Chimpanzee brain morphometry utilizing standardized MRI preprocessing and macroanatomical annotations
Figures
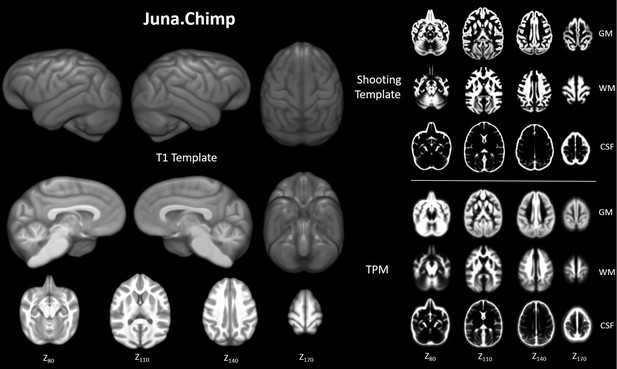
Juna.Chimp templates including the average.
T1- template, tissue probability maps (TPM), and Geodesic Shooting template. For Shooting templates and TPM axial slices are shown of gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF). All templates are presented at 0.5 mm resolution.
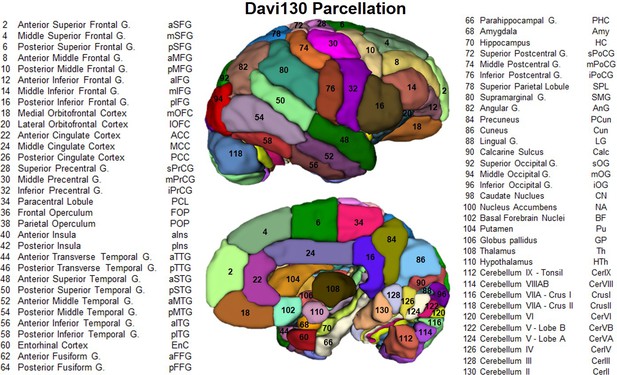
Lateral and medial aspect of the Davi130 parcellation right hemisphere.
Visible regions are numbered with Davi130 parcellation region numbers and correspond to names in the figure. Even numbers correspond to regions in the right hemisphere (as shown in the figure), while left hemisphere regions are odd numbers. A list of all Davi130 labels can be found at Figure 2—source data 1.
-
Figure 2—source data 1
Source file for Complete List of Davi130 Labels.
- https://cdn.elifesciences.org/articles/60136/elife-60136-fig2-data1-v2.docx
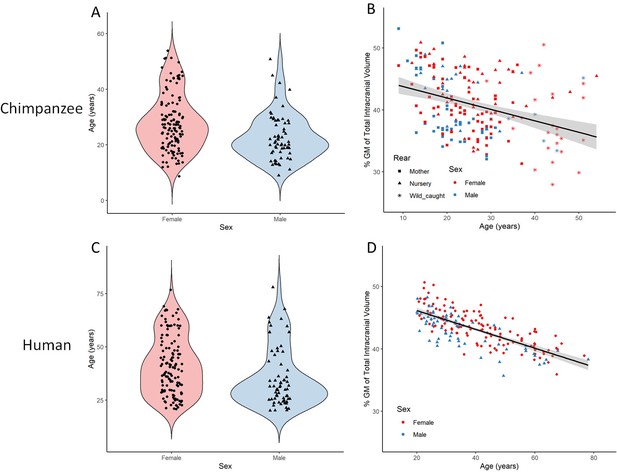
Total gray matter volume decline during aging in chimpanzees and matched human sample.
(A) Distribution of age and sex in the final sample of 194 chimpanzees. (B) Linear relationship between GM and age with standard error for chimpanzee sample. (C) Distribution of age and sex in the human (IXI) matched sample of 194 humans. (D) Linear relationship between GM and age with standard error for human sample. Figure 3—figure supplement 1 presents the age and sex distribution of the whole sample (n = 223). Figure 3—figure supplement 2 presents the age and sex distribution of the whole IXI sample (n = 496).
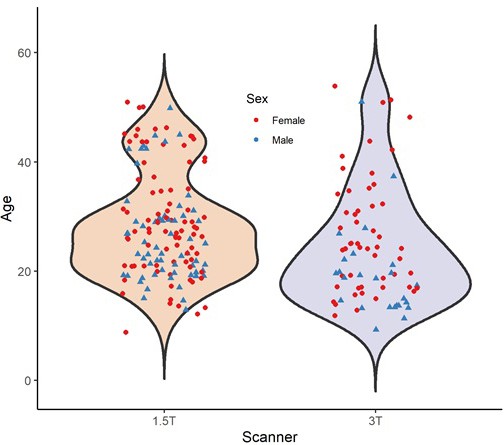
Age and sex distribution of complete chimpanzee (n = 223) sample separated by scanner field strength.

Age and sex distribution of complete IXI human sample (n = 496) separated by scanner field strength.
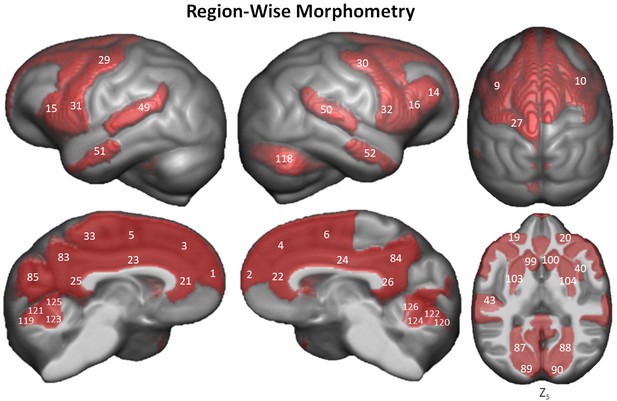
Region-wise morphometry in the Davi130 parcellation age regression.
Red regions represent Davi130 regions that remained significant at p≤0.05 following FWE correction (Holm method). The T-statistic and p-value for all Davi130 labels can be found in Figure 4—source data 1. 1 and 2 – aSFG, 3 and 4 – mSFG, 5 and 6 – pSFG, 9 and 10 – pMFG, 14 – mIFG, 15 and 16 – pIFG, 19 and 20 – lOFC, 21 and 22 – ACC, 23 and 24 – MCC, 25 and 26 – PCC, 27 – sPrCG, 29 and 30 – mPrCG, 31 and 32 – iPrCG, 33 – PCL, 40 – aIns, 43 – aTTG, 49 and 50 – pSTG, 51 and 52 – aMTG, 83 and 84 – PCun, 85 – Cun, 87 and 88 – LG, 89 and 90 – Calc, 97 and 98 – CN, 99 and 100 – NA, 103 and 104 – Pu, 118 – CrusII, 119 and 120 – CerVI, 121 and 122 – CerVB, 123 and 124 – CerVB, 125 and 126 – CerIV.
-
Figure 4—source data 1
Aging effect on gray matter in complete Davi130 Labels.
- https://cdn.elifesciences.org/articles/60136/elife-60136-fig4-data1-v2.docx
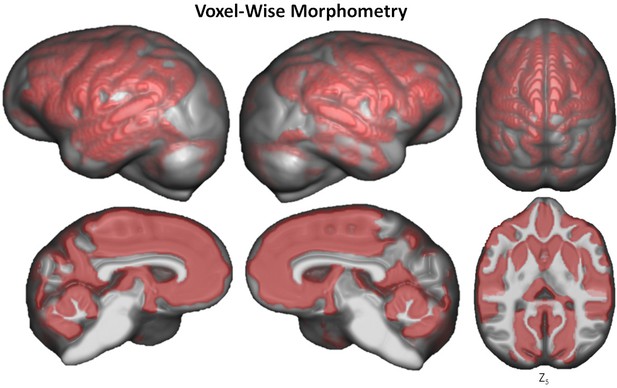
Voxel-based morphometry of aging on GM volume.
The significant clusters are found using TFCE with FWE correction at p≤0.05.

Voxel-based morphometry of aging on GM volume using TFCE with FWE correction at p≤0.05 without rearing as a covariate.
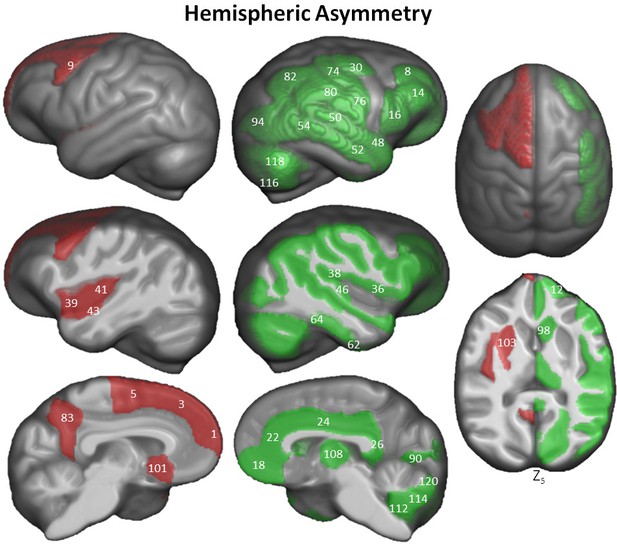
Hemispheric asymmetry of Davi130 regions within the chimpanzee sample.
Significant leftward (red) and rightward (green) asymmetrical regions are those with a p≤0.05 after FWE correction. The T-statistic and p-value for all Davi130 labels can be found in Figure 6—source data 1. 1 – aSFG, 3 – mSFG, 5 – pSFG, 8 – aMFG, 9 – pMFG, 12 – aIFG, 14 – mIFG, 16 – pIFG, 18 – mOFC, 22 – ACC, 24 – MCC, 26 – PCC, 30 – mPrCG, 36 – FOP, 38 – POP, 39 – aIns, 41 – pIns, 43 – aTTG, 46 – pTTG, 48 – aSTG, 50 – pSTG, 52 – aMTG, 54 – pMTG, 62 – aFFG, 64 – pFFG, 74 – mPoCG, 76 – iPoCG, 80 – SMG, 82, AnG, 83 – PCun, 90 – Calc, 94 – mOG, 98 – CN, 101 – BF, 103 – Pu, 108 – Th, 112 – CerIX, 114 – CerVIII, 115 –CrusI, 118 –CrusII, 120 – CerVI.
-
Figure 6—source data 1
Complete Davi130 labels hemispheric asymmetry.
- https://cdn.elifesciences.org/articles/60136/elife-60136-fig6-data1-v2.docx
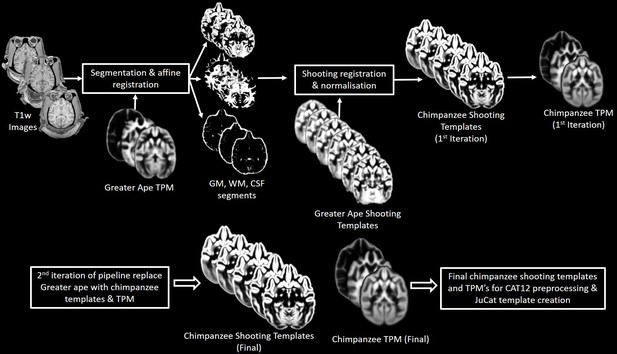
Workflow for creation of chimpanzee-specific shooting template and TPM, which can then be used in CAT12 structural preprocessing pipeline to create the Juna.Chimp template.
The resulting chimpanzee-shooting template, TPM and CAT atlas establishes the robust and reliable base to segment and spatially normalize the T1w images utilizing CAT12’s processing pipeline (Dahnke and Gaser, 2017; Gaser et al., 2020).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | CAT12 | http://www.neuro.uni-jena.de/cat/ | RRID:SCR_019184 | |
| Software, algorithm | NCBR | http://www.chimpanzeebrain.org/ | RRID:SCR_019183 | |
| Software, algorithm | MATLAB | http://www.mathworks.com/products/matlab/ | RRID:SCR_001622 | |
| Software, algorithm | SPM | http://www.fil.ion.ucl.ac.uk/spm/ | RRID:SCR_007037 | |
| Software, algorithm | RStudio | http://www.rstudio.com/ | RRID:SCR_000432 | |
| Software, algorithm | 3D Slicer | http://slicer.org/ | RRID:SCR_005619 |





