A variant-centric perspective on geographic patterns of human allele frequency variation
Figures
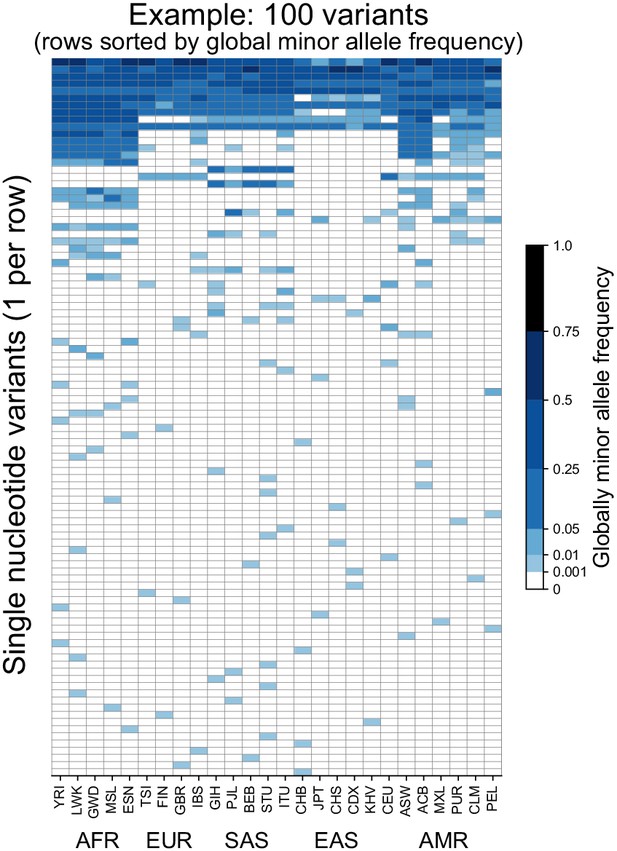
Allele frequencies at 100 randomly chosen variants from Chromosome 22.
Frequencies of the globally minor allele are shown across 26 populations (columns) from the 1KGP for 100 randomly chosen variants from Chromosome 22. Note that the allele frequency bin spacing is nonlinear to capture variation at low as well as high frequencies. Populations are ordered by broad geographic region (horizontal labels, see Figure 2A for legend). Definitions of abbreviations for the 26 1KGP populations are given in Supplementary file 1.
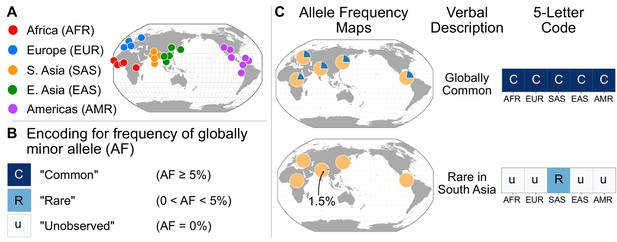
A simple coding system to represent geographic distributions of variants.
(A) Regional groupings of the 26 populations in the 1KGP Project. (B) Legend for minor allele frequency bins. (C) Two examples of how a verbal description of an allele frequency map can be communicated equivalently with a five-letter code (yellow signifies the major allele frequency, blue signifies the minor allele frequency in the pie charts).
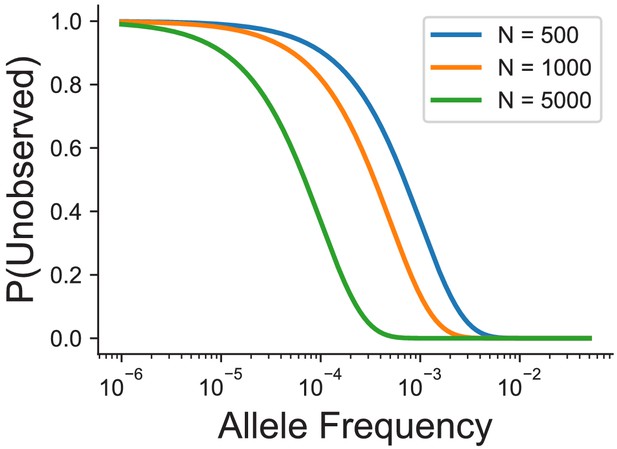
Probability of not observing a variant at a given allele frequency and sample size in number of individuals.
We have assumed that allele frequencies follow Hardy-Weinberg equilibrium, and the probability of no observations of an allele is calculated using the binomial distribution.
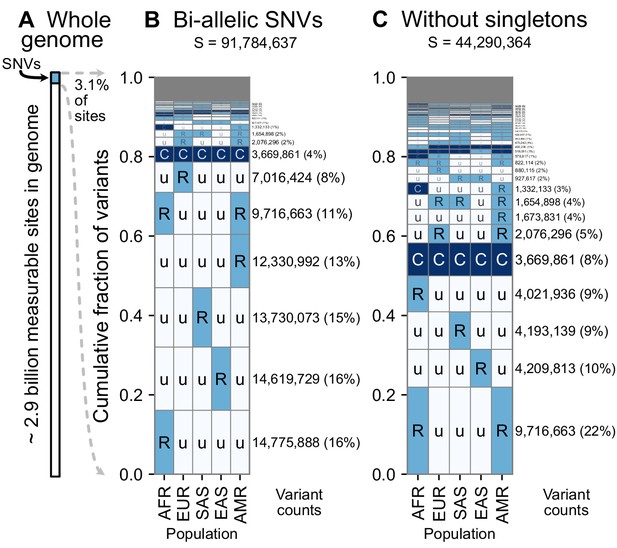
A summary of geographic distributions in human SNVs.
(A) We observe variants at ~3.1% of the measurable sites in the reference human genome (GRCh38). A measurable site is one at which it is possible to detect variation with current sequencing technologies (currently approximately 2.9 Gb out of 3.1 Gb in the human genome; ). (B and C) The relative abundance of different geographic distributions for 1KGP variants, (B) including singletons, and (C) excluding singletons. In panels B and C, the right-hand rectangles show the number and percentage of variants that fall within the corresponding geographic code on the left-hand side; distribution patterns are sorted by their abundance, from bottom-to-top. See Figure 2 for an explanation of the five-letter ‘u’, ’R’, ’C’ codes. The proportion of the genome with variants that have a given geographic distribution code can be calculated from the data above (for example, with the ‘Ruuuu’ code, as 17% × 3.1% = 0.53%). The gray box represents geographic distribution codes whose abundances are too rare to effectively display at the given figure resolution.
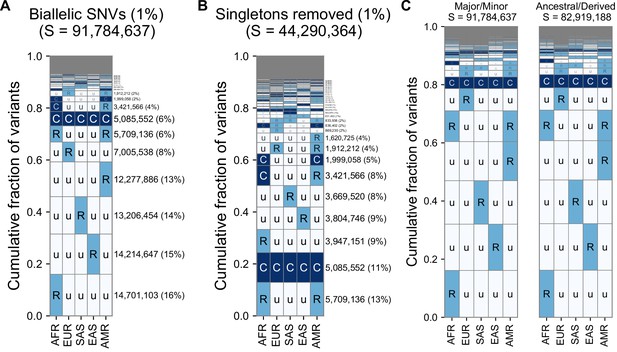
Alternate versions of the GeoVar plots with an alternateallele frequency threshold and tracking derived versus minor allele frequencies.
(A) The relative abundance of geographic distribution codes within the ~92 million variants when using an MAF of 1% as the distinction between ‘common’ (‘C’), and ‘rare’ (‘R’). The right-hand panel shows the percentage of variants that fall within the geographic code represented on the left-hand side; distribution patterns are sorted by their abundance, from bottom-to-top. (B) The abundance of geographic distribution codes for ~44 million non-singleton variants using an MAF of 1% as the boundary between ‘common’ (‘C’), and ‘rare’ (‘R’). (C) Comparison for the abundance of geographic distribution codes when polarizing to the ancestral and derived allele (using build 38) versus major/minor allele. We only include positions where an ancestral allele is supported by at least two outgroups. At 96.6% of variants (80,068,013/82,919,198), the minor allele is also the derived allele. Human ancestral allele calls for GRCh38 based on an eight primate EPO alignment from Ensembl (see key resources table), using only ancestral allele calls supported by at least two outgroup species.
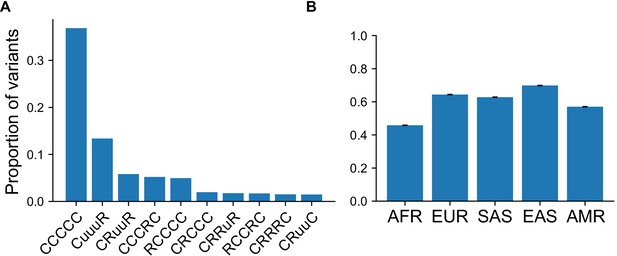
Proportion of variants with specific GeoVar patterns conditional on an allele being common in at least one continental group.
(A) Top 10 categories when conditioning on the variant being ‘common’ (MAF >5%) in at least one continental group. Conditioned on a variant being common in a single g, 37.3% of variants are categorized as ‘globally common’ or ‘CCCCC’. (B) The proportion of variants that fall within the ‘globally common’ or ‘CCCCC’ geographic distribution code conditional on the variant being common (MAF >5%) in the specific continental group.
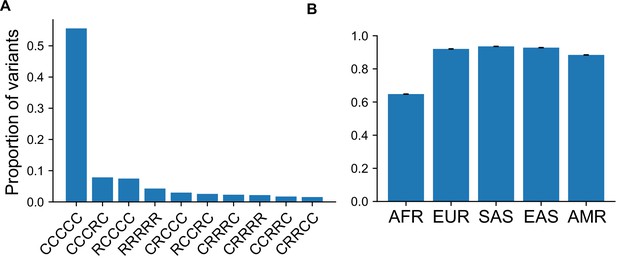
Proportion of variants with specific GeoVar patterns conditional on an allele being `globally widespread'.
(A) The proportion of variants that fall within a given geographic distribution code conditional on the variant being ‘globally widespread’, that is, a category that has no unobserved ('u') codes. We note that 55.6% of variants conditioned on being globally widespread are also globally common (‘CCCCC’). In terms of absolute numbers, variants that are common in at least one population (S = 9,958,838) that are also globally widespread (S = 6,322,767) comprise ~63% of the total when conditioning on being common in at least one population. When conditioning on variants common only in regions outside Africa (S = 7,544,648), the percentage of globally widespread variants (S = 6,179,781) increases to ~82%. (B) The proportion of variants that fall within a ‘globally present’ category, defined as categories that contain no unobserved (‘u’) codes, conditional on the variant being common (MAF >5%) in the specific continental group.
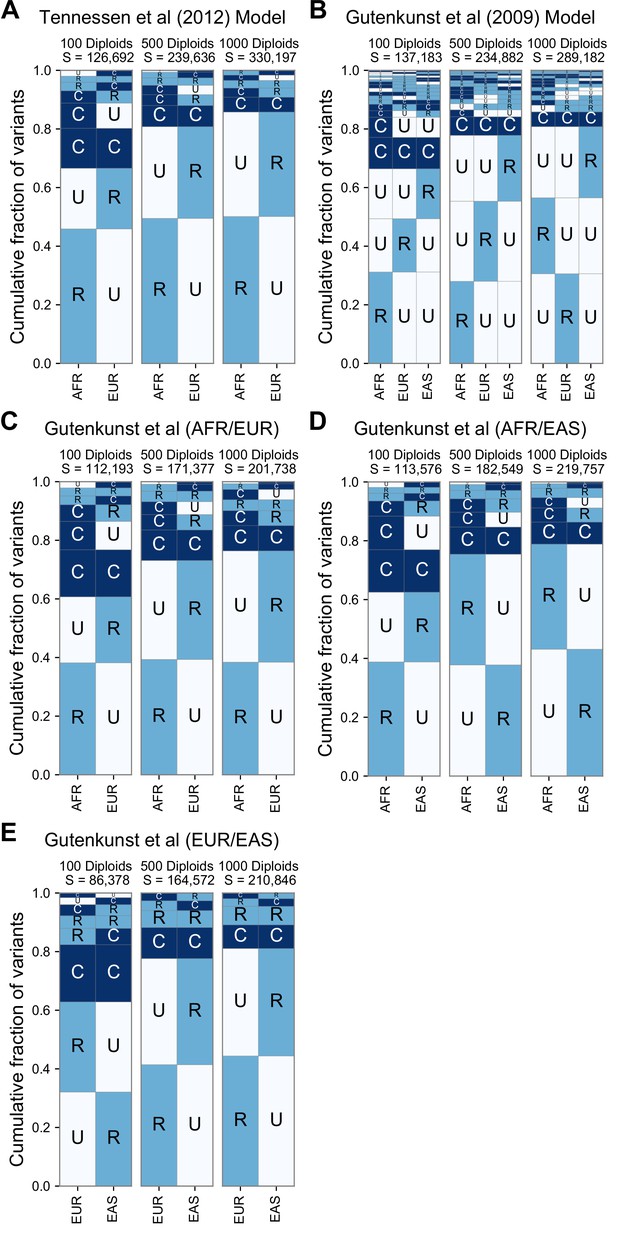
GeoVar plots derived from simulations of two published models of human demography.
(A) Gutenkunst et al., 2009, (B–E) Tennessen et al., 2012. For each model, we used stdpopsim (Adrion et al., 2020) to simulate 10 replicates of SNVs equivalent to 5% of Chromosome 22. For each model we simulated three different sample sizes per population, the first with 100 diploid samples, 500 diploid samples, and 1000 diploid samples. The panels with n = 500 diploid samples per population most closely match the sampling within the 1KGP (nAFR = 504, nEUR = 503, nEAS = 504). Both models replicate the qualitative prevalence of the ‘localized rare’ (‘RU’) and ‘globally common’ (‘CC’) patterns that we see in the 1KGP data. With higher sample sizes we find an increased proportion of localized rare (‘RU’) patterns, due to increased detection power. Panels (C–E) show specific pairwise comparisons of populations in the model of Gutenkunst et al., 2009 to compare against the two-population model of Tennessen et al., 2012. Panels (A) and (C) show that, when restricted to AFR/EUR comparisons, the two models predict very similar patterns. The prevalence of localized rare and globally common patterns is reproduced across all comparisons, as is the dependence on sample size. The EUR/EAS comparison (E) shows a larger number of ‘RR’ patterns, presumably reflecting the more recent divergence of those populations.
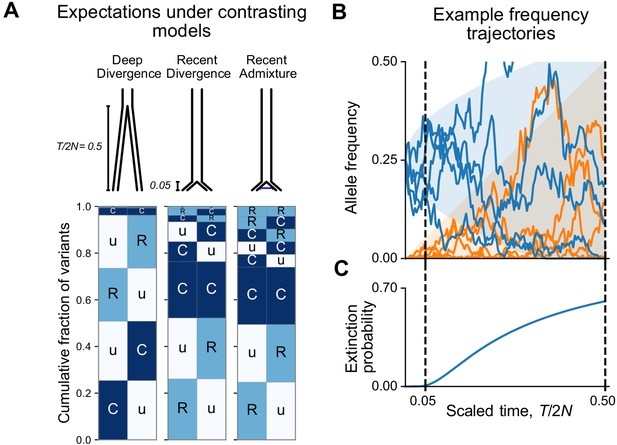
Allele frequency patterns depend on the time since population divergence and levels of admixture.
(A) Expected geographic distribution code abundances in a sample of 100 diploid individuals from each of two populations, for deep divergence ( = 0.5, = 0), recent divergence without admixture ( = 0.05, = 0), and recent divergence with admixture ( = 0.05, = 0.02). (B) Simulated allele frequency time series for mutations starting at 25% frequency (blue) and new mutations entering the population since the split (orange). (C) The probability of extinction of a mutation starting at 25% frequency (see Appendix 2).
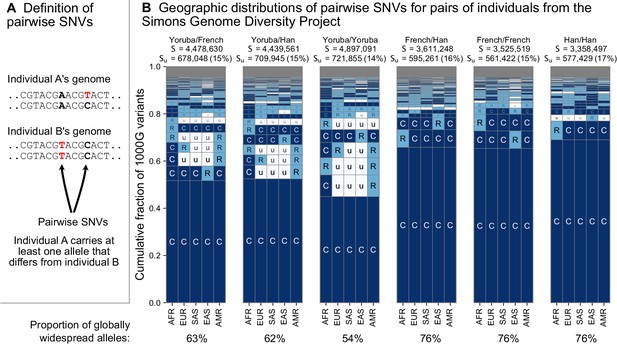
The geographic distributions of SNVs between pairs of individuals.
(A) Definition of a pairwise SNV. (B) The abundance of geographic distribution codes for different pairs of individuals from the SGDP dataset. Above each plot, we show the total number of variants that differ between each individual (S) and the number that were unobserved completely in the 1KGP data (SU). Across the bottom, we show the proportion of variants with globally widespread alleles for each pair. We calculate this as the fraction of variants with no ‘u’ encodings over the total number of variants (S). (Note: by doing so, we make the assumption that if a variant is not found in the 1KGP data it is not globally widespread). For this analysis, as in Mallick et al., 2016, we include only autosomal biallelic SNVs for variants that pass ‘filter level 1’.
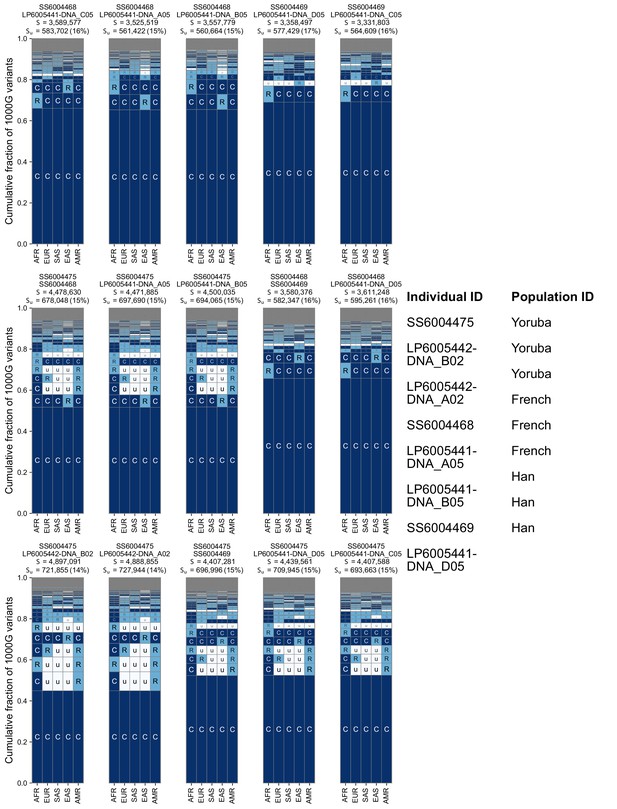
Additional examples of geographic distribution codes for pairwise variants from different pairs of sampled individuals in the SGDP.
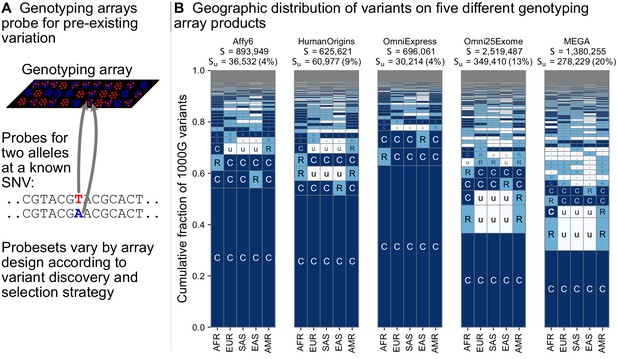
Geographic distribution for variants found on genotyping array products.
(A) Genotyping arrays consist of probes for a fixed set of variants chosen during the design of the array product. (B) For each array product, we extracted the genomic position of variants found on the array and kept variants that are also found within the 1KGP to highlight their geographic distributions. The arrays considered are the Affymetrix 6.0 (Affy6) genotyping array, the Affymetrix Human Origins array (HumanOrigins), the Illumina HumanOmniExpress (OmniExpress) array, the Illumina Omni2.5Exome, and the Illumina MEGA array. This plot is analogous to Figure 3B but rather than calculating frequencies with the five regional groupings, we compute them within each of the 26 1KGP populations. The total number of variants represented is the same as in Figure 3B (S = 91,784,367). See Figure 2 for an explanation of the ‘u’,’R’,’C’ codes.
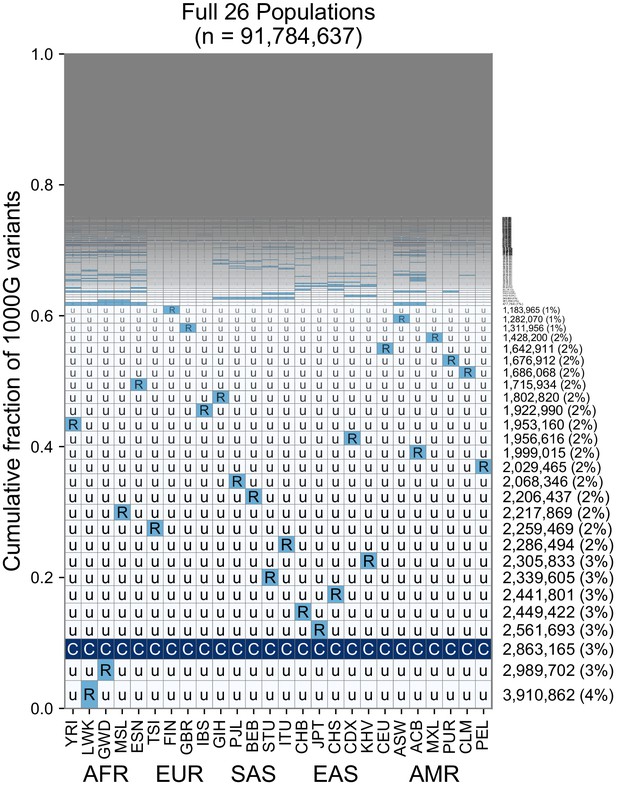
A finer-scale summary of geographic distributions in human SNVs from the 1KGP.
This plot is analogous to Figure 3B but rather than calculating frequencies with the five regional groupings, we compute them within each of the 26 1KGP populations. The total number of variants represented is the same as in Figure 3B (S = 91,784,367). See Figure 2 for an explanation of the ‘u’,’R’,’C’ codes.
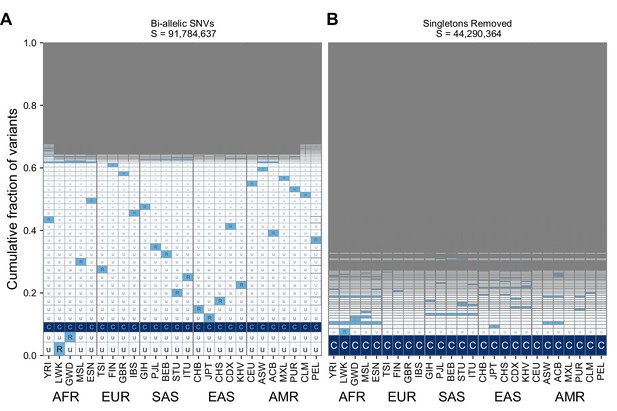
The geographic distribution of variants across all 26 populations (for legend see Supplementary file 1) in the 1KGP both with singletons included (A) and removed (B).
Regional groupings are provided on the bottom to reflect our choices for population groupings throughout the main paper.
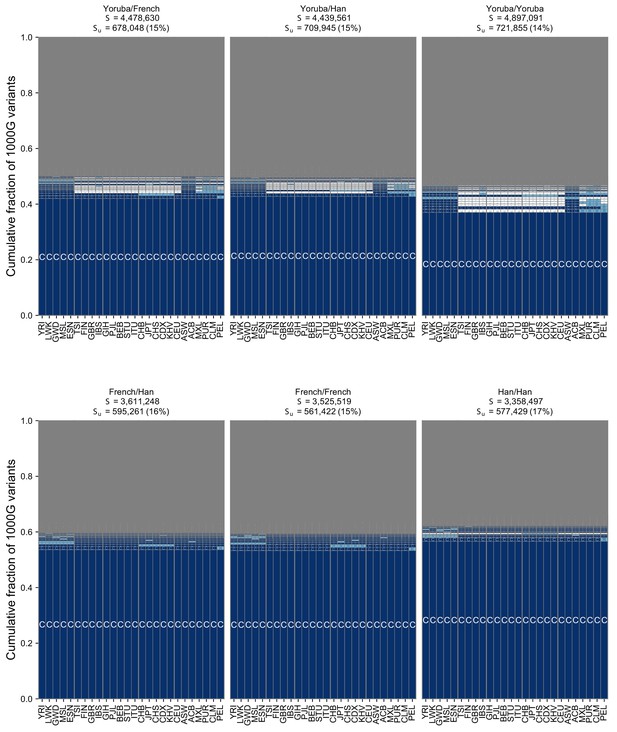
The geographic distribution of pairwise SNVs across pairs of individuals from the Simons Genome Diversity Project using the full set of 26 populations from the 1KGP.
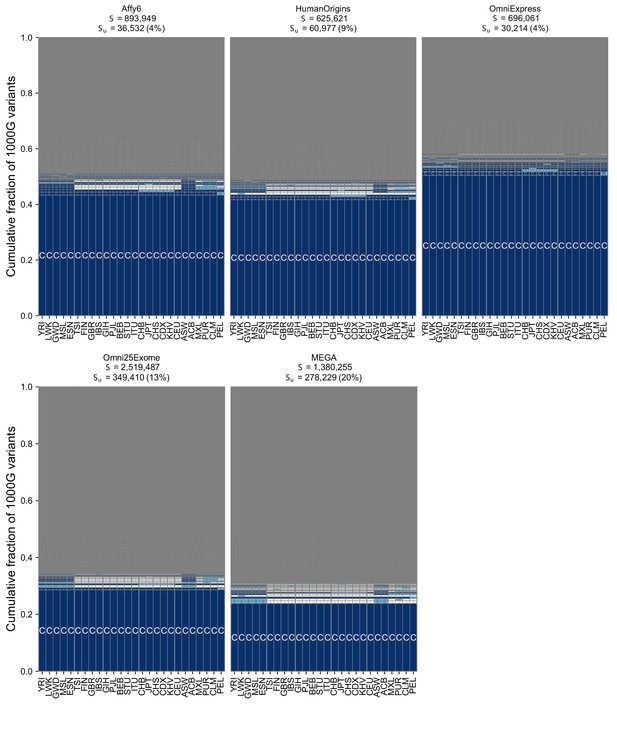
The geographic distribution of SNVs on genotyping s using the full set of 26 populations from the 1KGP.
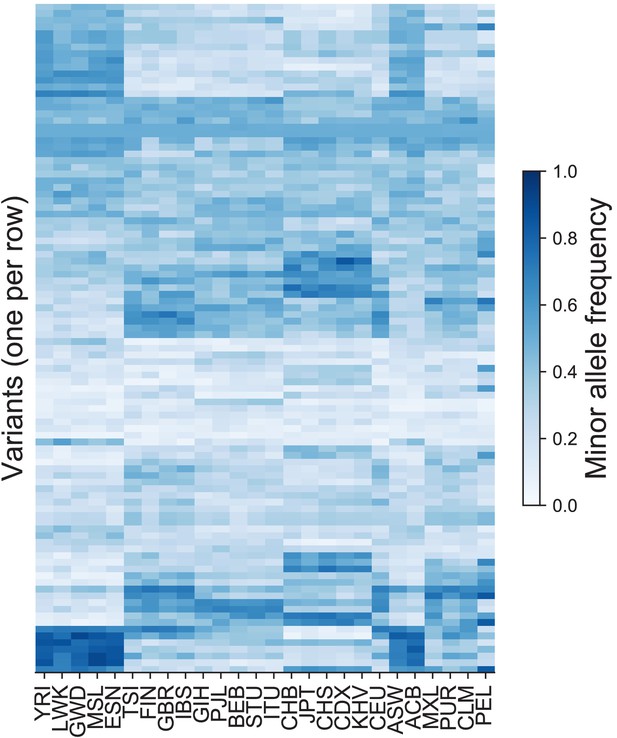
The minor allele frequencies of 300 variants in each of the 26 original population labels in the 1KGP.
The variants were chosen at random from among those on Chromosome 22 that have MAF >5% in all 26 populations. For example, the top row represents an allele that has higher frequency in several African and admixed American populations. Variants are ordered based on hierarchical clustering on the Euclidean distance between minor allele frequency profiles across all populations.
Tables
Additional files
-
Supplementary file 1
Table S1.
Population abbreviations and groupings used in the study.
- https://cdn.elifesciences.org/articles/60107/elife-60107-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60107/elife-60107-transrepform-v2.docx





