Microbiota-derived short chain fatty acids modulate microglia and promote Aβ plaque deposition
Figures
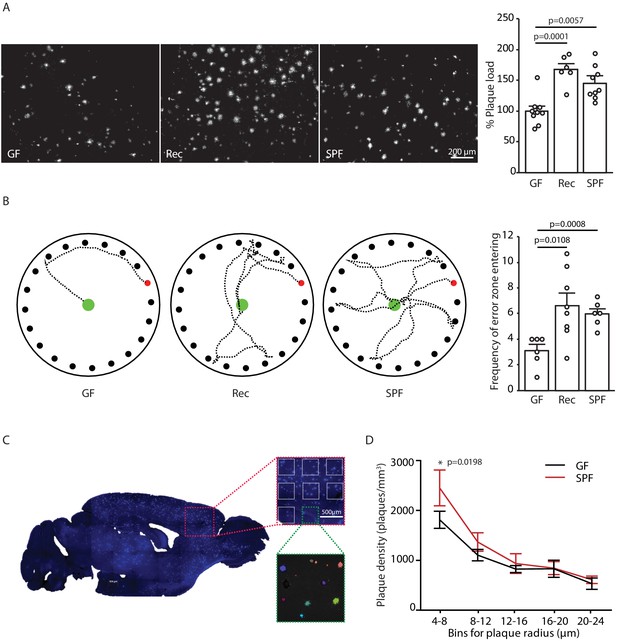
Germ-free APPPS1 mice show reduced Alzheimer’s disease (AD) pathology.
(A) Representative images and analysis of brain cortices from 5 months old germ-free (GF), naturally recolonized (Rec) and conventionally colonized (specific pathogen-free [SPF]) APPPS1 mice immunostained for Aβ (clone 6E10). Quantification of parenchymal plaque load reveals significantly reduced Aβ burden in GF mice compared to the Rec and SPF groups. Values are expressed as percentages of amyloid plaque area and normalized to GF group (ANOVA; n(GF) = 9, n(Rec) = 6, n(SPF) = 9). (B) Representative results and quantification from Barnes maze behavioral analysis of 5 months old GF, Rec and SPF APPPS1 mice. Quantification of frequency of error zone entering in the Barnes maze reveals a better performance in GF mice compared to Rec and SPF mice (ANOVA; n(GF) = 6, n(Rec) = 8, n(SPF) = 6). (C) Sagittal overview image indicating the analysis ROIs in the frontal cortex (blue: Methoxy-X04-positive plaques) and representative image demonstrating segmentation of Methoxy-X04 fluorescence intensity into individual plaques. The images show maximal intensity projections. Individual plaques are labeled with different colors. (D) Frequency distribution of plaque radius in 5 months old GF and SPF APPPS1 mice (two-way ANOVA; n(GF) = 5, n(SPF) = 5). All data are derived from at least three individual experiments and presented as mean ± SEM.
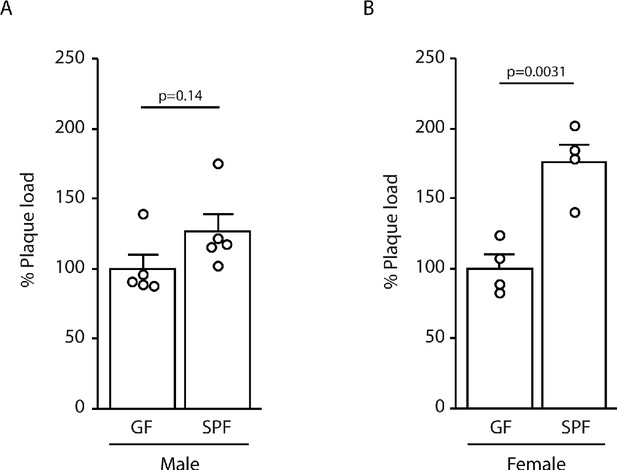
Sex-specific analysis of amyloid plaque load.
Plaque load of animals depicted in Figure 1A was individually analyzed in male (A; n = 5 per group) and female (B; n = 4 per group) mice, revealing a similar trend for increased plaque load in specific pathogen-free (SPF) mice compared to germ-free (GF) mice. Values are expressed as percentages of amyloid plaque area and normalized to GF group (unpaired T-test; at least three individual experiments).
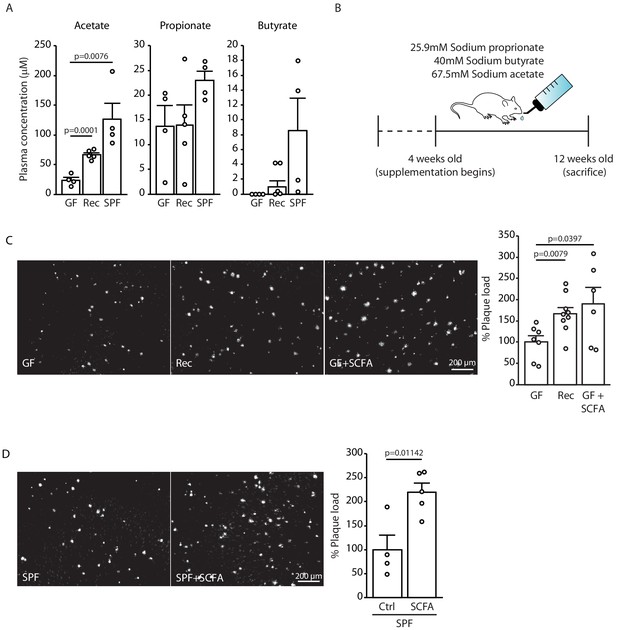
Short chain fatty acids (SCFA) are mediators of Aβ plaque deposition.
(A) Plasma SCFA concentrations of acetate, butyrate, and propionate were quantified using GC/MS-based metabolomics analysis, showing an increase in specific pathogen-free (SPF) compared to germ-free (GF) mice (ANOVA; n(GF) = 4, n(Rec) = 5, n(SPF) = 4; two individual experiments). (B) Experimental plan for SCFA supplementation in GF mice. Four weeks old GF mice have been treated with SCFA in drinking water for 8 weeks. (C) Representative images from GF (control-treatment), Rec, and GF supplemented with SCFA mice showing a significant increase in Aβ plaque load upon SCFA administration (ANOVA; n(GF) = 7, n(Rec) = 9, n(GF+SCFA) = 6; three individual experiments). (D) SPF APPPS1 mice were supplemented with control or SCFA in drinking water for 4 weeks (from 8 to 12 weeks of age). Histological analysis revealed a significantly increased plaque load in SCFA-supplemented SPF mice compared to control treatment (unpaired T-test; n(Ctrl) = 4, n(SCFA) = 5; one individual experiment). All data in this figure are presented as mean ± SEM.
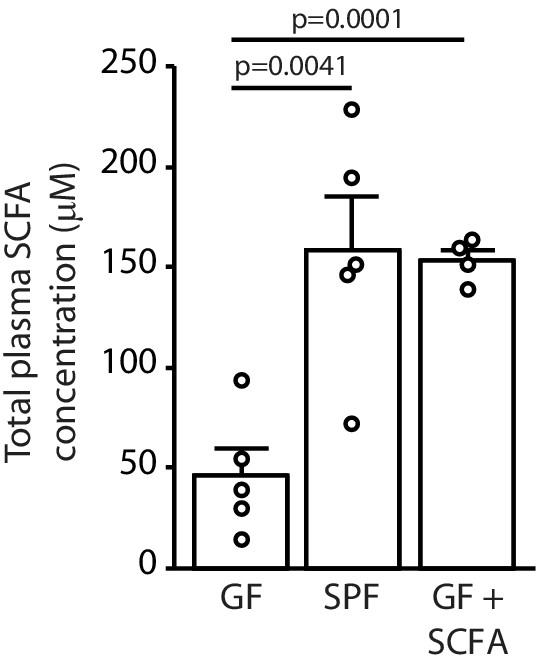
Total short chain fatty acids (SCFA) concentrations.
Total plasma SCFA concentrations of acetate, butyrate, and propionate in specific pathogen-free (SPF), germ-free (GF), and GF+SCFA treated mice (ANOVA; n(GF) = 5, n(SPF) = 5, n(GF+SCFA) = 4; two individual experiments). Data are presented as mean ± SEM.
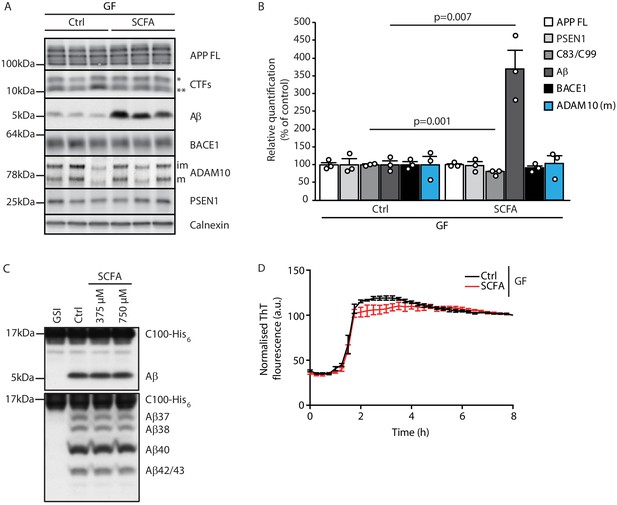
Short chain fatty acids (SCFA) mildly increase amyloidogenic processing.
(A) Western blot analysis and (B) its densitometry quantification of 3 months old brain homogenates of control (Ctrl)- and SCFA-supplemented germ-free (GF) APPPS1 animals. The Aβ level is significantly increased in SCFA group in comparison to Ctrl, despite unaffected APP FL levels. APP CTFs show a decreased C83 (**) to C99 (*) ratio. We could not detect alterations in protein levels of secretases involved in APP processing (ADAM10, BACE1, and γ-secretase/PSEN1). m = ADAM10 mature form; im = ADAM10 immature form. Data represent mean ± SEM (unpaired T-test; n(Ctrl) = 3, n(SCFA) = 3). (C) Upper panel: γ-Secretase reconstituted into lipid vesicles was incubated at 37°C together with the C99-based substrate C100-His6 in the presence of increasing doses of a SCFA mixture (375 and 750 μM final concentration of total SCFA of an equimolar mixture of Na-acetate, Na-propionate, and Na-butyrate) for 24 hr. Production of Aβ was analyzed by immunoblotting. γ-Secretase inhibitor (GSI) L-685,458 (0.4 μM) was used as a negative control. No alterations in Aβ levels were detected in the presence of SCFA. Lower panel: Qualitative analysis of individual Aβ species via Tris‐Bicine‐Urea SDS‐PAGE reveals that SCFA treatment does not alter the ratio among the different Aβ species (Aβ37-38-40-42/43) suggesting no direct effects on modulation of γ-secretase cleavage. (D) Aggregation kinetics of monomeric Aβ40 recorded by the increase in fluorescence of Thioflavin T incubated with either 30 mM NaCl (Ctrl) or 30 mM SCFA mixture do not show any significant difference, suggesting that SCFA do not directly modify Aβ fibrillarization. Data points represent mean ± SD from three independent experiments.
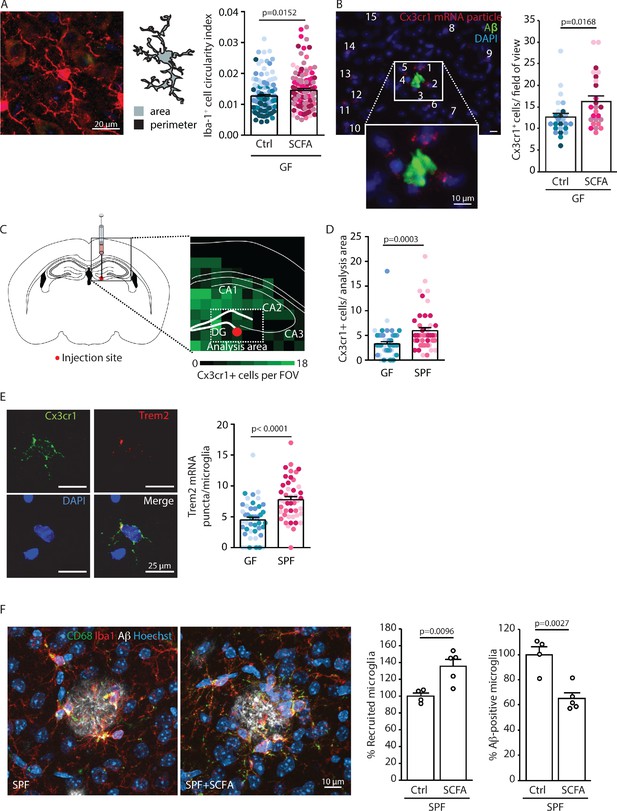
Short chain fatty acids (SCFA) modulate microglia.
(A) Morphological analysis of microglia shows an increase in the circularity index, indicating a more activated phenotype, in SCFA- compared to control-treated germ-free (GF) APPPS1 mice. Iba1 (red) has been used as microglial marker. For each group, each shade of color represents the microglia from a single mouse (U test; n(Ctrl) = 6, n(SCFA) = 5). (B) smFISH analysis of microglial cells (red, Cx3cr1 mRNA particles) surrounding amyloid plaques (green, anti-Aβ clone 2D8) shows an increased number of Cx3cr1-positive cells (>4 puncta) clustering around Aβ plaques in SCFA- compared to control-supplemented GF APPPS1 mice. DAPI (blue) was used as nuclear dye (U test; n(Ctrl) = 5, n(SCFA) = 5 mice, 5 FOV per mouse). (C) Experimental outline for APPPS1 brain homogenate injection into a WT mouse brain showing the injection site (red dot). (D) smFISH analysis of Cx3cr1-positive cells (>4 puncta) surrounding the APPPS1 brain homogenate injection site (analysis area relative to injection site for all brains) showing an enhanced recruitment of microglial cells in specific pathogen-free (SPF) versus GF WT mice. (E) APPPS1 brain homogenate injection induces higher microglial activation in SPF in comparison to GF WT mice as shown by the higher amount of Trem2 mRNA puncta (red) per Cx3cr1-positive (green) microglia. DAPI (blue) was used as nuclear dye. In D and E, each different shade of color represents the microglia from a single mouse (U test, n(GF) = 3, n(SPF) = 3; two individual experiments, 120 analyzed images (with multiple microglia) per mouse). (F) Microglial recruitment and Aβ plaque uptake were histologically quantified in SCFA- and control-supplemented SPF APPPS1 mice. Although significantly more CD68-positive (green) and Iba1-positive (red) microglial cells were located at Aβ plaques (white, anti-Aβ clone 3552), the number of Aβ-positive microglia was significantly reduced in the SCFA- compared to the control-treated group. Values are expressed as percentages and normalized to the control-treated group (unpaired T-test; n(Ctrl) = 4, n(SCFA) = 5; one individual experiment).
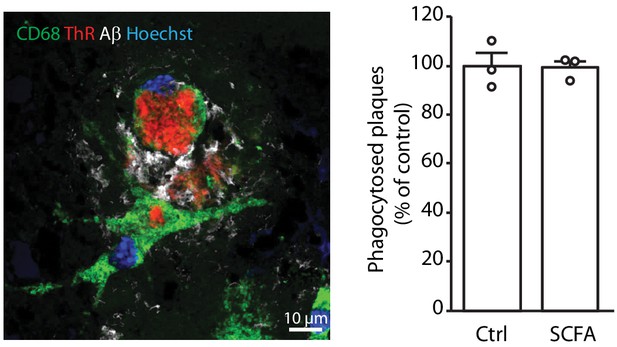
Ex vivo amyloid plaque clearance assay.
Primary WT microglia treated with short chain fatty acids (SCFA; 250 µM) do not show alteration in their phagocytic capacity toward Aβ plaques in comparison to the control-treated cells (Ctrl). CD68 (green) was used to visualize microglia. Amyloid plaques were visualized using both Thiazine red (ThR, red, fibrillar Aβ) and an anti-Aβ antibody (3552, white, total Aβ). DAPI (blue) was used as nuclear dye. Values are expressed as percentages of phagocytosed Aβ plaques normalized to the control-treated group and presented as mean ± SEM (unpaired T-test; n(Ctrl) = 3, n(SCFA) = 3; three independent experiments with two technical replicates per experiment).
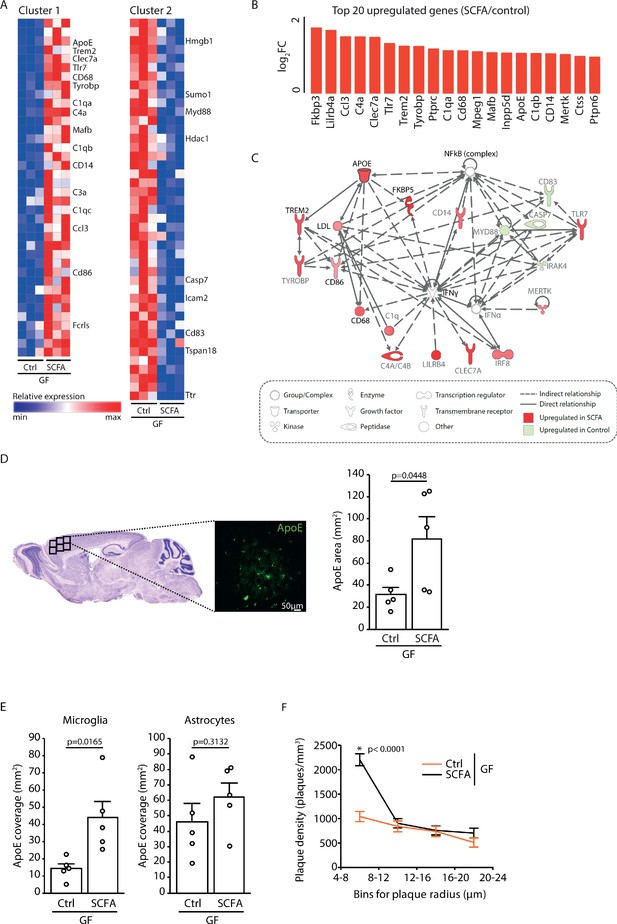
Increased ApoE expression marks microglial activation upon short chain fatty acids (SCFA) supplementation.
(A) Heatmap of Nanostring transcriptomic analysis from control- and SCFA-supplemented germ-free (GF) APPPS1 mice at 3 months of age. Row values were scaled using unit variance scaling. Genes previously associated with microglial function have been annotated on the heatmap. Three mice per group have been analyzed. (B) Top 20 upregulated genes in SCFA- versus control-treated samples. Most of transcriptome hits have been previously associated with microglial activation. (C) Functional gene interaction network analysis using Ingenuity Pathway Analysis. Genes are colored based on fold-change values determined by RNA-Seq analysis, where red indicates an increase in SCFA- and green in control-treated animals. Network analysis revealed upregulation of the ApoE-TREM2 axis as one of the principal biological pathways upregulated by SCFA. (D) Representative sagittal brain section indicating location of the analyzed region of interest in the frontal cortex and representative image showing ApoE (green) distribution. Quantification of ApoE signal showed a SCFA-dependent increase of ApoE expression (unpaired T-test; n(Ctrl) = 5, n(SCFA ) = 5; three individual experiments). (E) Quantification of ApoE colocalization (absolute coverage area in mm2) with microglia and astrocytes in control- and SCFA-supplemented GF APPPS1 mice at 3 months of age (unpaired T-test; n(Ctrl) = 5, n(SCFA) = 5; two individual experiments). (F) Analysis of Methoxy-X04-stained brain sections showed a specific increase in plaques of smaller sizes (4–8 µm radius) in 3 months old SCFA- compared to Ctrl-supplemented GF APPPS1 mice (unpaired T-test per bin; n(Ctrl) = 5, n(SCFA) = 5; three individual experiments). Data represent mean ± SEM.
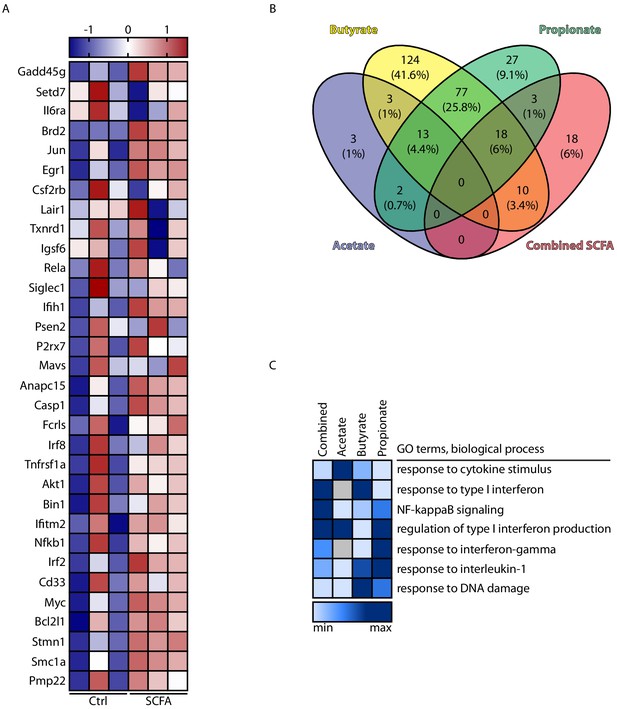
Effect of individual short chain fatty acids (SCFA) on microglial polarization.
Primary microglial cells isolated from specific pathogen-free (SPF) WT mice were cultured and treated with the individual SCFA acetate, butyrate, and propionate, the combined SCFA or the respective control (NaCl). Effect of SCFA on microglial polarization was tested by Nanostring analysis of the microglial transcriptome as in Figure 5 (n = 3 samples per group; data is combined from two individual experiments comparing combined SCFA with Ctrl and individual SCFA to Ctrl, respectively). (A) Heatmap of significantly regulated genes between control and combined SCFA treatment. Row values were scaled using unit variance scaling. (B) Venn diagram for common genes that were significantly regulated (in comparison to the respective salt control) for the individual and combined SCFA treatments. (C) Heatmap for scores of biological processes from gene ontology (GO) analysis (combined scores of p-value and z-score per GO term). Color code indicates min to max values per GO term, and gray color indicates not regulated pathway.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | APPPS1 | doi:10.1038/sj.embor.7400784 | MGI:3765351 | C57BL/6 background |
| Antibody | Anti-IBA1 (guinea pig polyclonal) | SYSY | Cat# 234 004, RRID:AB_2493179 | IHC (1:500) |
| Antibody | Anti-IBA1 (rabbit polyclonal) | Wako | Cat#:019–19741, RRID:AB_839504 | IHC (1:500) |
| Antibody | Anti-CD68 (rat monoclonal) | Bio-Rad | Cat#:MCA1957G, RRID:AB_324217 | IHC (1:500), WB (1:1000) |
| Antibody | Anti-Amyloid Y188 (rabbit monoclonal) | Abcam | Cat#:1565–1, RRID:AB_562042 | WB (1:1000) |
| Antibody | Anti-Amyloid beta 3552 (rabbit polyclonal) | doi:10.1523/JNEUROSCI.5354–05.2006 | IHC (1:500), WB (1:2000) | |
| Antibody | anti-beta-Amyloid, 6E10 (mouse monoclonal) | BioLegend | Cat# 803002, RRID:AB_2564654 | IHC (1:500) |
| Antibody | Anti-Presenilin 1 (NT1) (mouse monoclonal) | BioLegend | Cat# SIG-39194–500, RRID:AB_10720504 | WB (1:1000) |
| Antibody | Anti-Human Adam10 (mouse monoclonal) | R and D Systems | Cat# MAB1427, RRID:AB_2223057 | WB (1:1000) |
| Antibody | Anti-BACE1 (rabbit monoclonal) | Epitomics | Cat# 2882–1, RRID:AB_2061494 | WB (1:1000) |
| Antibody | Anti-Calnexin (rabbit monoclonal) | Stressgen | Cat# ADI-SPA-860, RRID:AB_10616095 | WB (1:1000) |
| Chemical compound, drug | Sodium butyrate 98% | Sigma Aldrich | 303410 | In vitro: 250 μM In vivo: 40 mM |
| Chemical compound, drug | Sodium propionate ≥ 99.0% | Sigma Aldrich | P1880 | In vitro: 250 μM In vivo: 25.9 mM |
| Chemical compound, drug | Sodium acetate anhydrous ≥ 99% | Sigma Aldrich | S2889 | In vitro: 250 μM In vivo: 67.5 mM |
| Antibody | Anti-Amyloid beta 2D8 (rat polyclonal) | DOI: 10.1016/j.nbd.2007.04.011 | WB (1:1000) IHC (1:300) | |
| Antibody | Anti-P2Y12 (rabbit polyclonal) | AnaSpec | Cat# 55043A, RRID:AB_2298886 | IHC (1:100) |
| Antibody | Anti-GFAP (rabbit polyclonal) | Dako | Cat# Z0334, RRID:AB_10013382 | IHC (1:200) |
| Antibody | Anti-APOE (mouse monoclonal) | DOI 10.1172/JCI96429 | Clone HJ6.3B | IHC (1:50) |
| Chemical compound, drug | Thiazine red | VWR Chemicals | 27419.123 | IHC (2 μM) |
| Chemical compound, drug | Hoechst 33342 | ThermoFisher | H3570 | IHC (1:2000) |





