CDK1 controls CHMP7-dependent nuclear envelope reformation
Figures
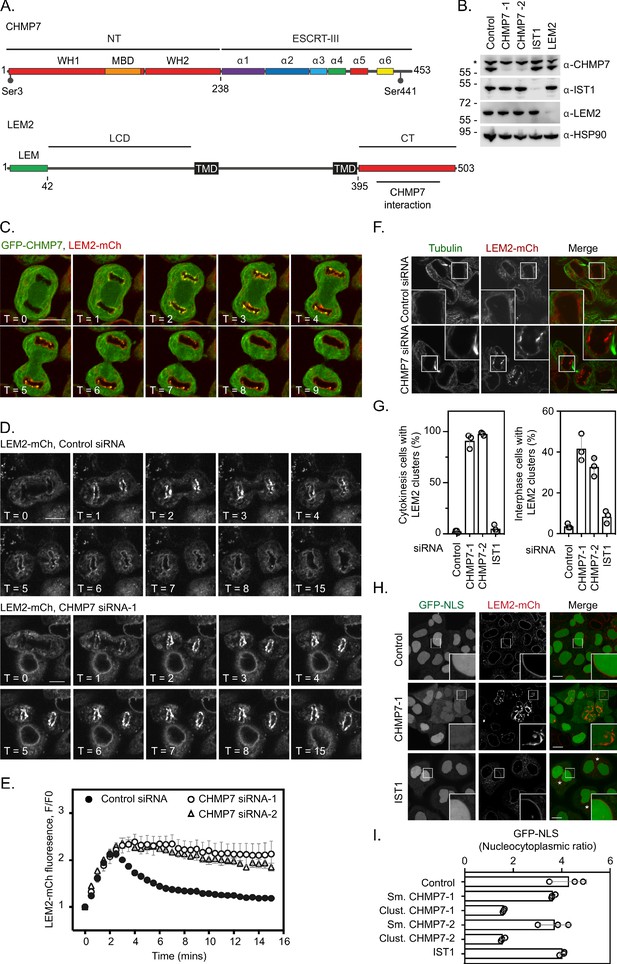
ESCRT-III controls LEM2 dynamics during nuclear envelope reformation.
(A) Cartoon depicting CHMP7 and LEM2 secondary structural elements. CHMP7 contains an ESCRT-II like N-terminus (NT) comprising tandem Winged Helix (WH) domains and a Membrane Binding Domain (MBD) required for ER localisation. Mitotically phosphorylated Serine residues identified in this manuscript are highlighted. LEM2 comprises an N-terminal LEM domain, a low-complexity domain (LCD) that has been shown to promote phase separation, two transmembrane (TM) domains that result in its insertion into the INM, and a C-terminal (CT) WH domain containing a region required for interaction with CHMP7 (415–485). (B) Resolved lysates from HeLa cells that had been transfected with the indicated siRNA were examined by western blotting with antibodies raised against CHMP7, IST1, LEM2, or HSP90, asterisk marks non-specific band. (C) HeLa cells stably expressing both GFP-CHMP7 and LEM2-mCh were imaged live during mitotic exit. Images representative of 4/4 (GFP-CHMP7, LEM2-mCh) Videos analysed. (D, E) HeLa cells stably expressing LEM2-mCh were treated with Control or CHMP7-targetting siRNAs and imaged live during mitotic exit. LEM2-mCh fluorescence levels in a region of interest (ROI) around the chromatin discs were quantified in E from 25 (Control) or 18 (CHMP7 siRNA-1) or 14 (CHMP7 siRNA-2) individually imaged cells acquired over three independent experiments, mean ± S.E.M. presented. Significance was calculated using a one-way ANOVA, with Dunnett’s post-hoc test, p < 0.0001 for both CHMP7 oligos compared to control siRNA. (F, G) HeLa cells stably expressing LEM2-mCh were treated with Control or CHMP7-targetting siRNA, fixed and processed for immunofluorescence. The number of cells displaying aberrant LEM2 clusters during cytokinesis or in interphase were quantified from 300 cells per condition, examined over three independent experiments and presented as mean ± S.E.M. (G). Significance was calculated using a 1-way ANOVA, with Dunnett’s post-hoc test; CHMP7 siRNA-1 p < 0.0001 (interphase), p = < 0.0001 (cytokinesis); CHMP7 siRNA-2 p = 0.0001 (interphase), p = < 0.0001 (cytokinesis); IST1 siRNA p = 0.456 (interphase, n.s.), p = 0.765 (cytokinesis, n.s.). In all panels, time in minutes and a scale bar of 10 μm are reported. (H, I) HeLa cells stably expressing GFP-NLS and LEM2-mCh were transfected with the indicated siRNA and imaged live. The nucleocytoplasmic ratio was quantified from cells treated with the indicated siRNA from three independent experiments and presented as mean ± S.E.M. (control, 295 cells; CHMP7 siRNA-1, 315 cells; CHMP7 siRNA-2, 195 cells; IST1 siRNA 215 cells). In CHMP7-depleted cells, the population was split into those cells displaying smooth LEM2 at the NE (Sm.) or those in which the LEM2 was clustered (Clust.). Significant breakdown in nucleocytoplasmic compartmentalisation was observed in the population of CHMP7-depleted cells displaying clusters (p = <0.0001 for CHMP7 siRNA-1 or CHMP7 siRNA-2 compared to controls, one-way ANOVA, with Dunnett’s post-hoc test).
-
Figure 1—source data 1
CHMP7-dependent dissolution of LEM2 clusters during M-exit.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig1-data1-v2.xlsx
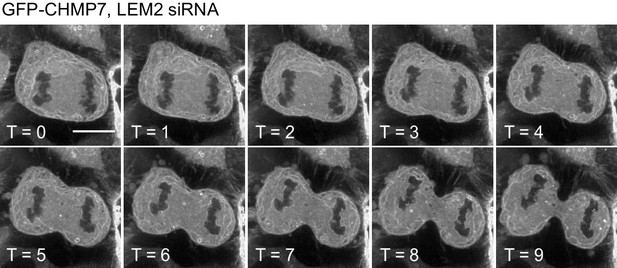
LEM2 is needed for CHMP7 recruitment to the reforming NE.
HeLa cells stably expressing GFP-CHMP7 and treated with LEM2-targetting siRNA were imaged live during mitotic exit. Images representative of 9/9 (LEM2-siRNA) Videos analysed, as described by Gu et al., 2017.
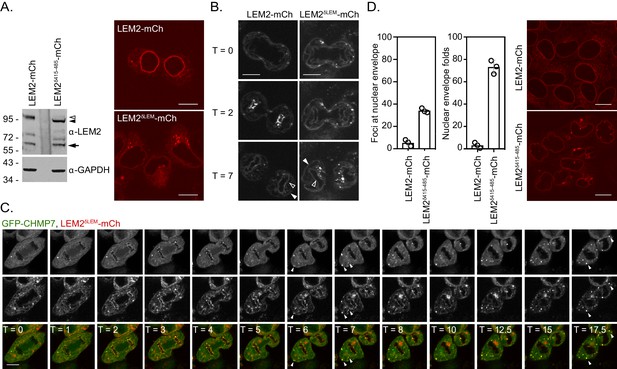
Activities within LEM2 controlling its assembly at the nuclear envelope.
(A) HeLa cells stably expressing LEM2-mCh or LEM2δLEM-mCh were lysed, resolved by SDS-PAGE and analysed by western blotting with anti-LEM2 or anti-GAPDH antisera endogenous LEM2, LEM2-mCh or LEM2δLEM-mCh indicated by arrow, open arrowhead or closed arrowhead, respectively. Alternatively, cells were imaged live. Images of LEM2δLEM-mCh representative of 35/40 cells. (B) Cells from A were imaged during mitosis. In addition to a failure to enrich at the reforming nuclear envelope, note the failure of LEM2δLEM-mCh to be retained at the INM (open arrowheads) relative to the peripheral ER (closed arrowheads). Images representative of 8/9 imaged cells. (C) HeLa cells stably expressing GFP-CHMP7 and LEM2δLEM-mCh were imaged live through mitosis. GFP channel presented in top row, mCh channel presented in middle row, merge presented in bottom row. Arrowheads indicated inappropriate extra-nuclear clustering of GFP-CHMP7 and LEM2δLEM-mCh. Note the limited NE enrichment of LEM2δLEM-mCh during nuclear envelope reformation and its sequestration in extranuclear clusters at the end of mitosis, rather than at the INM. (D) HeLa cells stably expressing LEM2-mCh or LEM2δ415-485-mCh were scored for nuclear morphology and LEM2 clustering defects. Mean ± S.D. from three independent experiments (LEM2-mCh, n = 775; LEM2δ415-485-mCh, n = 1009). Statistical differences between the two lines were analysed by an unpaired two-tailed T-test; nuclear envelope clusters, p = 0.0003; nuclear envelope folds, p = 0.0007.
-
Figure 1—figure supplement 2—source data 1
Nuclear envelope morphology defects in the presence of LEM2 lacking the CHMP7-binding region.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig1-figsupp2-data1-v2.xlsx
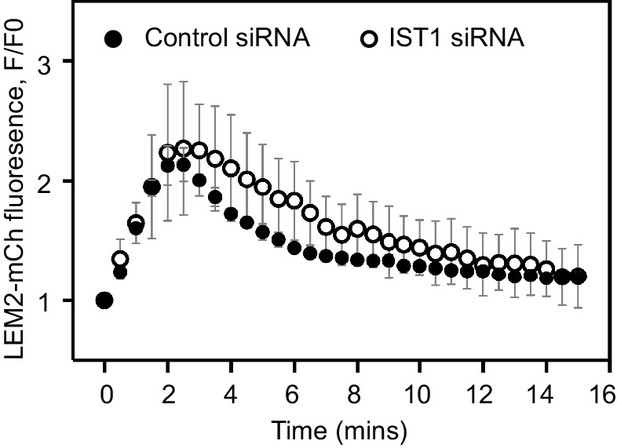
IST1 is dispensable for LEM2 cluster dissolution during M-exit HeLa cells stably expressing LEM2-mCh were treated with Control or IST1-targetting siRNA and imaged live during mitotic exit, as per Figure 1E.
LEM2-mCh fluorescence levels in a ROI around the chromatin were quantified from 25 (Control) or 11 (IST1 siRNA) individually imaged cells. Significance was calculated using an unpaired two-tailed T-test, p = 0.528, mean ± SD presented.
-
Figure 1—figure supplement 3—source data 1
IST1 is dispensible for dissolution of LEM2 clusters during M-exit.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig1-figsupp3-data1-v2.xlsx
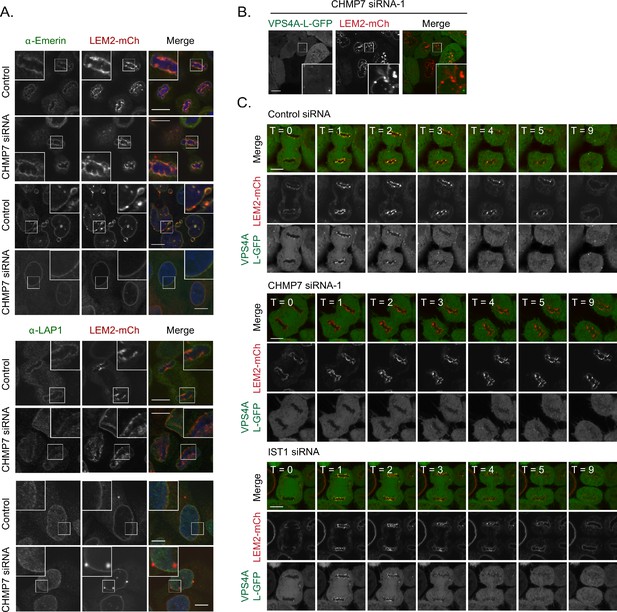
Analysis of proteins incorporated into LEM2-clusters formed in the absence of CHMP7.
(A) HeLa cells stably expressing LEM2-mCh were treated with control or CHMP7-1 targeting siRNA, fixed and stained with antisera raised against LAP1 or Emerin. Images representative of > 20 imaged cells in each case. (B, C) HeLa cells stably expressing VPS4A-L-GFP and LEM2-mCh were treated with the indicated siRNAs and imaged live. The clusters induced by CHMP7 depletion did not contain VPS4 (B). VPS4-L-GFP was recruited to the reforming NE in 16/16 Control siRNA-treated cell, 0/16 CHMP7-1 siRNA-treated cells, 2/11 CHMP7-2 siRNA-treated cells and 12/13 IST-1 siRNA-treated cells, collected in each case from three independent experiments.
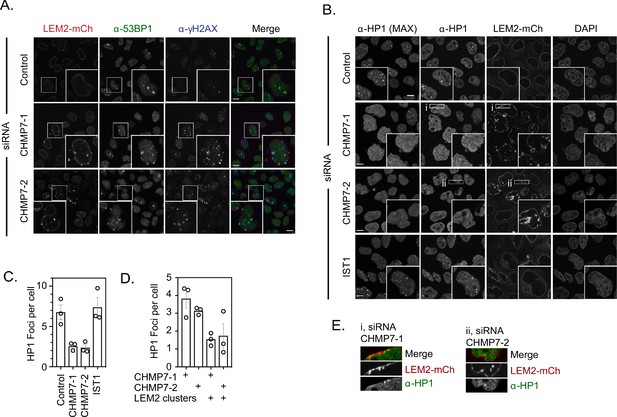
Effect of LEM2 clusters on genome integrity and organisation.
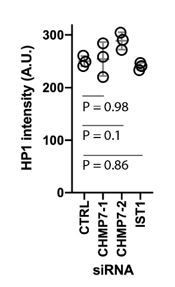
(A) HeLa cells stably expressing LEM2-mCh were transfected with Control, CHMP7-1, CHMP7-2 siRNA, fixed and stained with antisera raised against 53BP1 or γH2AX to demonstrate localisation of DNA damage markers to the nucleoplasmic side of LEM2-clusters. (B) HeLa cells stably expressing LEM2-mCh were transfected with Control, CHMP7-1, CHMP7-2 or IST1 siRNA, fixed and stained with antisera raised against HP1. Maximum projection and single slices of the HP1 channel were presented. (C) Quantification of data from B. HP1 foci per cell (Mean ± S.E.M). were scored from three independent experiments (Control, n = 297; CHMP7-1, n = 298; CHMP7-2, n = 311; IST1, n = 165 cells). Significance was calculated using a one-way ANOVA, with Dunnett’s post-hoc test, p = 0.013 (CHMP7-1), p = 0.010 (CHMP7-2), p = 0.894 (IST1). (D) Separation of data from CHMP7-depleted cells in C into cells with or without LEM2 clusters. E. Enlargement of region i and ii from CHMP7-depleted cells in B. In all panels, and a scale bar of 10 μm is depicted.
-
Figure 1—figure supplement 5—source data 1
Affect of CHMP7 depletion on the formation of heterochromatin-1 clusters.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig1-figsupp5-data1-v2.xlsx
Related to Figure 1C.
HeLa cells stably expressing GFP-CHMP7 and LEM2-mCh were imaged through mitotic exit. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 1—figure supplement 1B.
HeLa cells stably expressing GFP-CHMP7 were treated with LEM2 siRNA were imaged through mitotic exit. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 1—figure supplement 2C.
HeLa cells stably expressing LEM2-mCh or LEM2δLEM-mCh were imaged through mitotic exit. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 1D and E.
HeLa cells stably expressing LEM2 and treated with the indicated siRNA were imaged through mitotic exit. Frames were acquired at 30 s intervals and scale bar is 10 μm.
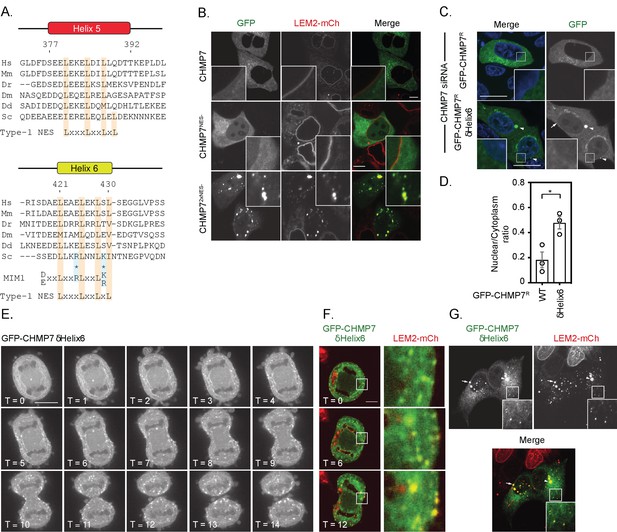
Deletion of a regulatory helix in CHMP7’s C-terminus decouples its assembly from the nuclear envelope during mitotic exit and causes inappropriate sequestration of LEM2 in interphase.
(A) Sequence alignment of CHMP7 Helix five and Helix six across phyla. Hs, Homo sapiens; Mm, Mus musculus; Dr, Danio rerio; Dm, Drosophila melanogaster; Dd, Dictyostelium discoideum; Sc, Saccharomyces cerevisiae. Type-1 Nuclear Export Sequence (NES) and MIM1 consensus sequence presented underneath; charge substituted residues in mammalian CHMP7 MIM1 indicated by asterisks. (B) HeLa cells stably expressing LEM2-mCh were transfected with plasmids encoding GFP-CHMP7, GFP-CHMP7 NES2-, GFP-CHMP7NES1- and NES2-, and imaged live. (C) HeLa cells transfected with CHMP7-targetting siRNA and transiently expressing GFP-CHMP7R or GFP-CHMP7R δHelix6 were fixed, stained with DAPI and imaged. GFP-CHMP7R δHelix6 was poorly exported from the nucleus, assembled aberrantly in the cytoplasm (arrowheads) and was not clearly enriched at the nuclear envelope (arrow). (D) Nucleocytoplasmic ratio of GFP-CHMP7 or GFP-CHMP7 δHelix6 from C. was quantified. Data presented as mean ± S.E.M from three independent experiments WT, n = 20, δHelix6, n = 117. Statistical significance was calculated with an unpaired two-tailed T-test, p = 0.023. (E) HeLa cells expressing GFP-CHMP7δHelix6 were imaged live during mitotic exit. Extra-nuclear envelope clustering of GFP-CHMP7δHelix6 was observed in 14/15 Videos acquired. (F, G) LEM2-mCh stable HeLa cells expressing GFP-CHMP7δHelix6 were imaged live during mitotic exit (images representative of 4/4 Videos acquired) or fixed and the localisation of LEM2-mCh was determined. Removal of LEM2-mCh from the INM and its sequestration in clusters observed in 18/18 imaged cells.
-
Figure 2—source data 1
Nucleocytoplasmic distribution of GFP-CHMP7 or GFP-CHMP7 delta-Helix6.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig2-data1-v2.xlsx
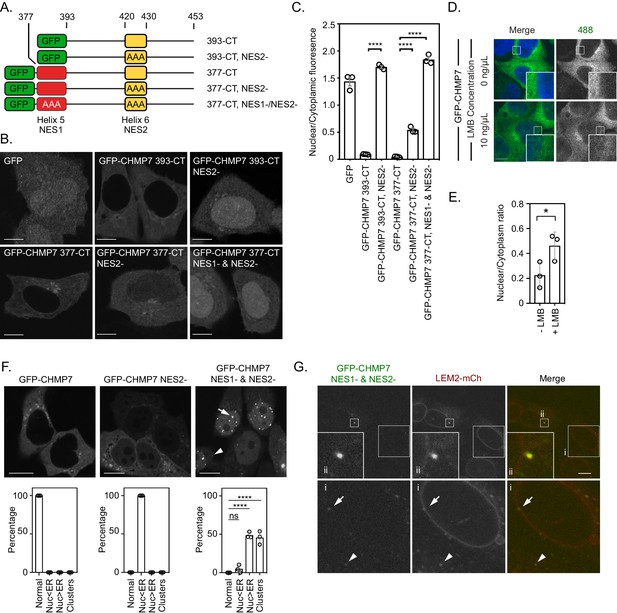
Nuclear export sequences (NESs) within CHMP7’s C-terminus protect against unregulated exposure to LEM2.
(A) Cartoon depiction of C-terminal fragments of CHMP7 used to analyse NESs. Colouring matches that in Figure 1A. (B) HeLa cells were transfected with the indicated GFP-tagged C-terminal fragments of CHMP7 with mutations in the NES in Helix 5 (L380A, L384A, L387A; NES1-) or Helix 6 (L421A, L425A, L428A; NES2-) and imaged. (C) Nucleocytoplasmic fluorescence intensities of the indicated constructs were quantified from B. Data presented as mean ± S.E.M. from N = three independent experiments. Statistical significance was calculated using a one-way ANOVA with Tukey’s multiple comparison. (GFP, n = 44; GFP-CHMP7 393-CT, n = 72; GFP-CHMP7 393-CT NES2-, n = 90; GFP-CHMP7 377-CT, n = 106; GFP-CHMP7 377-CT NES2-, n = 125; GFP-CHMP7 377-CT NES1- and NES2-, n = 99; **** p < 0.0001). (D). (E) HeLa cells stably expressing GFP-CHMP7 were treated with Leptomycin B for 4 hr, fixed and imaged. Data presented as mean ± S.E.M from three independent experiments. Statistical significance was calculated with an unpaired two-tailed T-test, P = 0.05; n = 13 (Control), n = 14 (LMB). (F) HeLa cells were transfected with plasmids encoding GFP-tagged CHMP7 with mutations in the NES in Helix 6 (GFP-CHMP7NES2-) or Helix 5 and Helix 6 (GFP-CHMP7NES1-&NES2-). Data presented as mean ± S.E.M. from N = three independent experiments, GFP-CHMP7 n = 83; GFP-CHMP7NES2-, n = 107; GFP-CHMP7NES1-&NES2-, n = 189. Localisation was binned into four categories: Normal ER localisation; nuclear localisation, but dimmer than the ER (Nuc<ER); nuclear localisation, but brighter than the ER (Nuc>ER) and clusters; the majority of clusters were detected inside the nucleus (arrow) with occasional detection outside the nucleus (arrowhead). Statistical significance was calculated using a one-way ANOVA with Dunnett’s multiple comparison, **** p < 0.0001. (G) HeLa cells stably expressing LEM2-mCh were fixed 48 hr after transduction with retrovirus packaging GFP-CHMP7NES1-&NES2- (n = 55 cells from N = two independent experiments). In all panels, and a scale bar of 10 μm is depicted.
-
Figure 2—figure supplement 1—source data 1
Examining NES sequences within CHMP7's C-terminus.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig2-figsupp1-data1-v2.xlsx
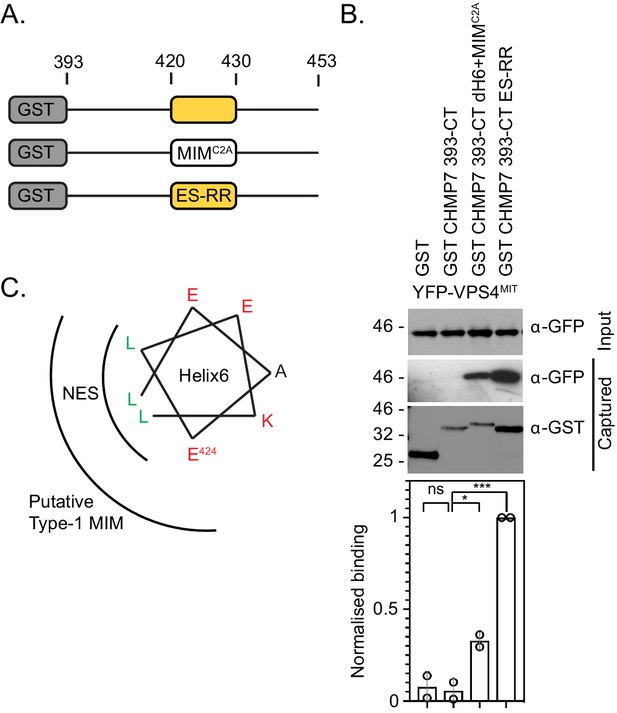
CHMP7 Helix6 does not contain a functional MIM.
(A) Cartoon depiction of C-terminal fragments of CHMP7 used to analyse NESs. Colouring matches that in Figure 1A. MIMC2A involves replacement of Helix6 with the MIM from CHMP2A; ES-RR involves charge reversal of E424R, S429R to restore the charge distribution of a type-1 MIM. (B) Glutathione-sepharose captured fractions and lysates from 293 T cells transiently expressing YFP-VPS4MIT and either GST, GST-CHMP7 393-CT, GST-CHMP7 393-CT+MIM, or GST-CHMP7 393-CT ES-RR were examined by western blotting with antisera raised against GST or GFP (N = 3) and quantified by infrared imaging (N = 2). Mean ± S.D. presented, p-values generated with one-way ANOVA with Tukey’s multiple comparison. CHMP7 393-CT δH6 + MIMC2A restored binding to YFP-VPS4MIT (p = 0.038). CHMP7 393-CT ES-RR (E424R, S429R) was created to restore the Type-1 MIM and permitted binding to YFP-VPS4MIT (p = 0.003). (C). Helical wheel displaying overlapping sequences of NES2 and the putative type-1 MIM within CHMP7 Helix6.
-
Figure 2—figure supplement 2—source data 1
CHMP7's C-terminus does not bind to VPS4's MIT domain.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig2-figsupp2-data1-v2.xlsx
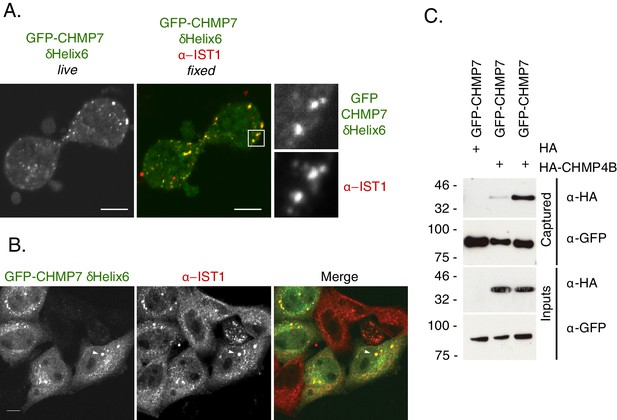
Deletion of CHMP7’s Helix6 results in its precocious assembly.
(A) HeLa cells expressing GFP-CHMP7δHelix6 were or imaged live, fixed and stained with antisera raised against IST1 and reimaged. Co-localisation observed with endogenous IST1 in 4/4 correlative live and fixed cell imaging attempts. (B) Additionally imaged interphase cells from A demonstrated that the GFP-CHMP7δHelix clusters that persisted into interphase remained associated with IST1. In all panels, a scale bar of 10 μm is depicted. (C) GFP-trap captured fractions from 293 T cells transiently expressing HA-CHMP4B and GFP-CHMP7 or GFP-CHMP7 δHelix6 were examined by western blotting with antisera raised against GFP or HA (N = 4).
Related to Figure 2E.
HeLa cells expressing GFP-CHMP7δHelix6 were imaged throughout mitotic exit.
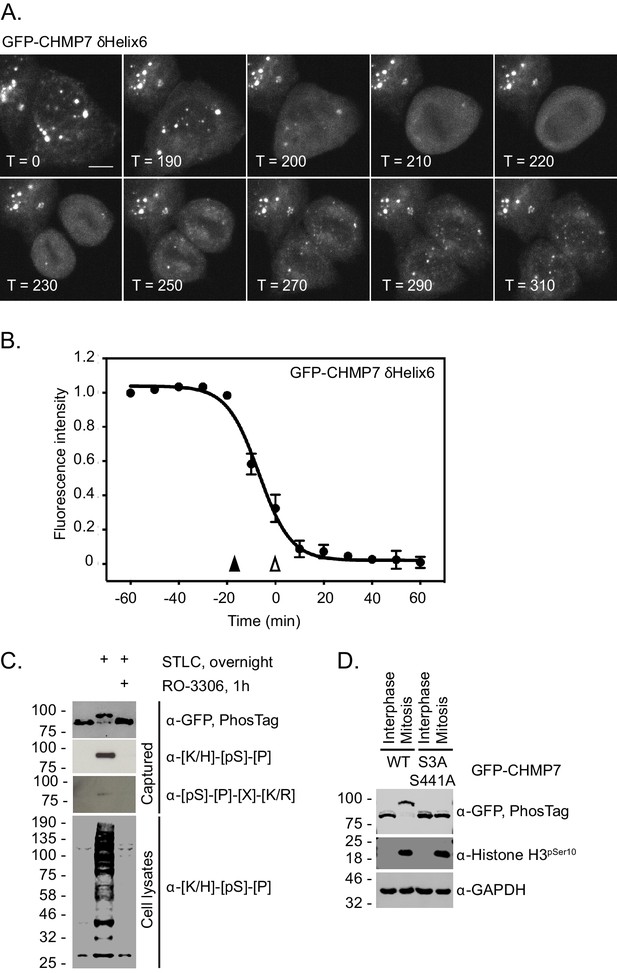
CHMP7 is phosphorylated by CDK1 upon mitotic entry.
(A, B) HeLa cells expressing GFP-CHMP7 δHelix6 were imaged live (A) and the intensity of GFP-CHMP7 δHelix6 puncta was quantified during M-phase (B). Quantification of 59 cells from nine experiments. Fluorescence traces normalised to metaphase onset (open arrowhead); nuclear envelope breakdown indicated by closed arrowhead. (C) Lysates of HeLa cells stably expressing GFP-CHMP7 and subject to the indicated synchronisations were immunoprecipitated using GFP-trap resin and resolved using normal or PhosTag SDS-PAGE. Inputs and captured fractions were examined by western blotting with antisera raised against GFP, or CDK1 substrate consensus sequences ([K/H]-[pS]-[P] or [pS]-[P]-[X]-[R/K]). Western blots representative of 6 PhosTag immunoprecipitations. (D) Lysates of HeLa cells stably expressing GFP-CHMP7 or GFP-CHMP7 S3A, S441A and subject to the indicated synchronisations were immunoprecipitated using GFP-trap resin and resolved using normal or PhosTag SDS-PAGE. Inputs and captured fractions were examined by western blotting with antisera raised against GFP, Histone H3 pSer10 or GAPDH (N = 3).
-
Figure 3—source data 1
GFP-CHMP7 delta-Helix6 cluster disassembly during M-phase.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig3-data1-v2.xlsx
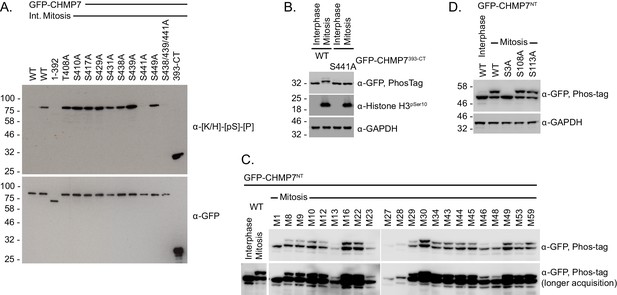
Mapping mitotic phosphorylation of CHMP7 to Ser3 and Ser441.
(A) 293 T cells expressing the indicated GFP-CHMP7 constructs were cultured asynchronously or synchronised in prometaphase with STLC. GFP-tagged proteins were immunoprecipitated and examined by western blotting with antisera raised against GFP and the CDK1 substrate consensus sequence [K/H]-[pS]-[P]. N = 3. (B) HeLa cells stably expressing GFP-CHMP7 393-CT or GFP-CHMP7 393-CT S441A were cultured asynchronously or synchronised prometaphase with STLC, then were lysed, resolved by normal or PhosTag SDS-PAGE and examined by western blotting with antisera raised against GFP, pSer10 Histone H3 or GAPDH (N = 3). (C, D) 293 T cells expressing the indicated GFP-CHMP7NT proteins were cultured asynchronously or synchronised in prometaphase with STLC, then were lysed, resolved by normal or PhosTag SDS-PAGE and examined by western blotting with antisera raised against GFP or GAPDH (N = 3). In C, Ser/Thr containing mutants selected from a previously generated panel of alanine-scanning GFP-CHMP7NT mutations (Olmos et al., 2016) were assessed for a mobility shift in mitosis. Mobility shift was compromised by M1, M27, and M28, although M27 and M28 were poorly expressed. M1 encodes the mutations W2A, S3A, P4A, E5A; M27 encodes the mutations R106A, E107A, S108A, D109A; M28 encodes the mutations F110A, M111A, A112G, S113A. Individual mutations of S3A, S108A, and S113A from M1, M27, and M28 revealed the site of phosphorylation to be Ser 3 (D).
Related to Figure 3A.
HeLa cells expressing GFP-CHMP7δHelix6 were imaged throughout mitotic exit. Frames were acquired at 30 s intervals and scale bar is 10 μm.
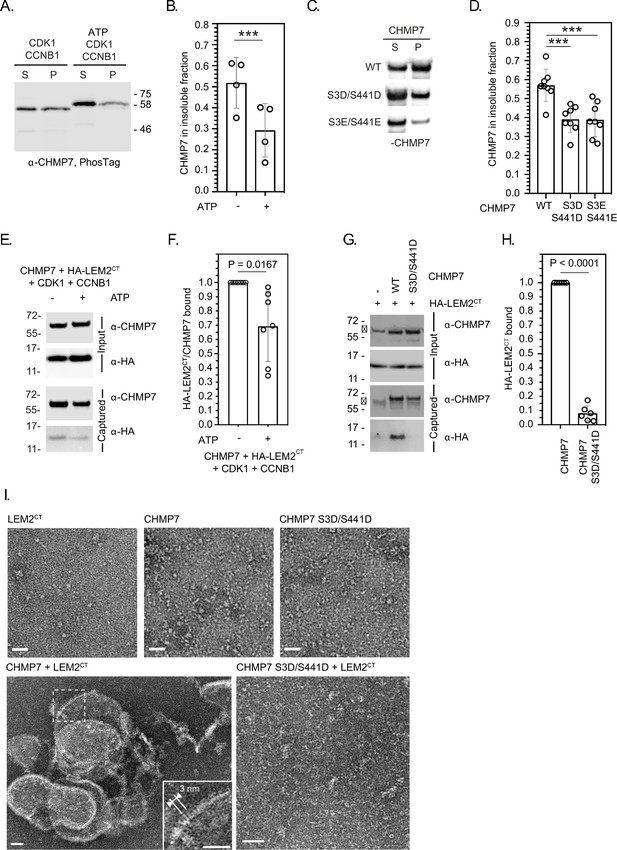
CDK phosphorylation suppresses CHMP7 sedimentation, interaction with LEM2 and LEM2-stimulated polymerisation.
(A) Recombinant CHMP7 was incubated with recombinant CDK1 and CCNB1 in the presence or absence of ATP and sedimented by ultracentrifugation. CHMP7 was recovered from pellet and supernatant fractions, respectively, resolved by PhosTag SDS-PAGE and analysed by western blotting using antisera raised against CHMP7. (B) Data from A presented as mean ± S.D., N = 4, *** p = 0.0007 by paired two-tailed T-test. (C) Recombinant CHMP7, CHMP7 S3D/S441D, or CHMP7 S3E/S441E were sedimented by ultracentrifugation, recovered from pellet and supernatant fractions, and analysed by western blotting with antisera raised against CHMP7. (D) Data from C presented as mean ± S.D., N = 8, *** p = 0.0004. (E) Recombinant CHMP7 was incubated with recombinant CDK1 and CCNB1 in the presence or absence of ATP. Recombinant HA-LEM2CT was added and CHMP7 was captured on magnetic anti-CHMP7 Dynabeads. Captured and input fractions were resolved by SDS-PAGE and examined by western blotting with anti-CHMP7 or anti-HA antisera. (F) Data from G are presented as mean ± S.D., N = 7, p = 0.0156 by paired two-tailed T-test. (G) Recombinant HA-LEM2CT was incubated alone, with CHMP7 or CHMP7 S3D/S441D. CHMP7 was captured on magnetic anti-CHMP7 Dynabeads. Captured and input fractions were resolved by SDS-PAGE and examined by western blotting with anti-CHMP7 or anti-HA antisera. Please note non-specific bands detected by CHMP7 antibody (*). (H) Data from G are presented as mean ± S.D., N = 6, p < 0.0001 by paired two-tailed T-test. (I) Recombinant LEM2CT, CHMP7, or CHMP7 S3D/S441D were examined by negative stain electron microscopy, either alone or in the combinations indicated. When incubated alone, no examples of polymerisation were observed. Images representative of N = three independent experiments. Scale bar is 25 nm.
-
Figure 4—source data 1
Influence of CDK1-phosphorylation on the CHMP7/LEM2 interaction.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig4-data1-v2.xlsx
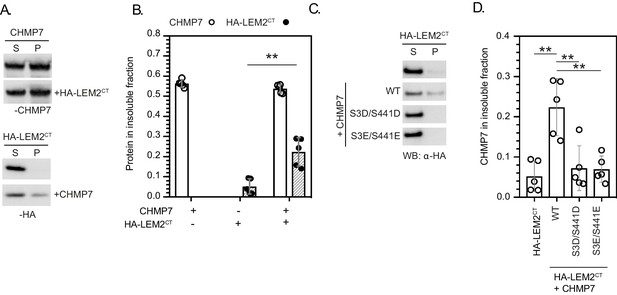
CDK phosphorylation on CHMP7 Ser3 and Ser441 suppresses CHMP7 sedimentation and capture of LEM2CT.
(A, B) Recombinant CHMP7 was incubated with or without an equimolar concentration of recombinant LEM2CT. Insoluble and soluble CHMP7, LEM2CT, and CHMP7-LEM2CT complexes were recovered from pellet (P) and supernatant (S) fractions respectively, resolved by SDS-PAGE and analysed by western blotting with anti-CHMP7 or anti-HA antisera. Data presented a mean ± S.D., N = 5, ** p = 0.0014 by two-tailed T-test. (C). HA-LEM2CT was subjected to the sedimentation assay described in Figure 4, either alone, or in combination with recombinant CHMP7 (WT, S3D/S441D. S3E/S441E). Supernatant and pellet fractions were examined by western blotting with antisera raised against HA. (D). The degree of HA-LEM2CT sedimentation from C was quantified (N = 5). Data presented as mean ± S.D., significance calculated with one-way ANOVA with Dunnett’s multiple comparison test. Compared to CHMP7 + HA-LEM2CT; HA-LEM2CT alone, p = 0.0002; CHMP7 S3D/S441D + HA-LEM2CT, p = 0.0008; CHMP7 S3E/S441E + HA-LEM2CT, p = 0.0007.
-
Figure 4—figure supplement 1—source data 1
Examination of the CHMP7 polymer by negative stain EM.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig4-figsupp1-data1-v2.xlsx
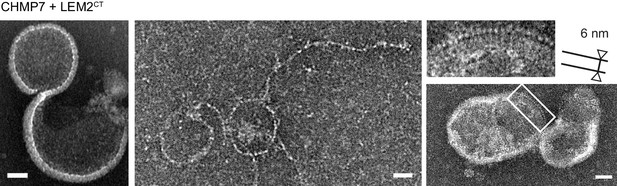
Electron microscopy of the CHMP7/LEM2CT polymer.
Examples of the polymer obtained through incubation of CHMP7 and LEM2CT. These include curved polymers with periodic densities as well as circular or hemispherical assemblies. The edge of these assemblies show a rod-shaped feature of length ~6 nm packed with a ~3 nm spacing. Scale bar is 25 nm.
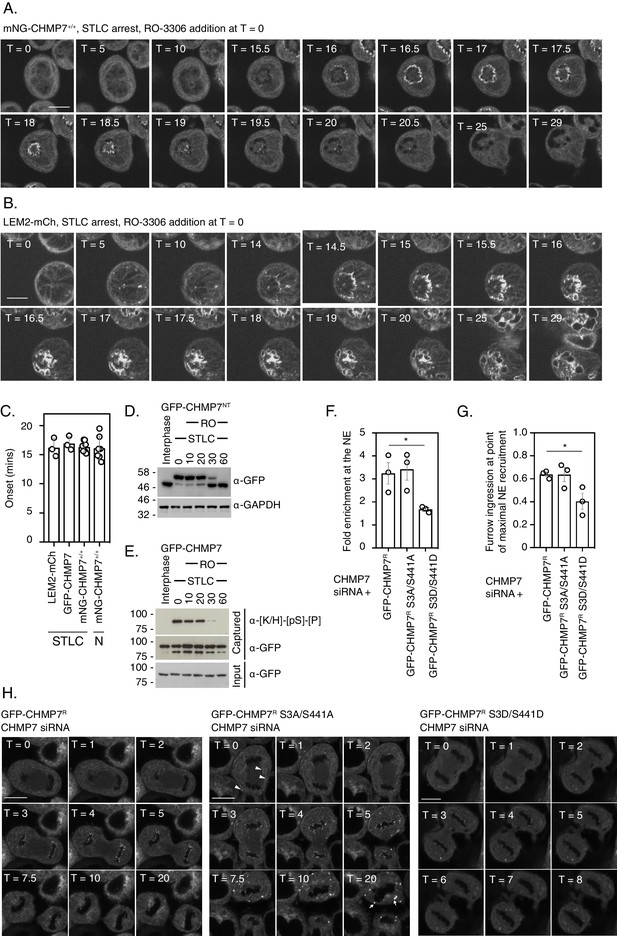
Dephosphorylation kinetics of CHMP7 during mitotic exit.
(A, B) CAL-51 cells homozygously edited to express mNG-CHMP7 (A) or HeLa cells stably expressing LEM2-mCh (B) were synchronised to prometaphase with STLC and then released through addition of RO-3306. Cells were imaged live through synchronised M-exit. (C) Quantification of assembly onset (assembly duration presented in Figure 5—figure supplement 1D) post RO3306 release. mNG-CHMP7+/+ (onset 16.3 ± 0.88 mins; duration 3.1 ± 0.44 mins; N = 9, n = 104) or LEM2-mCh (onset 16.2 ± 2.22 mins; duration 4.1 ± 0.44 mins; N = 3, n = 80). Additional quantification of the above parameters from HeLa cells stably expressing GFP-CHMP7 (onset 16.9 ± 1.21 min; duration 3.3 ± 0.13 min; N = 3, n = 97) and subject to STLC arrest and RO3306 release (from Figure 4—figure supplement 1) and CAL-51 cells homozygously edited to express mNG-CHMP7 (onset 16.1 ± 2.02 min; duration 4.2 ± 2.19 min; N = 7, n = 39) and subject to nocodazole arrest and RO3306 release (from Figure 5—figure supplement 1) are also presented in C. A one-way ANOVA with Tukey’s multiple comparisons revealed no significant differences between the datasets. (D). (E) HeLa cells stably expressing GFP-CHMP7NT (D) or GFP-CHMP7 (E) were either untreated or arrested in mitosis through STLC inhibition and forced out of mitosis using RO3306 for the indicated times. Lysates in D, reporting Ser3 dephosphorylation were resolved by PhosTag or normal SDS-PAGE and examined with antisera raised against GFP or GAPDH respectively. Lysates from E, reporting Ser441 dephosphorylation were immunoprecipitated using GFP-trap resin and both inputs and captured fractions were examined by western blotting with antisera raised against phosphorylated CDK1 substrates and GFP (N = 3). (F-H) CHMP7-depleted HeLa cells stably expressing GFP-CHMP7R, GFP-CHMP7R S3A/S441A, or GFP-CHMP7R S3D/S441D were imaged live during mitotic exit (H). In F, the fold-enrichment of GFP-signal at the reforming nuclear envelope was calculated from three independent experiments, presented as mean (N = 3) ± S.E.M. (WT, n = 24; S3A/S441A, n = 27, n.s. (p = 0.92), S3D/S441D, n = 52, p = 0.049; one-way ANOVA with Dunnett’s multiple comparisons). In G, the degree of furrow ingression (midzone diameter/daughter cell diameter) at the point of maximal GFP-CHMP7 recruitment was used as a proxy of progression through M-exit and was calculated from three independent experiments, presented as mean (N = 3) ± S.E.M. (WT, n = 24; S3A/S441A, n = 24, n.s. (p = 0.99), S3D/S441D, n = 46, p = 0.042; one-way ANOVA with Dunnett’s multiple comparisons). In H, for cells stably expressing GFP-CHMP7R S3A/S441A, arrowheads depict precocious clustering of CHMP7 in the peripheral ER during anaphase that develop into larger clusters during mitotic exit. In all panels, time in minutes and a scale bar of 10 μm are reported.
-
Figure 5—source data 1
Dynamics of CHMP7 assembly at the reforming nuclear envelope.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig5-data1-v2.xlsx
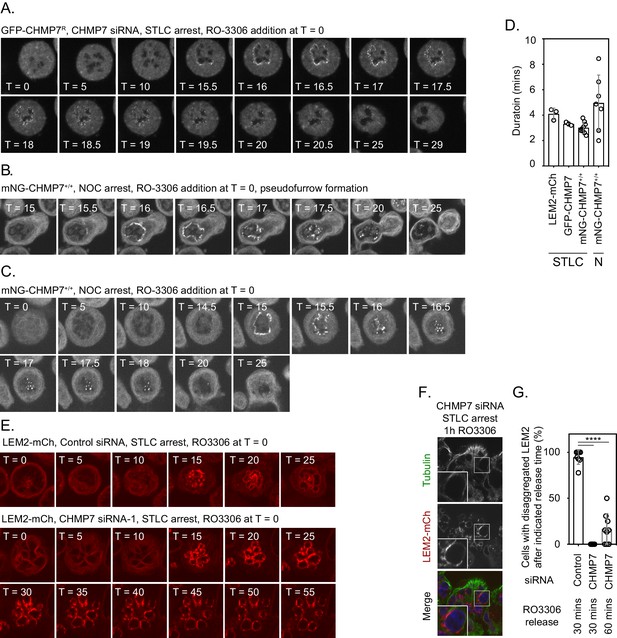
Analysis of the kinetics of GFP-CHMP7 and LEM2-mCh assembly at the reforming NE during monopolar mitotic exit.
(A) HeLa cells stably expressing GFP-CHMP7R and treated with CHMP7 siRNA were synchronised in prometaphase with STLC and then released through addition of RO-3306. Cells were imaged live through synchronised M-exit. Data were quantified in Figure 5C and D. (B, C) CAL-51 cells homozygously edited to express mNG-CHMP7 were synchronised to in prometaphase with nocodazole and then released from M-phase through addition of RO-3306. Cells were imaged live through synchronised M-exit. Note persistent assembly in the absence of microtubules (C) Recruitment observed in 245/258 cells (N = 3) and furrow-proximal initiation of assembly in 13/14 cells forming a pseudofurrow (B). (D) Quantification of assembly duration time (assembly onset time presented in Figure 5C) post RO3306 release from Figure 5A, Figure 5B and A-C. mNG-CHMP7+/+ (onset 16.3 ± 0.88 mins; duration 3.1 ± 0.44 mins; N = 9, n = 104) or LEM2-mCh (onset 16.2 ± 2.22 mins; duration 4.1 ± 0.44 mins; N = 3, n = 80). (E) HeLa cells stably expressing LEM2-mCh were transfected with Control or CHMP7-1 siRNA, arrested in prometaphase with STLC and released via RO3306 treatment. (F) Cells from E were fixed at the indicated time after RO3306 treatment, stained with antisera raised against Tubulin and imaged. (G). Cells from E were imaged live and the percentage of cells displaying LEM2-mCh clusters at the indicated time after RO3306 release was quantified Control siRNA, 30 min post RO3306 release, N = 8, n = 92; CHMP7 siRNA, 30 min post RO3306 release, N = 9, n = 98; CHMP7 siRNA 60 min post RO3306 release, N = 9, n = 98. Significance was calculated using a one-way ANOVA with Tukey’s multiple comparisons, **** p < 0.0001. In all panels, time in minutes and a scale bar of 10 μm is depicted.
-
Figure 5—figure supplement 1—source data 1
Kinetics of CHMP7 and LEM2 assembly, and LEM2 cluster dissolution, at the reforming monopolar NE.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig5-figsupp1-data1-v2.xlsx
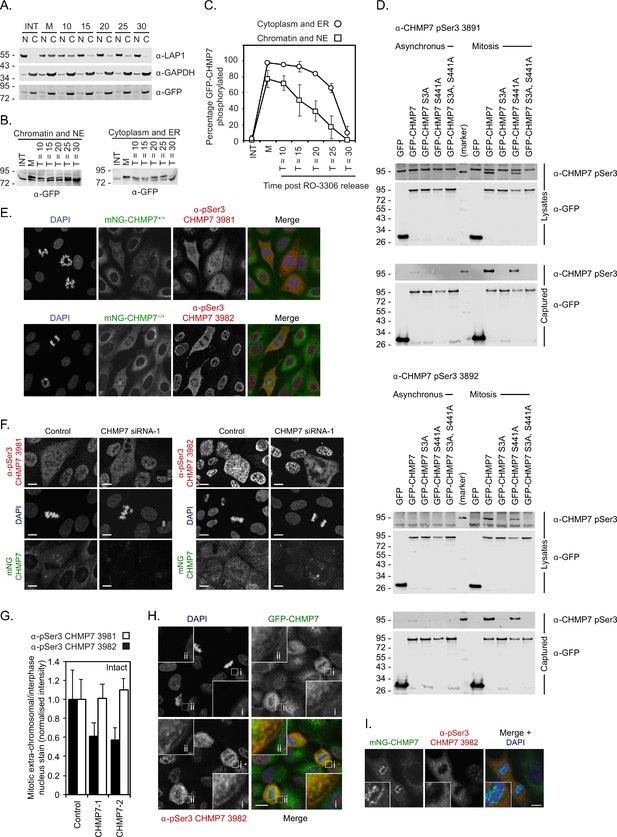
During M-exit, the nuclear-associated pool of CHMP7 is dephosphorylated in advance of the cytosolic/ER-associated pool.
(A) HeLa cells stably expressing GFP-CHMP7, either asynchronous or arrested at prometaphase with STLC and released from M-arrest for the indicated times with STLC were separated into chromatin-associated or non-chromatin-associated fractions. Fractions were resolved and examined by western blotting with antibodies raised against LAP1, GAPDH, or GFP. (B) Chromatin-associated or non-chromatin-associated fractions from the indicated timepoints were resolved by PhosTag SDS-PAGE and examined by western blotting with antisera raised against GFP. (C) Quantification of the phosphorylated fraction in B by densitometry (N = 4) revealed advanced dephosphorylation of the minor pool of CHMP7 associated with chromatin. (D) Lysates or GFP-trap immunoprecipitated fractions of HeLa cells stably expressing GFP, GFP-CHMP7, GFP-CHMP7 S3A, GFP-CHMP7 S441A, or GFP-CHMP7 S3A/S441A and cultured either asynchronously, or arrested in prometaphase with STLC, were resolved by SDS-PAGE and examined with antisera raised against GFP, or pSer3 CHMP7 (α−3891 or α−3892). Both α−3891 and α−3892 could detect Ser3-phosphorylated CHMP7 in M-phase extracts and captured fractions. (E) Asynchronous cultures of CAL-51 cells edited to homozygously express mNG-CHMP7 were stained with DAPI and antisera raised against pSer3 CHMP7 (α−3891 or α−3892). Both antibodies detected an extra-chromosomal signal in M-phase cells but also illuminated the nuclei of interphase cells, a staining which we deemed to be non-specific. (F, G) Control or CHMP7 siRNA treated CAL-51 mNG-CHMP7 cells were stained with antisera raised against pSer3 CHMP7 (α−3891 or α−3892) and the fluorescence intensity of the mitotic extra-chromosomal signal relative to the interphase nuclear signal was calculated. The M-phase extra-chromosomal signal reported by α−3891 was insensitive to CHMP7 depletion, whereas the signal reported by α−3892 was partially sensitive to CHMP7 depletion. We concluded that α−3892 was capable of detecting endogenous pSer3 CHMP7 in M-phase. (H) Asynchronous cultures of HeLa cells stably expressing GFP-CHMP7 were stained with α−3892, revealing a detectable signal on the mitotic ER that overlapped with GFP-CHMP7. (I) CAL-51 cells edited to homozygously express mNG-CHMP7 were stained with DAPI and α−3892. Whilst α−3892 illuminated all M-phase cells, the presence of α−3892 immunoreactivity coincident with enriched mNG-CHMP7 at the reforming nuclear envelope was observed in 0/100 cells. In all panels, scale bar is 10 μm.
-
Figure 5—figure supplement 2—source data 1
Kinetics of CHMP7 dephopshorylation in chromatin-associated and extra-chromatin associated fractions during M-exit and siRNA sensitivity of the pSer3 CHMP7 antisera.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig5-figsupp2-data1-v2.xlsx
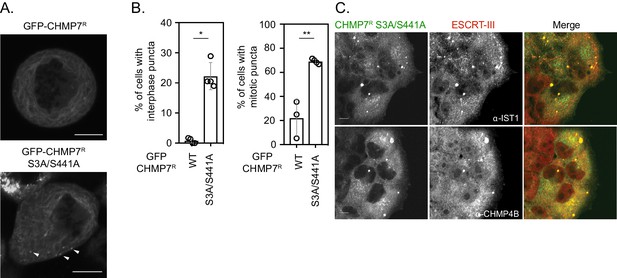
CDK phosphorylation on CHMP7 Ser3 and Ser441 suppresses CHMP7 assembly and inappropriate capture of ESCRT-III subunits during mitotic exit.
(A) HeLa cells stably expressing either GFP-CHMP7R or GFP-CHMP7R S3A/S441A were imaged during mitosis. Arrowheads depict precocious assembly of GFP-CHMP7R S3A/S441A in the peripheral ER. (B, C) Endogenous CHMP7 was depleted from HeLa cells stably expressing GFP-CHMP7R S3A/S441A. Cells were either fixed and stained with antisera raised against IST1 or CHMP4B and the number of cells exhibiting ESCRT-III puncta were quantified (B. Interphase; N = 4, n = 382 cells (GFP-CHMP7R); N = 4, n = 697 cells (GFP-CHMP7R S3A/S441A); p = 0.002, two-tailed T-test. Mitosis, N = 3, n = 94 (GFP-CHMP7R); N = 4, n = 80 (GFP-CHMP7R S3A/S441A); p = 0.03, two-tailed T-test. Data presented as mean ± S.D.).
-
Figure 5—figure supplement 3—source data 1
CDK1 phosphorylation of CHMP7 suppresses formation of clusters of CHMP7 that grow during M-exit.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig5-figsupp3-data1-v2.xlsx
Related to Figure 5A.
CAL-51 mNG-CHMP7+/+ were released from an STLC-induced mitotic arrest using RO3306. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 5—figure supplement 1A.
HeLa cells stably expressing GFP-CHMP7R and transfected with CHMP7 siRNA were released from an STLC-induced mitotic arrest using RO3306. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 5A.
HeLa cells stably expressing LEM2-mCh were released from an STLC-induced mitotic arrest using RO3306. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 5—figure supplement 1B.
CAL-51 mNG-CHMP7+/+ cells were released from a nocodazole-induced mitotic arrest using RO3306. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 5—figure supplement 1C.
CAL-51 mNG-CHMP7+/+ cells were released from a nocodazole-induced mitotic arrest using RO3306. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 5—figure supplement 1E.
HeLa cells stably expressing LEM2-mCh and transfected with control or CHMP7 siRNA-1 were released from an STLC-induced mitotic arrest using RO3306. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 5H.
HeLa cells stably expressing GFP-CHMP7R or GFP-CHMP7R S3A, S441A were transfected with CHMP7 siRNA-1 and were imaged live during mitotic exit. Frames were acquired at 30 s intervals and scale bar is 10 μm.
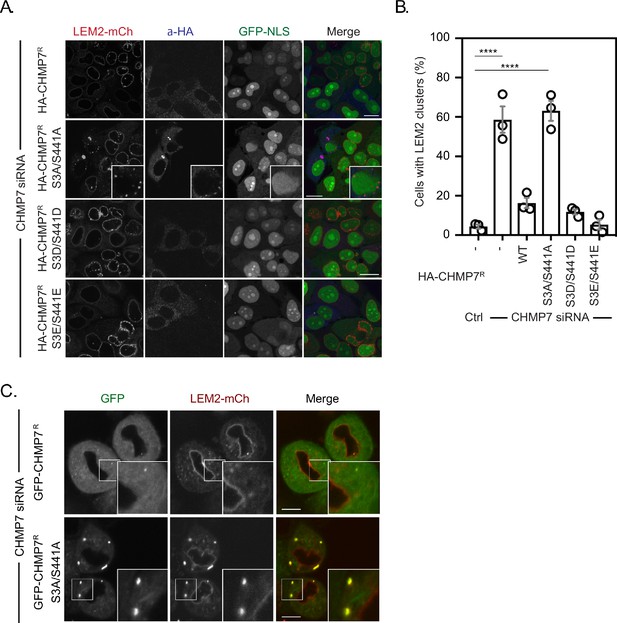
Mitotic CHMP7 phosphorylation suppresses inappropriate clustering of LEM2 during M-exit.
(A) HeLa cells stably expressing LEM2-mCh and GFP-NLS were treated with CHMP7-targetting siRNA and transfected with plasmids encoding the indicated HA-CHMP7R constructs, fixed and stained with antisera raised against HA. (B) The number of cells displaying extra-NE clusters of LEM2-mCh was quantified from A, presented as mean ± S.E.M from N = three independent experiments and the significance of LEM2 cluster formation was assessed by one-way ANOVA with Dunnett’s post-hoc test (Control, n = 416; CHMP7 siRNA, n = 916, p < 0.0001; CHMP7 siRNA + HA-CHMP7R n = 375, n.s., p = 0.159; CHMP7 siRNA + HA-CHMP7R S3A/S441A, n = 379, P < 0.001; CHMP7 siRNA + HA-CHMP7R S3D/S441D, n = 192, n.s., p = 0.529; CHMP7 siRNA + HA-CHMP7R S3E/S441E, n = 251, n.s., p = 0.999). (C) HeLa cells stably expressing LEM2-mCh and either GFP-CHMP7R or GFP-CHMP7R S3A/S441A were transfected with siRNA targeting CHMP7 and imaged during mitotic exit. Precocious clusters were observed in 15/49 (WT) and 12/15 (S3A/S441A) imaged cells. In all panels, time in minutes and a scale bar of 10 μm is depicted.
-
Figure 6—source data 1
CHMP7 phosphorylation prevents inappropriate LEM2 clusters forming in the peripheral ER during M-exit.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig6-data1-v2.xlsx
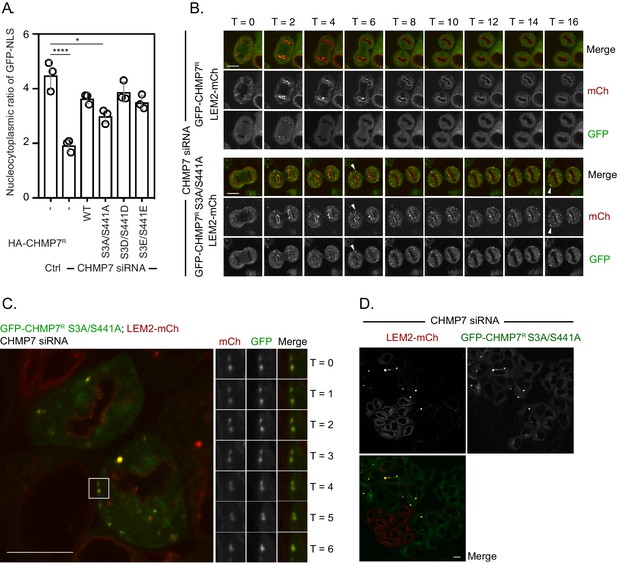
Failure to phosphorylated CHMP7 Ser3 and Ser441 during M-phase results in inappropriate clustering and capture of LEM2 in the peripheral ER that compromises its localisation to the INM.
(A) HeLa cells (from Figure 6A) stably expressing LEM2-mCh and GFP-NLS were treated with CHMP7-targetting siRNA and transfected with plasmids encoding the indicated HA-CHMP7R constructs, fixed and stained with antisera raised against HA. The nucleocytoplasmic ratio of GFP-NLS was quantified, presented as mean ± S.E.M. and the significance of nucleocytoplasmic compartmentalisation breakdown was assessed by one-way ANOVA with Dunn’s post-hoc test (Control, n = 416; CHMP7 siRNA, n = 916, p = 0.0038; CHMP7 siRNA + HA-CHMP7R n = 375, n.s., p = 0.968; CHMP7 siRNA + HA-CHMP7R S3A/S441A, n = 379, p = 0.0372; CHMP7 siRNA + HA-CHMP7R S3D/S441D, n = 192, n.s., p = 0.999; CHMP7 siRNA + HA-CHMP7R S3E/S441E, n = 251, n.s., p = 0.631). B-D. In (B), HeLa cells stably expressing LEM2-mCh and either GFP-CHMP7R or GFP-CHMP7R S3A/S441A were transfected with siRNA targeting CHMP7 and imaged during mitotic exit. Arrowheads in B depict precocious assembly of LEM2-mCh and GFP-CHMP7R S3A, S441A in the peripheral ER. Precocious clusters of LEM2-mCh and GFP-CHMP7R were observed in 9/31 imaged Videos Precocious clusters of LEM2-mCh and GFP-CHMP7R S3A/S441A were observed in 7/9 imaged Videos. In (C), an example of fusogenic behaviour of the LEM2-mCh and GFP-CHMP7 S3A/S441A-positive clusters is shown. Persistent fusion led to the generation of singular LEM2-mCh and GFP-CHMP7 S3A/S441A-positive cluster, depicted in D. In all panels, time in minutes and a scale bar of 10 μm is depicted.
-
Figure 6—figure supplement 1—source data 1
Nuclear envelope compartmenatlisation in the presence of phosphomutant or phosphomutant versions of CHMP7.
- https://cdn.elifesciences.org/articles/59999/elife-59999-fig6-figsupp1-data1-v2.xlsx
Related to Figure 6C.
HeLa cells stably expressing LEM2-mCh and GFP-CHMP7R S3A, S441A and transfected with CHMP7 siRNA-1 were imaged live during mitotic exit. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Related to Figure 6—figure supplement 1C and D.
HeLa cells stably expressing LEM2-mCh and GFP-CHMP7R S3A, S441A and transfected with CHMP7 siRNA-1 were imaged live. Frames were acquired at 30 s intervals and scale bar is 10 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CHMP7 | Ensembl | ENSG00000147457 | |
| Gene (Homo sapiens) | LEMD2 | Ensembl | ENSG00000161904 | Also called LEM2 |
| Strain, strain background (Escherichia coli) | NEB5a | NEB | C2987 | Cloning grade derivative of E. coli DH5a |
| Strain, strain background (Escherichia coli) | BL21Star (DE3) | Invitrogen | C601003 | Protein production strain of E. coli DH5a |
| Cell line (Homo sapiens) | HeLa | ATCC | STR profiled from Crick Cell Services | |
| Cell line (Homo sapiens) | CAL-51 | PMID:27618263 | STR profiled from Crick Cell Services | |
| Cell line (Homo sapiens) | 293T | ATCC | STR profiled from Crick Cell Services | |
| Cell line (Homo sapiens) | GP2-293 | Clontech | STR profiled from Crick Cell Services | |
| Cell line (Homo sapiens) | CAL-51 mNG-CHMP7+/+ | PMID:27618263 | STR profiled from Crick Cell Services | |
| Cell line (Homo sapiens) | HeLa GFP-CHMP7R | PMID:27618263 | STR profiled from Crick Cell Services | |
| Transfected construct (Homo sapiens) | pCMS28-EcoRI-NotI-XhoI | PMID:17556548 | Bicistronic retroviral packaging and exprssion vector expressing IRES-PuroR downstream of the MCS. GFP-fusions of CHMP7 were constructed in this vector | |
| Transfected construct (Homo sapiens) | pCMS28-EcoRI-NotI-XhoI-L-GFP | This study | A version of pCMS28 containing a LAP tag, based upon PMID:15644491 | |
| Transfected construct (Homo sapiens) | pMSCVneo EcoRI-NotI-XhoI | This study | A kind gift from Prof Martin-Serrano (King’s College London) Modified version of Clontech’s pMSCV-neo. LEM2-mCherry fusions were expressed from this vector. A retroviral packaging vector that encodes neomycin resistance from a second promoter. | |
| Transfected construct (Homo sapiens) | pLHCX-MCS-HA-CHMP7R | This study | Retroviral packaging and expression vector (a kind gift from Prof Tony Ng, King’s College London) into which HA-CHMP7Rwas inserted and mutagenised as necessary. | |
| Transfected construct (Homo sapiens) | pCR3.1 YFP/HA EcoRI-NotI-XhoI | PMID:14519844 | Mammalian expression vector for YFP- or HA-fusion proteins | |
| Transfected construct (Homo sapiens) | pCAGGS/GST | PMID:14519844 | Mammalian expression vector for GST-fusions proteins | |
| Transfected construct (Homo sapiens) | pCMVdR8.91 | A kind gift from Dr Caroline Goujon (IRIM CNRS, Montpellier) | Packaging plasmid used for lentivirus production. Mammalian expression of Gag-Pol driven by CMV promoter. | |
| Transfected construct (E. coli) | pET28a-NusA- EcoRI-NotI-XhoI | This paper | A version of pET28a comprising N-terminal tandem His6 affinity tag, an N-utilisation sequence A (NusA) solubilisation tag. | |
| Antibody | GAPDH (Mouse monoclonal) | Merck Millipore | Clone 6C5, MAB374 | (1:10,000) |
| Antibody | Calnexin (Rabbit polyclonal) | Abcam | ab22595 | (1:1000) |
| Antibody | Alpha-Tubulin (Mouse monoclonal) | Merck Millipore | Clone DM1A, CP06 | (1:500) |
| Antibody | CHMP2A (Rabbit polyclonal) | Proteintech | 10477–1-AP | (1:1000) |
| Antibody | IST1 (Rabbit polyclonal) | Proteintech | 51002–1-AP | (1:1000) |
| Antibody | CHMP7 (Rabbit polyclonal) | Proteintech | 16424–1-AP | (1:1000) |
| Antibody | LAP1 (Rabbit polyclonal) | Proteintech | 21459–1-AP | (1:1000) |
| Antibody | Emerin (Rabbit polyclonal) | Proteintech | 16424–1-AP | (1:1000) |
| Antibody | GFP (Mouse monoclonal) | Roche | Clone 7.1/13.1, 11814460001 | (1:1000) |
| Antibody | mCherry (Rabbit polyclonal) | Abcam | ab167453 | (1:1000) |
| Antibody | HA.11 (Mouse monoclonal) | Biolegend | Clone 16B12, 901502 | (1:1000) |
| Antibody | gH2AX (Mouse monoclonal) | Merck Millipore | Clone JBW301, 05–636 | (1:200) |
| Antibody | 53BP1 (Rabbit polyclonal) | Novus Biologicals | NB100-305 | (1:200) |
| Antibody | Phospho-CDK Substrate Motif [(K/H)pSP] (Rabbit monoclonal) | Cell Signaling Technology | 9477 | (1:1000) |
| Antibody | Phospho-MAPK/CDK Substrates [PXS*P or S*PXR/K] (Rabbit monoclonal) | Cell Signaling Technology | Clone 34B2, 2325 | (1:1000) |
| Antibody | Phospho-Histone H3 (Ser10) Antibody (Rabbit polyclonal) | Cell Signaling Technology | 9701 | (1:1000) |
| Antibody | LEM2 (Rabbit polyclonal) | Merck Millipore | HPA017340 | (1:1000) |
| Antibody | HSP 90α/β Antibody (Mouse monoclonal) | Santa Cruz Biotechnology | Clone F8, sc-13119 | (1:1000) |
| Antibody | CHMP7 pSer3 | This study | 3891 | (1:500) In-house produced rabbit polyclonal phospho S3 CHMP7 antisera. |
| Antibody | CHMP7 pSer3 | This study | 3892 | (1:500) In-house produced rabbit polyclonal phospho S3 CHMP7 antisera. |
| Chemical compound, drug | PhosTag Acrylamide | Fujifilm Wako Chemicals | AAL-107 | |
| Chemical compound, drug | RO-3306 | Merck Millipore | SML-0569 | |
| Chemical compound, drug | S-Trityl L-Cysteine | Merck Millipore | 164739 | |
| Chemical compound, drug | Nocodazole | Merck Millipore | M1404 | |
| Peptide, recombinant protein | CDK1:CyclinB1 complex | Merck Millipore | 14–450M-D | |
| Chemical compound, drug | HALT Protease Inhibitors | Fisher Scientific | 78429 | |
| Chemical compound, drug | Complete EDTA-free protease inhibitors | Merck Millipore | 11836170001 | |
| Chemical compound, drug | PhosSTOP phosphatase inhibitor tablets | Merck Millipore | PHOSS-RO | |
| Commercial assay, kit | GFP-Trap Magnetic Agarose | Chromotek | Gtma-20 | |
| Commercial assay, kit | Glutathione Sepharose 4B GST-tagged protein purification resin | GE Healthcare | 17075601 | |
| Commercial assay, kit | Dynabeads antibody coupling kit | Invitrogen | 14311D | |
| Sequence-based reagent | Control siRNA | Horizon Discovery | D-001810 | |
| Sequence-based reagent | CHMP7 siRNA-1 | Horizon Discovery, PMID:27618263 | GGGAGAAGATTGTGAAGTTdTdT | |
| Sequence-based reagent | CHMP7 siRNA-1 | Horizon Discovery, PMID:27618263 | GGAGGUGUAUCGUCUGUAUdTdT | |
| Sequence-based reagent | IST-1 siRNA | Horizon Discovery | M-020977 | |
| Sequence-based reagent | LEM2 siRNA | Horizon Discovery | M-017941 | |
| Other |
Additional files
-
Source data 1
Lane crops of blots used in this manuscript.
- https://cdn.elifesciences.org/articles/59999/elife-59999-data1-v2.zip
-
Source data 2
Scans of full blots used in this manuscript.
- https://cdn.elifesciences.org/articles/59999/elife-59999-data2-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59999/elife-59999-transrepform-v2.docx