rabbit complement. Results are presented as the percentage of plaque reduction determined by the ratio of the number of plaques counted in the sample wells relative to the ratio of plaques in wells containing the serum of an uninfected mouse used as a negative control. Results are expressed as mean +/-SD. Mice treated with anti-ICOSL mAb are represented as red squares and mice untreated are represented as empty circles. Two-tailed unpaired t-test was used to assess statistical differences between experimental groups. *p<0.05, **p<0.01.
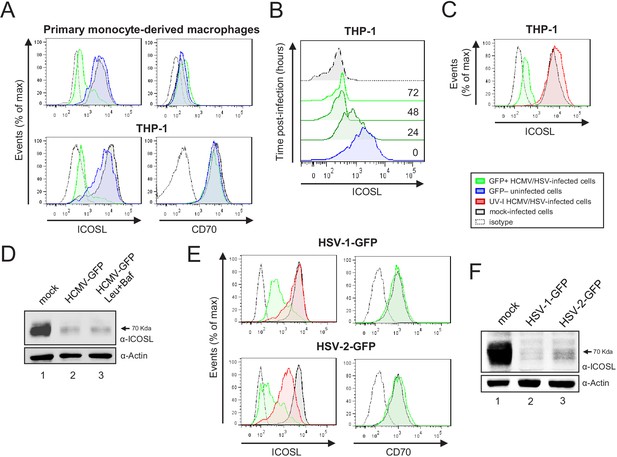
HCMV, HSV-1, and HSV-2 also limit cell surface expression of ICOSL on antigen-presenting cells (APCs).
(A) Primary monocyte-derived macrophages (upper panels) and PMA-treated THP-1 cells (bottom panels) were mock-infected or infected for 72 hr with HCMV-GFP at an moi of 10 and analyzed by flow cytometry for cell-surface expression of human ICOSL or CD70 using specific mAbs against each of these receptors. Gray histograms represent the expression of mock-infected cells, green histograms represent the expression on HCMV-infected (GFP+) cells, and blue histograms represent the expression on uninfected (GFP-) cells from the same culture. (B) PMA-treated THP-1 cells were mock-infected (time 0) or infected with HCMV-GFP as in A and analyzed by flow cytometry for surface expression of ICOSL at the different time points after infection indicated. (C) Same as in A, except that an moi of 20 was used, and THP-1 cells were also exposed for 72 hr to the same amount of HCMV-GFP UV-inactivated (red histogram). (D) Equal amounts of lysates from PMA-treated THP-1 cells mock-infected (lane 1) or infected for 72 hr at an moi of 20 with HCMV-GFP (lanes 2 and 3), and when indicated, treated with 250 μM leupeptin and 20 nM of bafilomycin A1 (lane 3), were lysed and analyzed by western blot with antibodies against ICOSL and actin, followed by anti-rabbit IgG-HRP (ICOSL) or anti-mouse IgG-HRP (actin). (E) PMA-treated THP-1 cells were mock-infected or infected with HSV-1-GFP, HSV-1-GFP UV-inactivated, HSV-2-GFP or HSV-2-GFP UV-inactivated at an moi of 100 (HSV-1) or 200 (HSV-2). Twenty-four hr later, the expression of human ICOSL and CD70 were analyzed as in A. Green histograms represent the expression of ICOSL on infected cells, red histograms represent the expression on UV-inactivated HSV infected cells, and gray histograms represent the expression on mock-infected cells. The isotype for the ICOSL antibody was used as a negative control (dotted lines). A representative experiment out of three performed is shown. (F) PMA-treated THP-1 cells were mock-infected (lane 1) or infected with HSV-1-GFP (lane 2) and HSV-2-GFP (lane 3) as indicated in E. Cell lysates were prepared, and subjected to western blot analysis using anti-human ICOSL or anti-actin mAbs as in D. The molecular weight of the band corresponding to ICOSL is indicated in D and E in kilodaltons on the right margin.
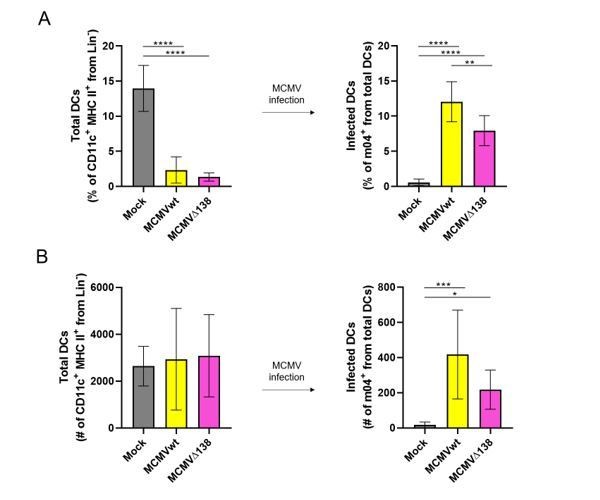
(A) Percentage (%) and (B) number (#) of total peritoneal DCs (live Lin- CD11c+ MHC II+) and MCMV-infected DCs (m04+ cells from total DCs).
BALB/c mice were i.p. infected with 106 PFU/mice of MCMVwt and MCMVΔm138. DCs were isolated from the peritoneal cavity 6 hr post-infection and analyzed by flow cytometry. Results represent two merged individual experiments (mock n=8, MCMVwt n=6, MCMVΔ138 n=7). One-way ANOVA test was used for statistical analysis. Graphs show mean with SEM as error bars. ****, p ≤0.0001; ***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-mouse CD8 (clone YTS 169.4) (Rat monoclonal) | Bio X cell West Lebanon, NH, USA | Cat#: BE0117 RRID:AB_10950145 | in vivo depletion (150 µg/injection) |
| Antibody | anti-mouse CD4 (clone GK1.5) (Rat monoclonal) | Bio X cell West Lebanon, NH, USA | Cat#: BE0003-3 RRID:AB_1107642 | in vivo depletion (150 µg/injection) |
| Antibody | anti-mouse NK.1.1 (clone PK136) (Rat monoclonal) | Bio X cell West Lebanon, NH, USA | Cat#: BE0036 RRID:AB_1107737 | in vivo depletion (250 µg/injection) |
| Antibody | anti-mouse ICOSL (clone HK5.3) (Rat monoclonal) | Bio X cell West Lebanon, NH, USA | Cat#: BE0028 RRID:AB_1107566 | in vivo (100 µg/injection) |
| Antibody | anti-m04 (clone m04.17) (Mouse monoclonal) | Center for Proteomics, Faculty of Medicine, University of Rijeka, Croatia PMID:31142589 | Stock 1 mg/mL FACS (1:100) | |
| Antibody | anti-m04 (clone m04.16) (Mouse monoclonal) | Center for Proteomics, Faculty of Medicine, University of Rijeka, Croatia PMID:31142589 | Stock 1 mg/mL FACS (1:100) | |
| Antibody | anti-HA (clone C2974) (Mouse monoclonal) | Sigma-Aldrich, St. Louis, MO, USA | Cat#: H9658 RRID:AB_260092 | WB (1:2000) |
| Antibody | anti-human IE1 (clone 8B1.2) (Mouse monoclonal) | Merck Millipore, Burlington, MA, USA | Cat#: MAB810R RRID:AB_11212266 | WB (1:2000) |
| Antibody | anti-human Fc IgG (clone 29.5) (Mouse monoclonal) | In house Department of Biomedical Sciences, University of Barcelona, Spain | Stock 1 mg/mL FACS (1:400) | |
| Antibody | anti-m138 (clone m138.1.120) (Mouse monoclonal) | In house Department of Biomedical Sciences, University of Barcelona, Spain | This manuscript | Stock 1 mg/mL FACS (1:100) IF (1:50) |
| Antibody | anti-mouse CD80 (clone 16-10A1) (Armenian hamster monoclonal) | Biolegend San Diego, CA, USA | Cat#: 104714 RRID:AB_313134 | FACS (1:200) |
| Antibody | anti-human ICOSL (clone MIH12) (Mouse monoclonal) | Thermo Fischer Scientific, Waltham, MA, USA | Cat#: 16-5889-82 RRID:AB_469129 | FACS (1:50) |
| Antibody | anti-mouse ICOSL (clone HK5.3) (Rat monoclonal) | Biolegend San Diego, CA, USA | Cat#: 107403 RRID:AB_345259 (Biotin) Cat#: 107405 RRID:AB_2248797 (PE) | FACS (1:100) IF (1:50) |
| Antibody | anti-human ICOSL (Rabbit polyclonal) | Elabscience Wuhan, Hubei, China | Cat#: E-AB-15519 | WB (1:1000) |
| Antibody | anti-mouse ICOSL (clone 599841) (Rat monoclonal) | R and D Systems, Minneapolis, MN, USA | Cat#: MAB158 RRID:AB_10719415 | IF (1:500) |
| Antibody | anti-m123/IE1 (MCMV) (clone IE1.01) (Mouse monoclonal) | Center for Proteomics, Faculty of Medicine, University of Rijeka, Croatia | Cat#: HR-MCMV-12 | Stock 1 mg/mL IF (1:100) FC (1:100) WB (1:1000) |
| Antibody | anti-HA−Agarose conjugate (Mouse monoclonal) | Sigma-Aldrich, St. Louis, MO, USA | Cat#: A2095 RRID:AB_257974 | |
| Cell line (M. musculus) | NIH3T3 | ATCC, Manassas, VA, USA | Cat#: CRL-1658 RRID:CVCL_0594 | |
| Cell line (M. musculus) | DC2.4 | Merck Millipore, Burlington, MA, USA | Cat#: SCC142 RRID:CVCL_J409 | |
| Cell line (M. musculus) | MEFs p53 | Dr. Jay Nelson, Health Sciences University, Oregon, USA | ||
| Cell line (M. musculus) | SVEC4-10 | ATCC, Manassas, VA, USA | Cat#: CRL-2181 RRID:CVCL_4393 | |
| Cell line (M. musculus) | IC-21 | ATCC, Manassas, VA, USA | Cat#: TIB-186 RRID:CVCL_3726 | |
| Cell line (M. musculus) | NS1 | ECACC, Public Health England, Salisbury, UK | Cat#: 85011427 RRID:CVCL_2155 | |
| Cell line (M. musculus) | 300.19 | Dr. Michel Streuli, Dana Farber Cancer Institute, MA, USA | ||
| Cell line (Homo-sapiens) | THP-1 | ATCC, Manassas, VA, USA | Cat#: TIB-202 RRID:CVCL_0006 | |
| Cell line (Homo-sapiens) | HFF | ATCC, Manassas, VA, USA | Cat#: SCRC-1041 RRID:CVCL_3285 | |
| Cell line (Homo-sapiens) | HEL299 | ATCC, Manassas, VA, USA | Cat#: CCL-137 RRID:CVCL_2480 | |
| Cell line (Cercopithecus aethiops) | Vero | ECACC, Public Health England, Salisbury, UK | Cat#: 84113001 RRID:CVCL_0059 | |
| Cell line (Cercopithecus aethiops) | COS-7 | ATCC, Manassas, VA, USA | Cat#: CRL-1651 RRID:CVCL_0224 | |
| Strain, strain background- (Mouse cytomegalovirus) | wild type (BAC-derived strain pSM3fr) | PMID:10400809 | ||
| Strain, strain background- (Mouse cytomegalovirus) | BAC-derived strain pSM3fr-GFP | PMID:12660946 | ||
| Strain, strain background- (Mouse cytomegalovirus) | Mutant MCMV-GFPΔ15 | PMID:16831899 | Lacks ORFs m128-m138 | |
| Strain, strain background- (Mouse cytomegalovirus) | Mutant MCMV-GFPΔm138 | PMID:16831899 | Lacks ORF m138 | |
| Strain, strain background- (Mouse cytomegalovirus) | Mutant MCMV-GFPΔ1 | Brune et al., 2006 | Lacks ORFs m1-m17 | |
| Strain, strain background- (Mouse cytomegalovirus) | Mutant MCMV-m138ΔIg2 | PMID:16831899 | Lacks Ig2 domain of m138 | |
| Strain, strain background- (Mouse cytomegalovirus) | Mutant MCMV-m138ΔIg2/Ig3 | PMID:16831899 | Lacks Ig2 and Ig3 domains of m138 | |
| Strain, strain background- (Human cytomegalovirus) | HCMV-GFP (strain TB40/E) | PMID:18198366 | ||
| Strain, strain background (M. musculus) | MAXI mice | Central Animal Facility, Faculty of Medicine, University of Rijeka, Croatia PMID:22046127 | ||
| Strain, strain background (M. musculus) | BALB/c mice | Central Animal Facility, Faculty of Medicine, University of Barcelona, Spain and Central Animal Facility, Faculty of Medicine, University of Rijeka, Croatia | ||
| Strain, strain background (M. musculus) | C57BL/6 mice | Central Animal Facility, Faculty of Medicine, University of Rijeka, Croatia | ||
| Strain, strain background (M. musculus) | M25-II mice | Central Animal Facility, Faculty of Medicine, University of Rijeka, Croatia PMID:22876184 | ||
| Strain, strain background (M. musculus) | C57BL/6 SCID | Central Animal Facility, Faculty of Medicine, University of Rijeka, Croatia | ||
| Chemical compound, drug | Amaxa Cell Line Nucleofactor Kits R and V | Lonza, Basel, Switzerland | Cat#: VCA-1001 and VCA-1003 | |
| Chemical compound, drug | G418 | Invivogen, Toulouse, France | Cat#: ant-gn-1 | 1.2 mg/mL |
| Chemical compound, drug | leupeptin | Sigma-Aldrich, St. Louis, MO, USA | Cat#: L2884 | 250 μM |
| Chemical compound, drug | bafilomycin A1 | Sigma-Aldrich, St. Louis, MO, USA | Cat#: B1793 | 20 nM |
| Chemical compound, drug | Biotin | Sigma-Aldrich, St. Louis, MO, USA | Cat#: B5161 | 2 mg/million of cells |
| Chemical compound, drug | Protein G Sepharose 4 Fast Flow | GE Healthcare, Chicago, IL, USA | Cat#: 17-0618-01 | |
| Chemical compound, drug | Tissue-Tek O.C.T. Compound | Sakura, Torrance, CA, USA | Cat#: 4583 | |
| Chemical compound, drug | PNA biotin | Sigma-Aldrich, St. Louis, MO, USA | Cat#: L6135 | |
| Chemical compound, drug | Brefeldin A | eBioscience, Thermo Fischer Scientific, Waltham, MA, USA | Cat#: 00-4506-51 | 1 µg/ml |
| Chemical compound, drug | LysoTracker Red DND-99 | Invitrogen, Thermo Fischer Scientific, Waltham, MA, USA | Cat#: L7528 | |
| Chemical compound, drug | Intracellular Fixation and Permeabilization Buffer Set | eBioscience, Thermo Fischer Scientific, Waltham, MA, USA | Cat#: 88-8824-00 | |
| Chemical compound, drug | Foxp3/ Transcription Factor Fixation/Permeabilization | eBioscience, Thermo Fischer Scientific, Waltham, MA, USA | Cat#: 00-5521-00 | |
| Cell Isolation kit | CD4+ T Cell Isolation Kit, mouse | Miltenyi Biotec, Bergisch Gladbach, Germany | Cat#: 130-104-454 | |
| Recombinant DNA reagent | pGEM-T (plasmid) | Promega, Madison, WI, USA | Cat#: A3600 | |
| Recombinant DNA reagent | pDisplay (plasmid) | Invitrogen, Thermo Fischer Scientific, Waltham, MA, USA | Cat#: V660-20 | |
| Recombinant DNA reagent | pEGFP-N3 (plasmid) | Clontech, Takara, Tokio, Japan | Cat#: 6080–1 | |
| Recombinant DNA reagent | pCMV6-mouse ICOSL (plasmid) | Origene, Rockville, MD, USA | Cat#: MR204667 | |
| Sequence-based reagent | m138PstIFor: 5’-CTGCAGGCATCAATTACCTGCGTGCCAG-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | m138PstIRev: 5’-CTGCAGTTACGTGTGACGTACGCAACC-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | m138BamFor: 5’-GGATCCATGGCGCCTTCGACGCTGATC-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | m138BamRev: 5’-GGATCCCGTGTGACGTACGCAACCCGG-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | SOEm138∆Ig1Rev: 5’-AGTCCCGGTGGAGTCGGTGATCAGTTGCGT-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | SOEm138∆Ig1For: 5’-ACGCAACTG ATCACCGACTCCACCGGGACT-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | SOEm138∆Ig2Rev: 5’-CATCAGCG AC CGGCCAGT CCCGGTGGAGTC-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | SOEm138∆Ig2For: 5’-GACTCCACC GGGACTGGCCGGTCGCTGATG-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | SOEm138∆Ig2/3Rev: 5’-CTGAG GGGACGTGACAGTCCCGGTGGAGTC-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | SOEm138∆Ig2/3For: 5’-GACTC CACCGGGACTGTCACGTCCCCTCAG-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | ICOSLmurineSalIFor: 5’-GTCGACAGAGACTGAAGTCGGTGCAATG-3’ | Sigma-Aldrich, St. Louis, MO, USA | ||
| Sequence-based reagent | ICOSLmurineNotIRev: 5’-GCGGCC GCTTAGGCGTGGTCTGTAAGTTCA-3’ | Sigma-Aldrich, St. Louis, MO, USA |





