Common cell type nomenclature for the mammalian brain
Figures
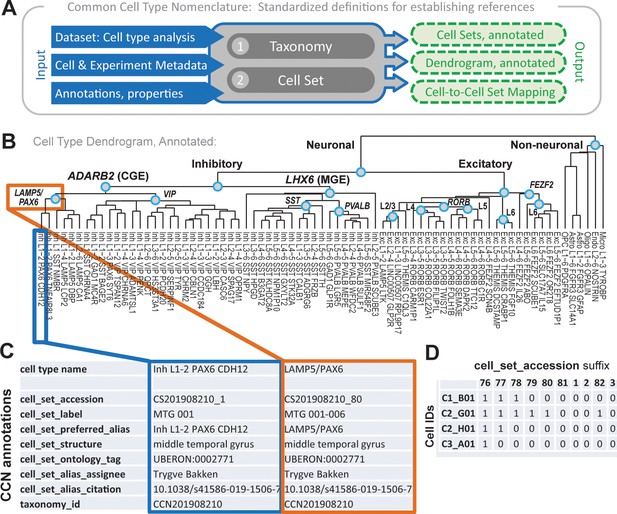
Overview of common cell type nomenclature (CCN) and application to human middle temporal gyrus (MTG).
(A) Schematic of CCN components and process. (B–D) Example outputs from the CCN. (B) Annotated dendrogram of cell types in human MTG, along with associated cell type names, reproduced from Hodge et al., 2019. Internal nodes with a term (teal circles) represent cell sets with preferred alias tags. (C) CCN annotations for a putative cell type (outlined in blue) and an internal node (outlined in orange) of this dendrogram. (D) Snippet of an output file from the CCN showing cell to cell set mappings as applied to human MTG.
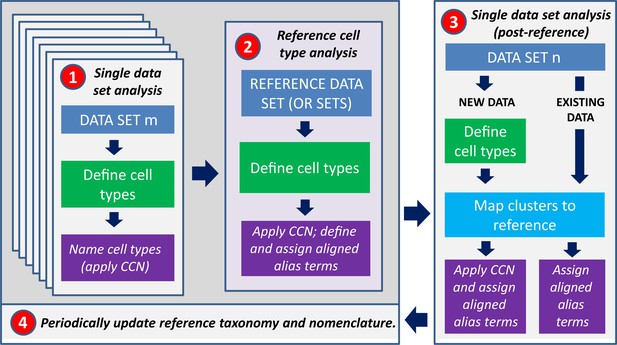
Workflow for assigning types to a given dataset with taxonomy.
(1) Cell type classification will initially be performed separately on all taxonomies. (2) One, some, or all of these datasets will be combined into a high-confidence reference taxonomy which can be used as a comparator for any related datasets, by (3) mapping existing and new datasets to the reference taxonomy. (4) The reference will periodically be updated as new datasets and taxonomies are generated.
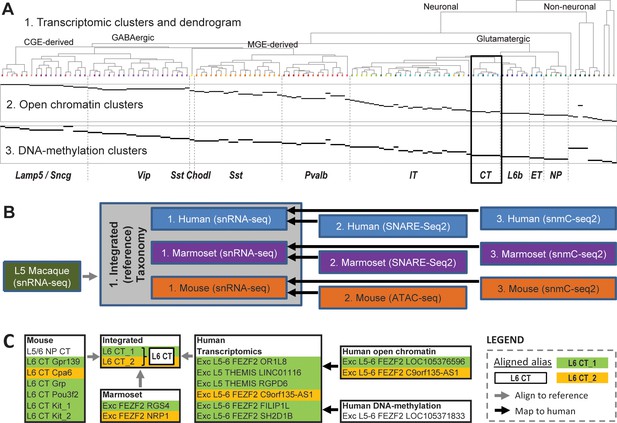
Series of multimodal, cross-species taxonomies in primary motor cortex (M1) demonstrates utility of nomenclature schema.
(A) Taxonomies based on transcriptomic (‘1’; top), open chromatin (‘2’; middle), and DNA methylation (‘3’; bottom) in human M1. Epigenomic clusters (‘2’, ‘3’; in rows) aligned to RNAseq clusters (‘1’) as indicated by horizontal black bars and are also assigned matching cell sets in the relevant taxonomies. Adapted from Bakken et al., 2020a. (B) Flow chart showing all 11 taxonomies generated for this project and their connections. The integrated (reference) taxonomy included nuclei collected using snRNA-seq from three species (gray box), with nuclei collected from layer five in macaque mapped to this space post hoc (gray line). Separately, epigenetics taxonomies from human, marmoset, and mouse were aligned to their respective transcriptomics taxonomies (black lines). This entire taxonomic structure is captured by the CCN (see Supplementary file 1). (C) An example mapping of corticothalamic (L6 CT) provisional cell types across the human and transcriptomics taxonomies using the CCN (black box in A). Preferred aliases for each taxonomy are used for clarity.
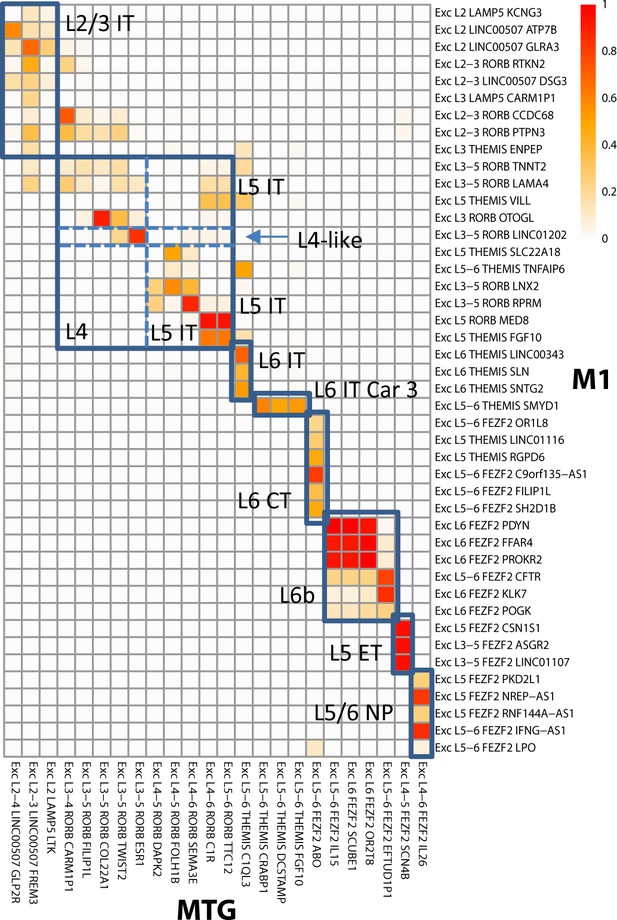
Alignment of glutamatergic cell sets in human middle temporal gyrus (MTG) to a reference primary motor cortex (M1) taxonomy.
Cluster overlap heatmap showing the proportion of nuclei from MTG clusters and the reference (M1) clusters that coalesce with a given aligned cluster. Cell sets corresponding to aligned aliases in the MTG and M1 taxonomies are labeled and indicated by blue boxes. Adapted from Bakken et al., 2020a.
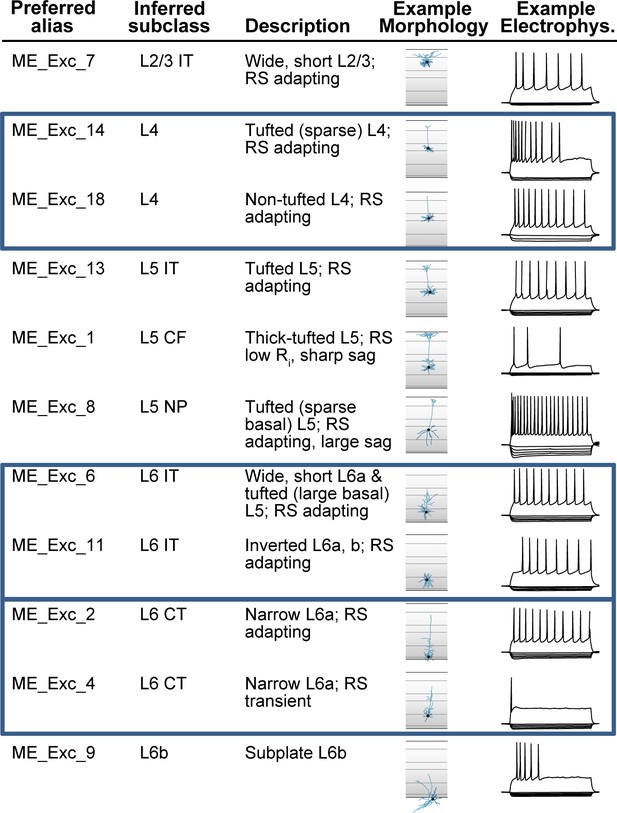
Application of common cell type nomenclature (CCN) to glutamatergic me-types in the mouse visual cortex.
Excitatory (glutamatergic) me-types from Gouwens et al., 2019 that have been incorporated into the nomenclature schema. Eleven of the original 20 excitatory me-types are shown as examples. Representative morphologies and electrophysiological responses are shown to illustrate the differences between types. The ‘inferred subclass’ calls perfectly map to cell set aligned aliases from the reference M1 taxonomy in Figure 3, except that L5 CF (corticofugal) is an additional alias for L5 ET, and cells sets corresponding to L4, L6 IT, and L6 CT (blue boxes) have been added to the taxonomy.
Tables
Glossary of terms.
Terminology used with the common cell type nomenclature (CCN), definitions for use, and examples of how terms are applied. Terms are presented in bold upon first use in the text. This glossary is intended to clarify use for the purposes of the CCN since some terms are open to multiple interpretations, and effective classification requires disambiguation. Asterisks denote terms that represent specific components of the CCN.
| Term | Definition | Example |
|---|---|---|
| Taxonomy | Set of quantitatively derived data clusters defined by a specific computational algorithm on a specific dataset(s). Taxonomies are given a unique label and can be annotated with metadata about the taxonomy, including details of the algorithms and relevant cell and cell set IDs. | Any clustering result in a cell type classification manuscript |
| Dataset | Feature information (e.g., gene expression) and associated metadata from a set of cells collected as part of a single project. | Gene expression from 6000 human MOp nuclei |
| Ontology | A structured controlled vocabulary for cell types. | Cell Ontology |
| Marker gene(s) | A gene (gene set) which, when expressed in a cell, can be used to accurately assign that cell to a specific cell set. | GAD2; PVALB; CHODL |
| Taxonomy ID* | An identifier uniquely tagging a taxonomy of the format CCN[YYYYMMDD][#]. | CCN201910120 |
| Cell | A single entry in a taxonomy representing data from a single cell (or cell compartment, such as the nucleus). Cells have metadata including a unique ID. | N/A |
| Cell set | Any tagged group of cells in a taxonomy. This includes cell types, groups of cell types, and potentially other informative groupings (e.g., all cells from one donor, organ, cortical layer, or transgenic line). Cell sets have several IDs and descriptors (as discussed below) and can also have other metadata. | A cell type; a group of cell types; all cells from layer two in MTG; all cells from donor X |
| Provisional cell type | Quantitatively derived data cluster defined within a taxonomy. This is a specific example of a cell set that is of high importance, as most other cell sets are groupings of one or more provisional cell types. Here, the term ‘cell type’ is synonymous with ‘provisional cell type.’ . | A cell type defined in a specific study |
| Dendrogram | A hierarchical organization of provisional cell types defined for a specific taxonomy. Dendrograms have a specific semantic and visualizable structure and include nodes (representing multiple provisional cell types) and leaves (representing exactly one). Not all taxonomies include a dendrogram (e.g., if the structure of cell sets is non-hierarchical). | N/A |
| Community structure | Non-hierarchical relationships between cell types defined as groups of cell types in a graph. | N/A |
| Cell set accession ID* | A unique ID across all tracked datasets and taxonomies. This tag labels the taxonomy and numbers each cell type. CS[taxonomy id]_[unique # within taxonomy] | CS201910120_1 |
| Cell set label* | An ID unique within a single taxonomy that is used for assigning cells to cell sets defined as a combination of multiple ‘provisional cell types’. | MTG 12 MTG 01–08 |
| Cell set alias* | Any cell set descriptor. It can be defined computationally from the data, or manually based on new experiments, prior knowledge, or a combination of both. Cell aliases beyond the ‘preferred’ or ‘aligned’ are defined as ‘cell set additional aliases’. | (Any ‘cell set aligned alias’); Interneuron 1; Rosehip |
| Cell set preferred alias* | The primary cell set alias (e.g., what cell types might be called in a publication). This can sometimes match the aligned alias, but not always, and can be left unassigned. | Inh L1-2 PAX6 CDH12; ADARB2 (CGE); Chandelier; [blank] |
| Cell set aligned alias* | Analogous to ‘gene symbol’. At most one biologically driven name for linking matching cell sets across taxonomies and with a reference taxonomy. | L2/3 IT 4; Pvalb 3; Microglia 2 |
| Cell set structure* | The location in the brain (or body) from where cells in the associated set were primarily collected. | Neocortex |
| Cell set ontology tag* | A tag from a standard ontology (e.g., UBERON) corresponding to the listed cell set structure. | UBERON:0001950 |
| Cell set alias assignee* | Person responsible for assigning a specific cell set alias in a specific taxonomy (e.g., the person who built the taxonomy or uploaded the data, or a field expert). | (First author of manuscript) |
| Cell set alias citation* | The citation or permanent data identifier corresponding to the taxonomy where the cell set was originally reported. | (Manuscript DOI); [blank] |
| Reference taxonomy | A taxonomy based on one or a combination of high-confidence datasets, to be used as a baseline of comparison for datasets collected from the same organ system. | Cross-species cortical cell type classification |
| Morpho-electric(ME) type | A provisional cell type defined using a combination of morphological and electrophysiological features. | ME_Exc_7 |
| Governing body | A forum of subject-matter experts to guide policy and manage change of the CCN and associated ontologies and databasing efforts. | N/A |
Proposed strategy for naming cortical cell types.
| Class | Format | Example |
|---|---|---|
| Glutamatergic | [Layer] [Projection] # | L2/3 IT 4 |
| GABAergic | [Canonical gene(s)] # | Pvalb 3 |
| Non-neuronal | [Cell class] # | Microglia 2 |
| Any class | [Historical name] # | Chandelier 1 |
Nomenclature for ‘Sst Chodl’ cell sets cited in Bakken et al., 2020a.
Relevant common cell type nomenclature (CCN) entities and taxonomy metadata, including the cell set additional alias that links to cell set labels from relevant transcriptomics taxonomies. All listed cell sets have a cell set structure of ‘primary motor cortex’ and a cell set ontology tag of ‘UBERON:0001384’.
| # | Cell set preferred alias | Cell set label | Cell set accession | Cell set aligned alias | Cell set additional alias |
|---|---|---|---|---|---|
| 1 | Inh L1-6 SST NPY | RNA-seq 040 | CS201912131_40 | Sst Chodl | |
| 2 | Inh L1-5 SST AHR | DNAm 12 | CS202002272_12 | RNA-seq 040, 046–047, 050–052, 068 in CCN201912131 | |
| 3 | Inh L1-6 SST NPY | ATAC-seq 08 | CS202002273_8 | Sst Chodl | RNA-seq 040 in CCN201912131 |
| 4 | Inh SST NPY | RNA-seq 01 | CS201912132_1 | Sst Chodl | |
| 5 | Sst Chodl | RNA-seq 028 | CS202002013_28 | Sst Chodl | |
| 6 | Sst Chodl | DNAm 09 | CS202002276_9 | Sst Chodl | RNA-seq 028 in CCN202002013 |
| 7 | Sst Chodl | ATAC-seq 10 | CS202002277_10 | Sst Chodl | RNA-seq 028 in CCN202002013 |
| 8 | Sst Chodl | Integrated 14 | CS202002270_14 | Sst Chodl | Long-range projecting Sst |
| # | Cell set alias assignee | Cell set alias citation | Taxonomy id | Species | Modality |
|---|---|---|---|---|---|
| 1 | Nikolas Jorstad | 10.1101/2020.03.31.016972 | CCN201912131 | Human | RNA-seq |
| 2 | Wei Tian | 10.1101/2020.03.31.016972 | CCN202002272 | Human | DNAm |
| 3 | Blue Lake | 10.1101/2020.03.31.016972 | CCN202002273 | Human | ATAC-seq |
| 4 | Fenna Krienen | 10.1101/2020.03.31.016972 | CCN201912132 | Marmoset | RNA-seq |
| 5 | Zizhen Yao | 10.1101/2020.02.29.970558 | CCN202002013 | Mouse | RNA-seq |
| 6 | Hanqing Liu | 10.1101/2020.02.29.970558 | CCN202002276 | Mouse | DNAm |
| 7 | Yang Li | 10.1101/2020.02.29.970558 | CCN202002277 | Mouse | ATAC-seq |
| 8 | Nikolas Jorstad | 10.1101/2020.03.31.016972 | CCN202002270 | All | RNA-seq |
Taxonomies with applied CCN.
Table showing the set of taxonomies included in Supplementary file 1. All taxonomies include the annotated nomenclature table. Asterisk (*) and carrot (^) indicate that the updated dendrogram and cell to cell set mapping files are also included for that taxonomy, respectively. CCN202002270 is the reference taxonomy presented in Figure 3B.
| Taxonomy id | Description | Reference |
|---|---|---|
| CCN201810310^* | Mouse VISp + ALM (from the Tasic et al., 2018) | Tasic et al., 2018 |
| CCN201908210^* | Human MTG (from the Tasic et al., 2018) | Hodge et al., 2019 |
| CCN201908211^* | Joint mouse/human analysis (slight modification from Hodge et al., 2019) | Hodge et al., 2019 |
| CCN201912130 | Human M1 taxonomy using 10× data | Bakken et al., 2020a |
| CCN201912131 | Human M1 taxonomy using Smart-seq and 10x data | Bakken et al., 2020a |
| CCN201912132 | Marmoset M1 taxonomy using 10× data | Bakken et al., 2020a |
| CCN202002013* | Mouse MOp BICCN taxonomy using multiple RNAseq datasets | Yao et al., 2020a |
| CCN202002270 | Cross species (integrated) transcriptomics taxonomy | Bakken et al., 2020a |
| CCN202002271 | Macaque transcriptomics taxonomy, layer 5/6 only | Bakken et al., 2020a |
| CCN202002272 | Human DNA methylation taxonomy | Bakken et al., 2020a |
| CCN202002273 | Human ATAC-seq taxonomy | Bakken et al., 2020a |
| CCN202002274 | Marmoset DNA methylation taxonomy | Bakken et al., 2020a |
| CCN202002275 | Marmoset ATAC-seq taxonomy | Bakken et al., 2020a |
| CCN202002276 | Mouse DNA methylation taxonomy | Yao et al., 2020a |
| CCN202002277 | Mouse ATAC-seq taxonomy | Yao et al., 2020a |
| CCN202005150^ | Mouse inhibitory neurons in VISp defined using electrophysiology, morphology, and transcriptomics | Gouwens et al., 2020 |
| CCN201906170 | Mouse neurons in VISp defined using electrophysiology and morphology | Gouwens et al., 2019 |
| CCN201805250 | Turtle pallium transcriptomics taxonomy | Tosches et al., 2018 |
Additional files
-
Supplementary file 1
Output files from applying the CCN on 17 taxonomies.
This file contains annotated cell sets from all 17 taxonomies shown in Table 4 along with annotated dendrograms and cell to cell set assignments for a subset of these taxonomies. This file is available on GitHub (https://github.com/AllenInstitute/nomenclature).
- https://cdn.elifesciences.org/articles/59928/elife-59928-supp1-v2.zip
-
Supplementary file 2
A set of aligned aliase in mammalian M1, reproduced from Bakken et al., 2020a.
These terms are also applicable to other cortical areas, representing a starting point for future cell type classification efforts and for ontology curation. InterLex identifiers are provided in parentheses when available (Adkins et al., 2020).
- https://cdn.elifesciences.org/articles/59928/elife-59928-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59928/elife-59928-transrepform-v2.pdf





