New insights into the mechanism of dynein motor regulation by lissencephaly-1
Figures
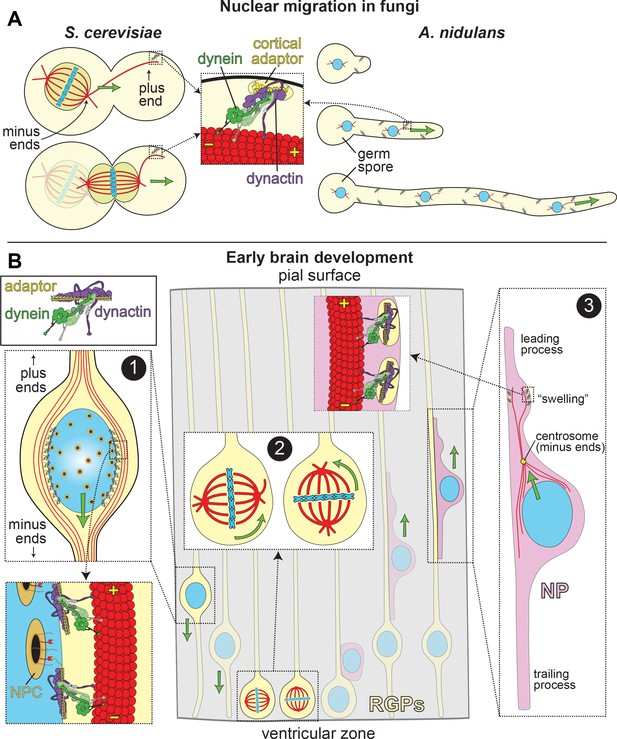
LIS1 and Dynein ensure appropriate transport and position of nuclei in various biological systems.
(A) In fungal model systems, cortically anchored dynein-dynactin complexes move the nucleus toward the mother-bud neck (the future site of cytokinesis) in budding yeast (left), or in the direction of the growing hyphal tip to ensure proper nuclear distribution in Aspergillus nidulans (right). Dynein-dynactin complexes are recruited and anchored to the cell cortex in budding yeast and A. nidulans by Num1 and ApsA, respectively, (Fischer and Timberlake, 1995; Heil-Chapdelaine et al., 2000; Veith et al., 2005), which exhibit significant degrees of similarity within their C-terminal pleckstrin homology domains (for membrane association; 50% identity/64% similarity) and their N-terminal coiled-coil-containing dynein-dynactin interacting regions (24% identity/41% similarity; note this has not been experimentally determined in A. nidulans) (Tang et al., 2012). (B) In the developing brain, LIS1 and dynein are critical for the three distinct illustrated processes: (Box 1) During interkinetic nuclear migration (INM), the nucleus is transported to the ventricular surface in highly elongated radial glial precursors (RGPs), where mitosis takes place. Dynein-dynactin complexes anchored to the nuclear envelope – through interactions between dynein adaptors (e.g., BicD2, in yellow) and nuclear pore complex components (e.g., RanBP2, in red; nuclear pore complex, or NPC, in orange) – transport the nucleus toward the minus ends of microtubules (in red), which are situated toward the ventricular surface (‘-' and ‘+' depict minus and plus ends of microtubules, respectively). (Box 2) The mitotic spindles (in red, with chromosomes shown in blue) in RGPs are actively rotated by cortical dynein-dynactin complexes. The orientation of the spindle dictates whether a dividing cell will generate two RGPs (horizontal orientation, left), or one RGP and a differentiated cell type (e.g., a neuronal precursor, or NP cell; vertical, right). (Box 3) Subsequent to their generation from RGPs, NPs ascend toward the pial surface of the developing brain, where the cortical plate resides. Evidence suggests that dynein is enriched in a dilation, or ‘swelling’ within the leading process. It has been proposed that dynein anchored at the cell cortex (by an unknown cortical receptor, in yellow) in this region pulls on astral microtubules emanating from the centrosome, thus moving the nucleus toward the leading process (see text) (Tsai et al., 2007). The manner by which the centrosome is linked to the nucleus is not entirely clear, but may rely on dynein and microtubules (Vallee et al., 2009).
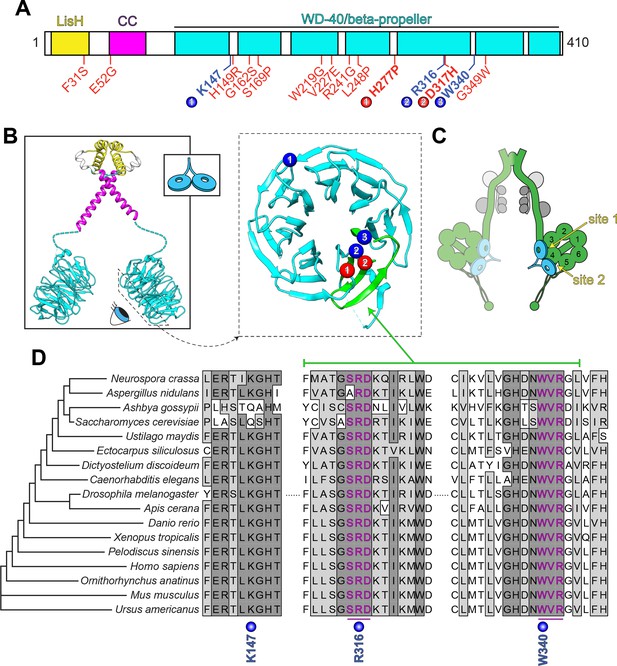
LIS1 structure and conservation.
(A) Domain architecture of human LIS1. LIS1-homology (LisH), coiled-coil (CC), and WD-40 domains indicated, along with missense mutations found in patients with lissencephaly (red; bolded residues depicted in panel B, right) and the three residues shown to be important for Pac1/LIS1-dynein interaction (blue; numbers correspond to numbered residues in panel B, right) (Gutierrez et al., 2017; Pandey and Smith, 2011; Toropova et al., 2014). (B) Model of full-length, dimeric LIS1 structure (adapted from Tarricone et al., 2004), with domains color-coded according to panel A (generated using PDB 1VYH and 1UUJ [Kim et al., 2004; Tarricone et al., 2004] and UCSF Chimera [Pettersen et al., 2004]). Inset shows cartoon depiction of LIS1 used throughout this review. The dynein-interacting surface of one of the beta-propellers of LIS1 is shown, along with the residues found to be important for dynein binding (blue), and the surface-exposed lissencephaly-causing mutations (red). The 5th blade of the beta-propeller that encompasses the highly conserved dynein-interacting residues is shown in green. (C) A cartoon schematic of the dynein complex (green, heavy chains with AAA+ domains numbered; gray, light, light-intermediate, and intermediate chains) with a LIS1 dimer bound to each motor head. Note that ‘site 1’ is a key site for the dynein-LIS1/Pac1 interaction (Gutierrez et al., 2017; Toropova et al., 2014), while ‘site 2’ has also been implicated in regulating dynein (DeSantis et al., 2017) (see text). It is also worth noting that multi-color single-molecule imaging experiments revealed that the majority of motile mammalian DDA or yeast dynein complexes, stably bind only one LIS1/Pac1 dimer (presumably bound to a single motor domain), although the binding of two LIS1 dimers to motile DDA or yeast dynein complexes has also been observed (Gutierrez et al., 2017; Marzo et al., 2020). (D) Sequence alignment of the dynein-interacting region of LIS1 (indicated in green in panel B, right) and its homologs from the indicated species. Note the presence of two small groups (three amino acids each) of nearly invariant residues (in magenta) that cluster around the two residues shown to be important for binding to dynein and PAF-AH (Gutierrez et al., 2017; Tarricone et al., 2004; Toropova et al., 2014).
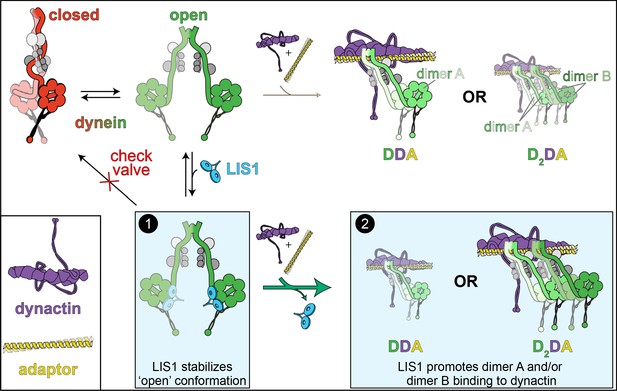
The ‘catalytic check valve’ model for LIS1 function.
The new model for LIS1 function posits the following mode of action: dynein stochastically switches from closed (phi conformation, red), to open states (green; accessory chains shown in gray for both states) (Marzo et al., 2020). Adopting the latter state is likely a rate-limiting step in assembly of dynein-dynein-adaptor (DDA) complexes (Zhang et al., 2017). LIS1/Pac1 preferentially binds dynein in the open state (Htet et al., 2020; Marzo et al., 2020), and acts as a ‘check valve’ that prevents the return of dynein to the phi conformation (Box 1). In so doing, LIS1/Pac1 promotes assembly of active, processive DDA (with only one dimer, dimer A) and D2DA complexes (with dimers A and B). The probability of D2DA forming may be the product of the probability of a DDA complex assembling times itself (pDDA x pDDA); however, it is possible that the addition of the 2nd dynein dimer (dimer B) occurs cooperatively due to the number of apparent contacts between the two dynactin-bound dynein dimers (Urnavicius et al., 2018), in which case the probability of D2DA assembly is greater than pDDA2. In either case, by increasing the probability of DDA complex assembly, addition of LIS1 also increases the probability of D2DA assembly, as has been observed in vitro (Elshenawy et al., 2020; Htet et al., 2020). In addition to stabilizing the open conformation, LIS1 also promote DDA/D2DA complex by an unknown mechanism that may involve LIS1-mediated dynein allostery, or linking dimer B to dimer A (Box 2; see text). It is interesting to note that LIS1 does not need to comigrate with dynein to affect DDA (or D2DA) motility; in fact, studies show that LIS1/Pac1 does not localize with dynein cargoes in cells, and may dissociate in a regulated manner prior to initiation of cargo transport (Jha et al., 2017; Lammers and Markus, 2015). In this way, LIS1 acts catalytically to increase DDA and D2DA assembly.
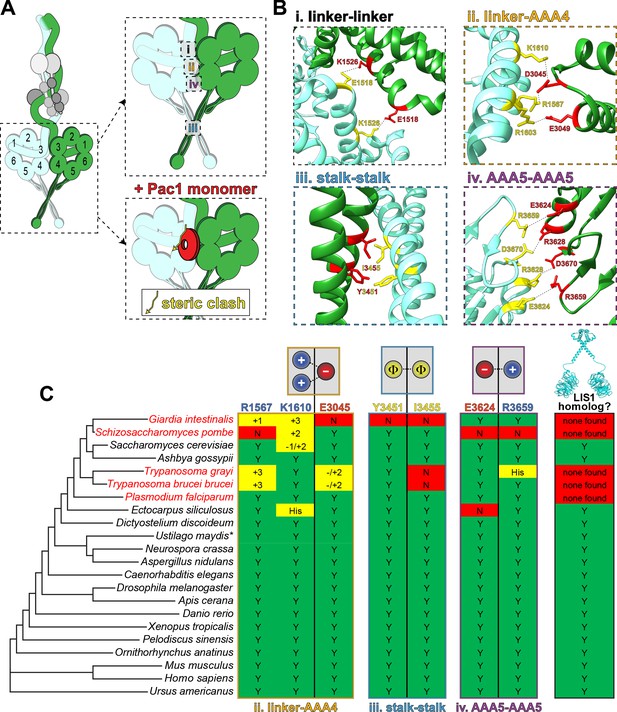
Dynein’s phi particle conformation and its potential coevolution with LIS1 function.
(A) A cartoon model for the dynein phi particle (green and aquamarine, heavy chains with AAA domains numbered; gray, accessory chains), with the four intermolecular contact points identified by cryoEM indicated. Bottom panel illustrates the steric clash (indicated by jagged yellow line) between a dynein-bound LIS1/Pac1 monomer (red, bound to the dynein motor domain in green), and the 2nd motor domain (in aquamarine; adapted from Marzo et al., 2020). (B) High resolution cryoEM structures of the four intermolecular contact points that stabilize the human dynein phi particle (adapted from Zhang et al., 2017). Note that regions ii, iii and iv have been experimentally validated, while region i has not yet been tested for its contribution to phi particle assembly (Marzo et al., 2020; Zhang et al., 2017). (C) Phylogenetic tree of cytoplasmic dynein heavy chains from the indicated species, and summary of the degree of their conservation (or lack thereof) for the three validated intermolecular contact points that stabilize the phi particle (only those sequences annotated as cytoplasmic dynein-1 homologs were selected for this analysis; see Figure 4—figure supplement 1 for sequence alignments of all four regions; *note that the dynein heavy chain from Ustilago maydis is encoded by two separate genes, dyn1 and dyn2, which encode approximately 2/3rd and 1/3rd of the motor, respectively [Straube et al., 2001]). The residues listed above (for the human dynein heavy chain) are color coded according to their chemical properties, with their mode of interaction depicted in the cartoons (blue, positively charged residues, ‘+”; red, negatively charged residues, ‘-‘; yellow, hydrophobic residues, ‘Φ”). Degree of conservation for each residue is indicated as ‘Y’ (green, yes, conserved), ‘N’ (red, not conserved), or with a +/- value (yellow) indicating a potentially conserved residue up to three positions N- or C-terminally situated, respectively. ‘His’ indicates the presence of a histidine in these respective positions, which has the potential to form a salt bridge depending on the local microenvironment. The column on the right indicates whether a clear LIS1 ortholog was identified by sequence alignment or genome annotation (‘Y’, homolog present). Species names indicated in red are those in which no LIS1 homolog was identified. Note that the five species with no apparent LIS1 homolog exhibit a low predicted propensity to adopt the phi particle, as determined by the sequence alignments.
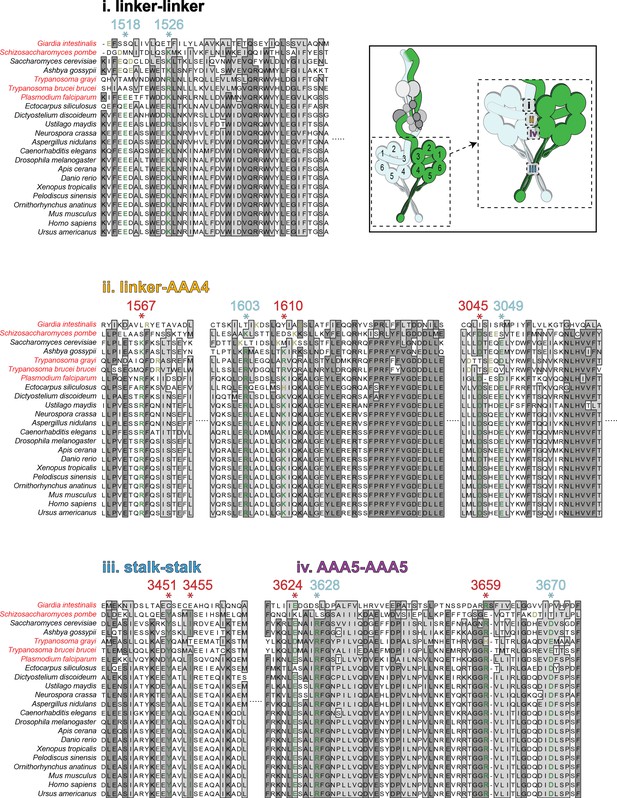
Sequence alignments of the intermolecular contact regions for cytoplasmic dyneins from disparate species.
Alignments of the four predicted intermolecular contact points that stabilize the phi particle conformation (listed in order from surfaces 1 through 4, as indicated). Cytoplasmic dynein-1 heavy chain sequences were identified based on sequence annotations. The numbered residues above each line (with *) denote the residues that mediate the intermolecular interactions (red and cyan depict residues experimentally validated, and those not yet tested, respectively; those in red were used to generate the table shown in Figure 4C). See Figure 4B for precise predictions of interacting residues. For each starred residue, a conserved residue in each ortholog is denoted in green, and potentially conserved residues (offset by up to three positions in the N- or C-terminal direction) are denoted in yellow. If no potentially conserved residue was identified, the residue is denoted in black. Species names in red indicate those species with no apparent LIS1 homolog.





