Caenorhabditis elegans methionine/S-adenosylmethionine cycle activity is sensed and adjusted by a nuclear hormone receptor
Figures
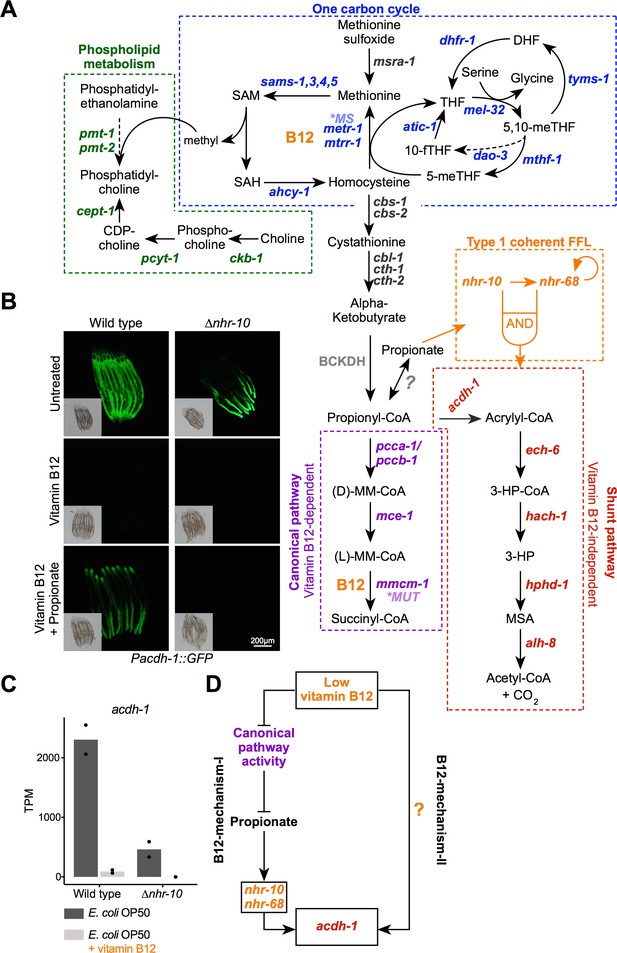
Two mechanisms of gene regulation by low vitamin B12 dietary conditions.
(A) Cartoon of vitamin B12-related metabolic pathways in C. elegans. CDP –cytidine 5’-diphosphocholine; DHF – dihydrofolate; 3-HP – 3-hydroxypropionate; 5,10-meTHF – 5,10-methylenetetrahydrofolate; 5-meTHF – 5-methyltetrahydrofolate; 10-fTFH –10-formyltetrahydrofolate; BCKDH – branched-chain α-ketoacid dehydrogenase complex; MM-CoA – methylmalonyl-coenzyme A; *MUT – human methylmalonyl-coenzyme A mutase; *MS – human methionine synthase; MSA – malonic semialdehyde; SAH – S-adenosylhomocysteine; SAM – S-adenosylmethionine; THF – tetrahydrofolate; FFL – feed forward loop. Dashed arrows indicate multiple reaction steps. (B) Fluorescence microscopy images of Pacdh-1::GFP reporter animals in wild type and ∆nhr-10 mutant background with different supplements as indicated. Insets show brightfield images. (C) RNA-seq data of acdh-1 mRNA with and without 20 nM vitamin B12 in wild type and ∆nhr-10 mutant animals (Bulcha et al., 2019). Datapoints show each biological replicate and the bar represents the mean. TPM – transcripts per million. p adjusted values are provided in Supplementary file 1. (D) Cartoon illustrating two mechanisms of gene regulation by low vitamin B12 dietary conditions.
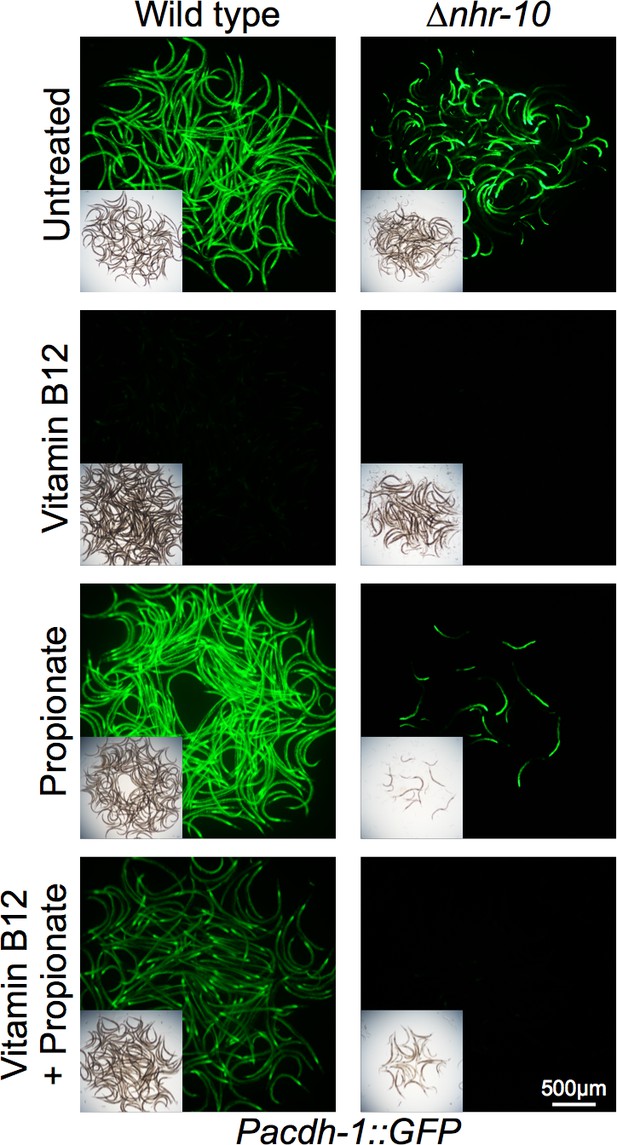
Fluorescent microscopy images of Pacdh-1::GFP animals in wild type and ∆nhr-10 mutant backgrounds with supplemented metabolites as indicated.
Insets show brightfield images.
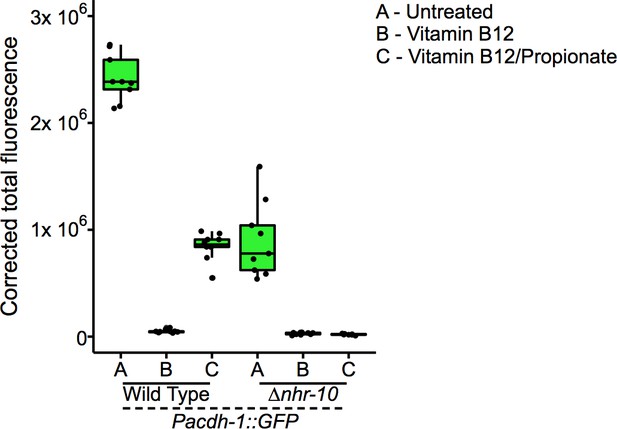
Boxplot showing median and interquartile range of normalized GFP intensity measurements of fluorescent images shown in Figure 1B.
GFP intensity measurements of fluorescent images shown in Figure 1B.
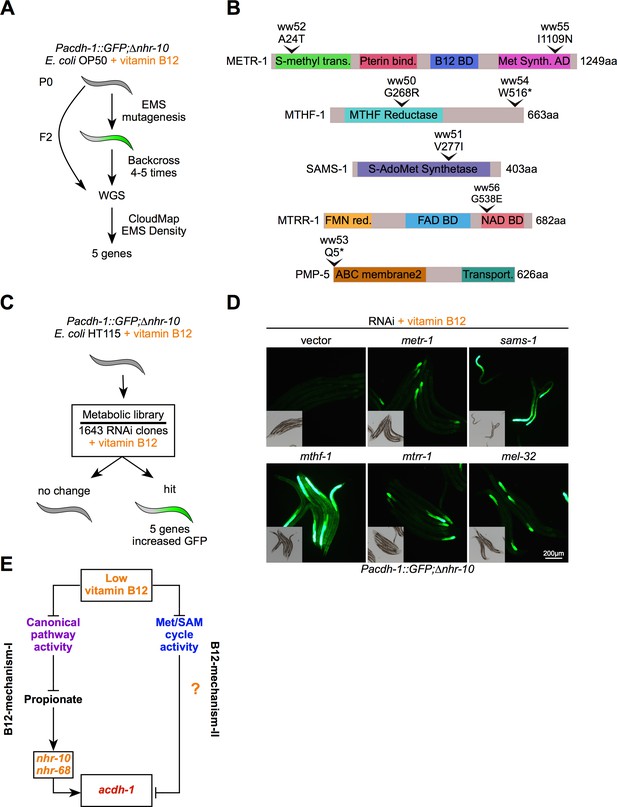
Met/SAM cycle perturbations activate B12-mechanism-II.
(A) Workflow for the EMS mutagenesis screen using Pacdh-1::GFP;∆nhr-10 reporter animals supplemented with 20 nM vitamin B12. WGS – whole genome sequencing. (B) To-scale cartoons of amino acid changes in the proteins encoded by the genes found in the forward genetic screen. (C) Workflow for RNAi screen using Pacdh-1::GFP;∆nhr-10 reporter animals supplemented with 20 nM vitamin B12. (D) Fluorescence microscopy images of Pacdh-1::GFP;∆nhr-10 reporter animals subjected to RNAi of the indicated metabolic genes. Insets show brightfield images. (E) Cartoon illustrating the activation of B12-mechanism-II by low vitamin B12 or genetic perturbations in the Met/SAM cycle.
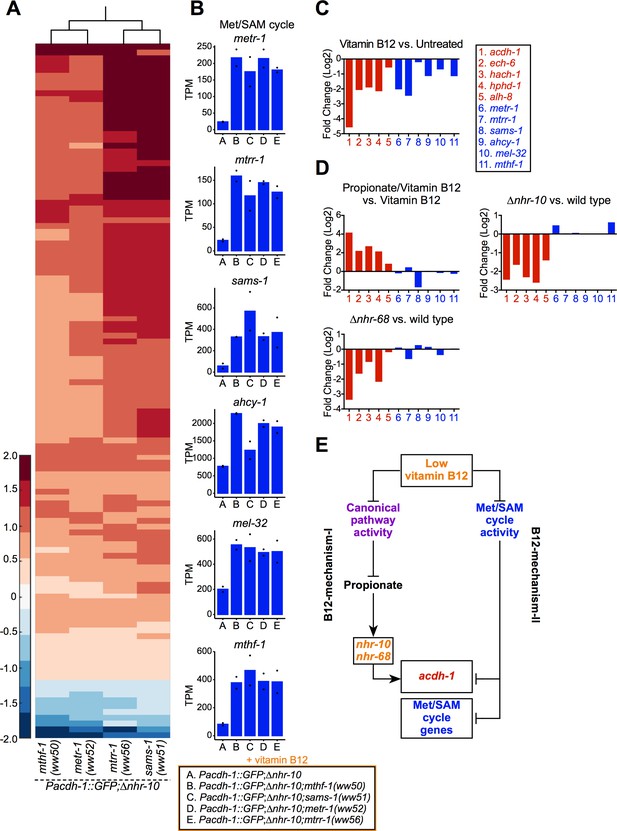
Genetic mutations in Met/SAM cycle genes activate Met/SAM cycle gene expression.
(A) Hierarchal clustering of log10-transformed fold change RNA-seq data. Changes are relative to the Pacdh-1::GFP;∆nhr-10 parent strain with the cutoffs of a fold change of 1.5 and a p adjusted value less than 0.01. Only genes that change in expression in all four Met/SAM cycle gene mutants are shown. (B) Bar graphs of Met/SAM cycle gene expression by RNA-seq in the four Met/SAM cycle mutants and in the Pacdh-1::GFP;∆nhr-10 parent strain. Datapoints show each biological replicate and the bar represents the mean. TPM – transcripts per million. p adjusted values are provided in Supplementary file 2. (C) RNA-seq comparison of Met/SAM cycle (blue) and propionate shunt (red) gene expression in response to 20 nM vitamin B12. Bar represents the mean of two biological replicates. p adjusted values are provided in Supplementary file 1. (D) Comparison of Met/SAM cycle and propionate shunt genes in response to 20 nM vitamin B12 plus 40 mM propionate or in nhr-10 and nhr-68 mutant animals. Bar represents the mean of two biological replicates. p adjusted values are provided in Supplementary file 1. (E) Cartoon illustrating met/SAM cycle gene activation in response to B12-mechanism-II.
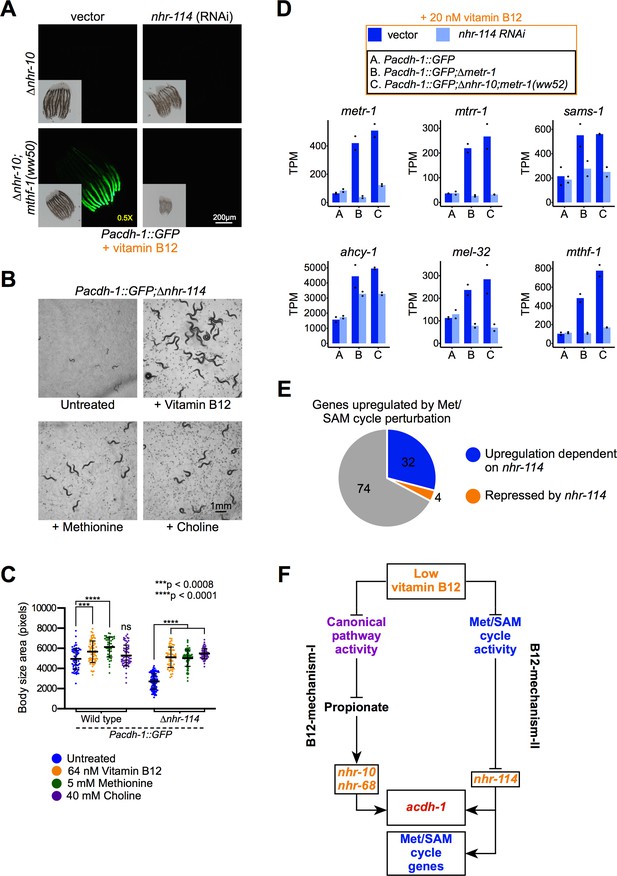
nhr-114 is required for B12-mechanism-II in response to Met/SAM cycle perturbations.
(A) nhr-114 RNAi reduces GFP expression in Pacdh-1::GFP;∆nhr-10 animals harboring Met/SAM cycle gene mutations. Insets show brightfield images. (B) ∆nhr-114 mutant growth and fertility phenotypes are rescued by vitamin B12, methionine or choline supplementation. Difference in exposure time is indicated in yellow. (C) Quantification of body size of wild type and ∆nhr-114 mutant animals that are untreated or supplemented with either vitamin B12, methionine, or choline. Trial two is shown in Figure 4—figure supplement 4. Statistical significance was determined by the Kruskal-Wallis test with post hoc comparison using Dunn’s multiple comparison test. (D) Bar graphs of Met/SAM cycle gene expression by RNA-seq in Pacdh-1::GFP, Pacdh-1::GFP;∆metr-1 and Pacdh-1::GFP;∆nhr-10;metr-1(ww52) animals treated with vector control or nhr-114 RNAi. Datapoints show each biological replicate and the bar represents the mean. TPM – transcripts per million. p adjusted values are provided in Supplementary file 3. (E) Pie chart showing portion of genes upregulated by Met/SAM cycle perturbation that are nhr-114 dependent. (F) Cartoon illustrating the requirement of nhr-114 in B12-mechanism-II.
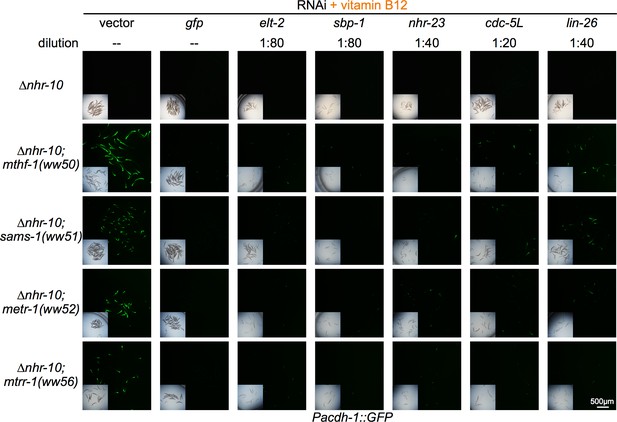
Diluted RNAi of indicated TFs repress Pacdh-1::GFP in met/SAM cycle mutants harboring a deletion in nhr-10.
Some TF RNAi experiments were diluted with vector control RNAi to circumvent strong deleterious phenotypes. Dilution ratios of clone to vector by volume are indicated per clone. Insets show brightfield images.
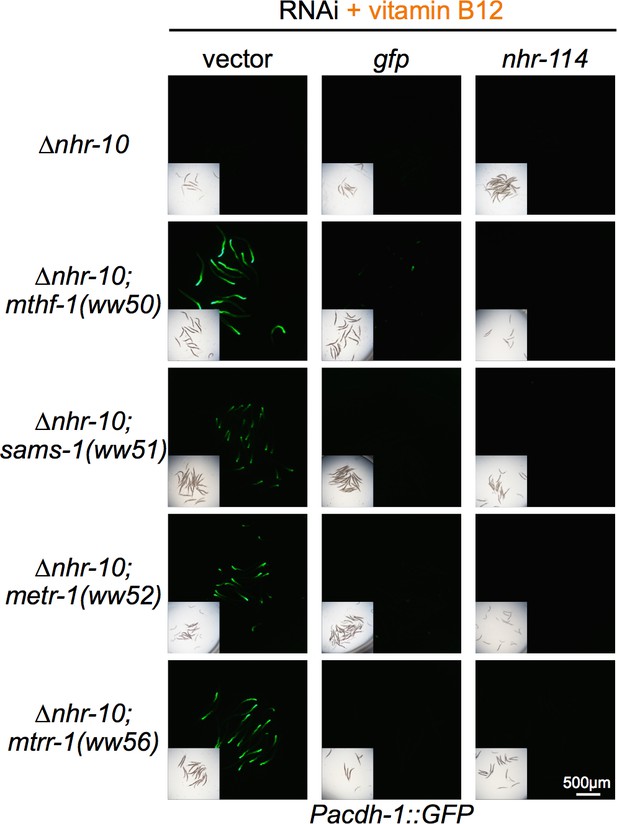
nhr-114 RNAi represses Pacdh-1::GFP expression in all Met/SAM cycle mutants.
Insets show brightfield images.
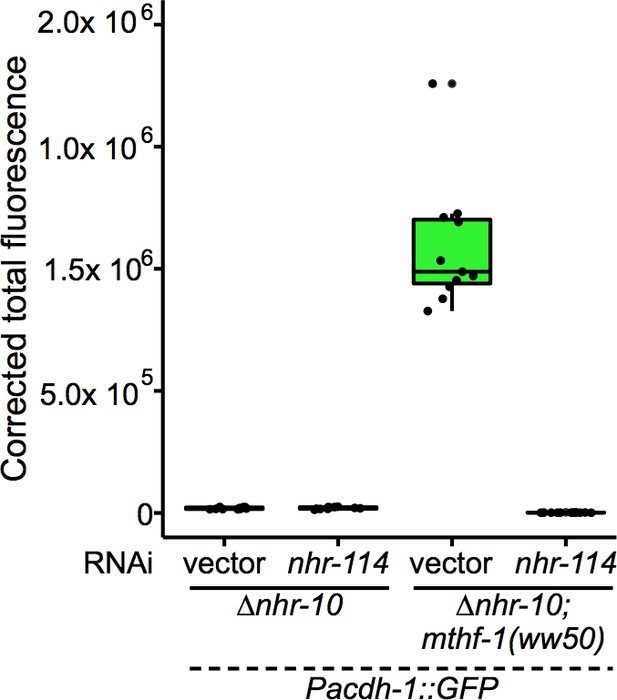
Boxplot showing median and interquartile range of normalized GFP intensity measurements of fluorescent images shown in Figure 4A.
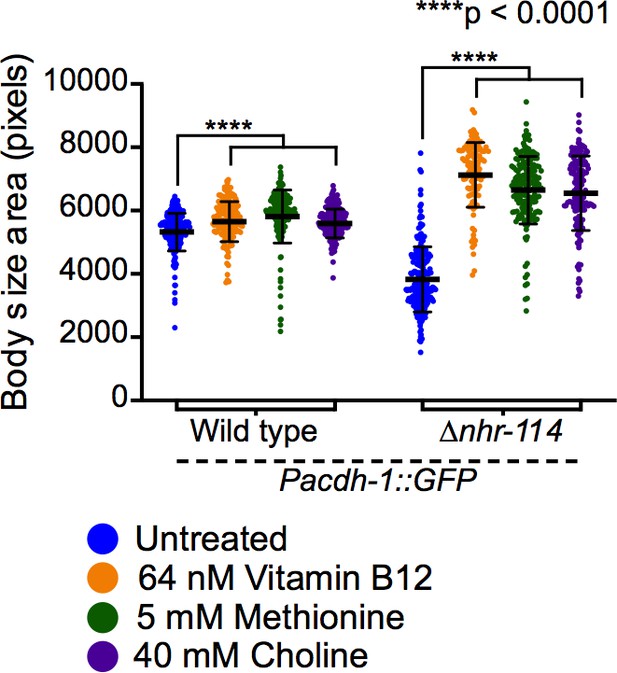
Replicate experiment of rescue of ∆nhr-114 mutant developmental phenotype by vitamin B12, methionine, and choline shown in Figure 4C.
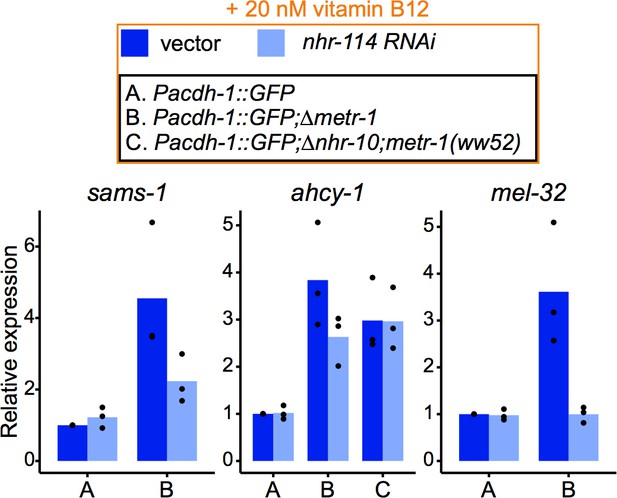
Relative mRNA level expression (fold change) as determined by qRT-PCR of Met/SAM cycle genes whose p adjusted values were high in RNA-seq shown in Figure 4D.
Datapoints show each biological replicate and bar represents the mean.
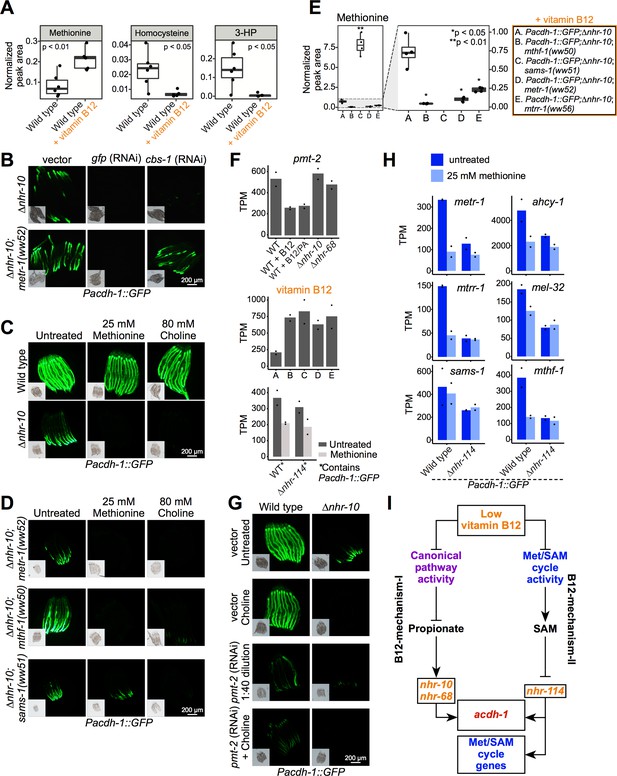
Methionine and choline supplementation suppress vitamin B12-mechanism-II.
(A) Box plots showing GC-MS data from wild type animals fed E. coli OP50 with or without supplemented 64 nM vitamin B12. Statistical significance determined by two-tailed t-test. 3-HP – 3-hydroxypropionate. (B) cbs-1 RNAi reduces GFP expression in Pacdh-1::GFP;∆nhr-10 and Pacdh-1::GFP;∆nhr-10 mutants harboring Met/SAM cycle gene mutations. (C) Methionine or choline supplementation represses GFP expression in Pacdh-1::GFP;∆nhr-10 animals. Insets show brightfield images. (D) Methionine or choline supplementation represses GFP expression in Pacdh-1::GFP;∆nhr-10;metr-1(ww52) and Pacdh-1::GFP;∆nhr-10;mthf-1(ww50) animals, while choline but not methionine supplementation represses GFP expression in Pacdh-1::GFP;∆nhr-10;sams-1(ww51) animals. This experiment was performed without vitamin B12 supplementation. (E) GC-MS quantification of methionine levels in Met/SAM cycle mutants and in the parental strain. Statistical significance was determined using one-way ANOVA with post-hoc comparison using Dunnett’s T3 test. (F) RNA-seq data of pmt-2 expression in each of the datasets. PA – propionic acid; X axis labels match legend in panel E. Datapoints show each biological replicate and the bar represents the mean. TPM – transcripts per million. p adjusted values are provided in Supplementary files 1, 2 and 4. (G) pmt-2 RNAi suppresses GPF expression in Pacdh-1::GFP;∆nhr-10 animals. pmt-2 RNAi experiments were diluted with vector control RNAi to circumvent strong deleterious phenotypes. (H) Bar graphs of RNA-seq data showing Met/SAM cycle gene expression in wild type and ∆nhr-114 animals with and without methionine supplementation. p adjusted values are provided in Supplementary file 4. (I) Cartoon illustrating B12-mechanism-II whereby low vitamin B12 reduces Met/SAM cycle activity, leading to the depletion of SAM and the activation of met/SAM cycle gene expression mediated by nhr-114.
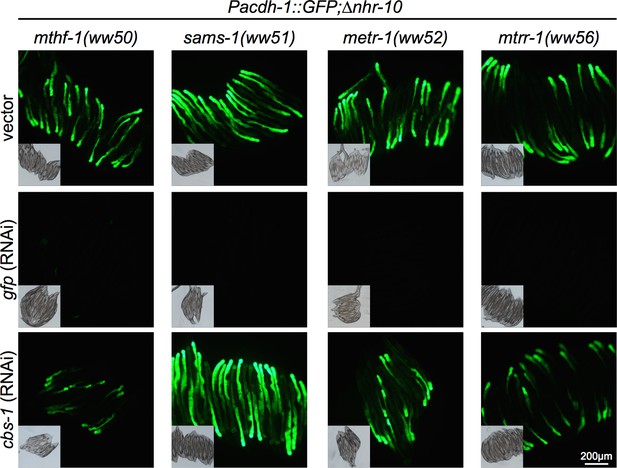
Met/SAM cycle point mutants expressing Pacdh-1::GFP treated with cbs-1 RNAi.
Insets are brightfield images.
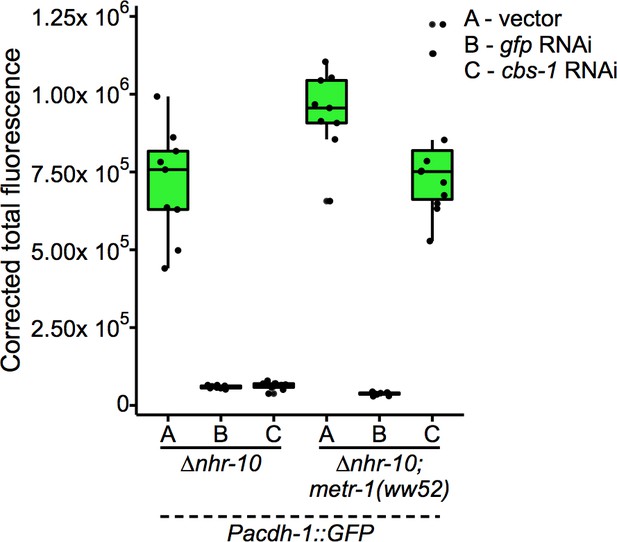
Boxplot showing median and interquartile range of normalized GFP intensity measurements of fluorescent images shown in Figure 5B.
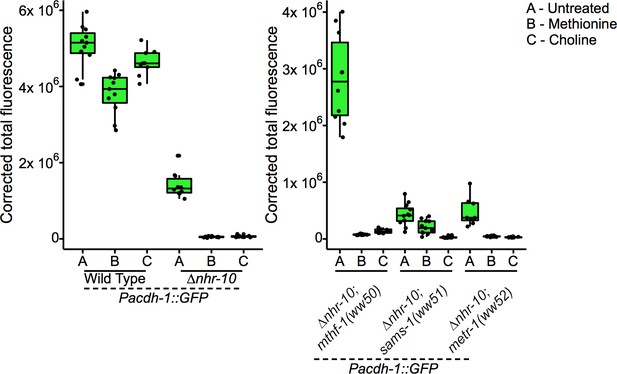
Boxplot showing median and interquartile range of normalized GFP intensity measurements of fluorescent images shown in Figure 5C and D.
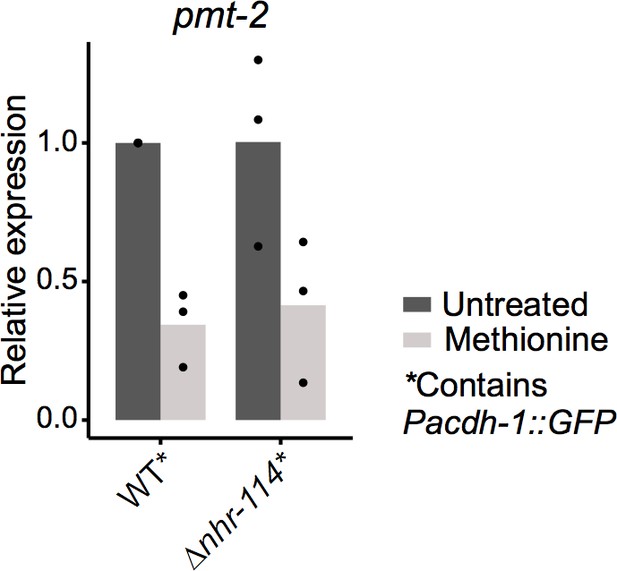
Relative mRNA level expression (fold change) as determined by qRT-PCR of pmt-2 whose p adjusted value was high in RNA-seq shown in Figure 5F.
Datapoints show each biological replicate and bar represents the mean.
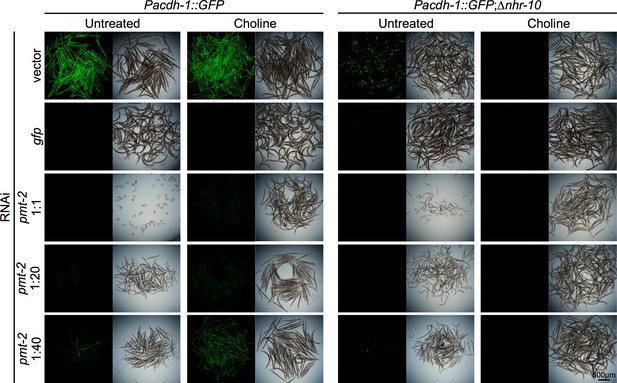
Wild type or ∆nhr-114 expressing Pacdh-1::GFP with pmt-2 RNAi supplemented with and without choline.
Dilutions by volume of pmt-2 RNAi with vector control are indicated. Insets show brightfield images.
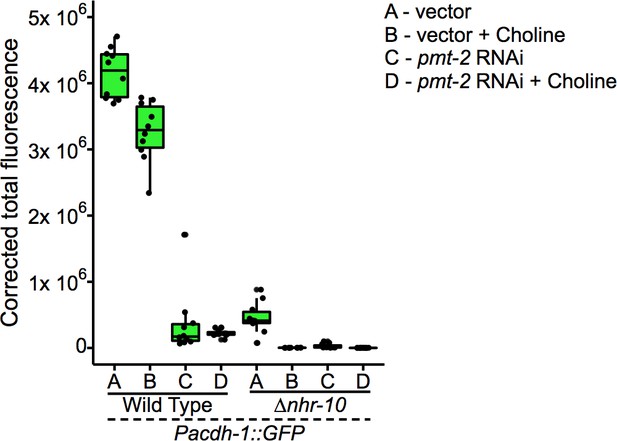
Boxplot showing median and interquartile range of normalized GFP intensity measurements of fluorescent images shown in Figure 5G.
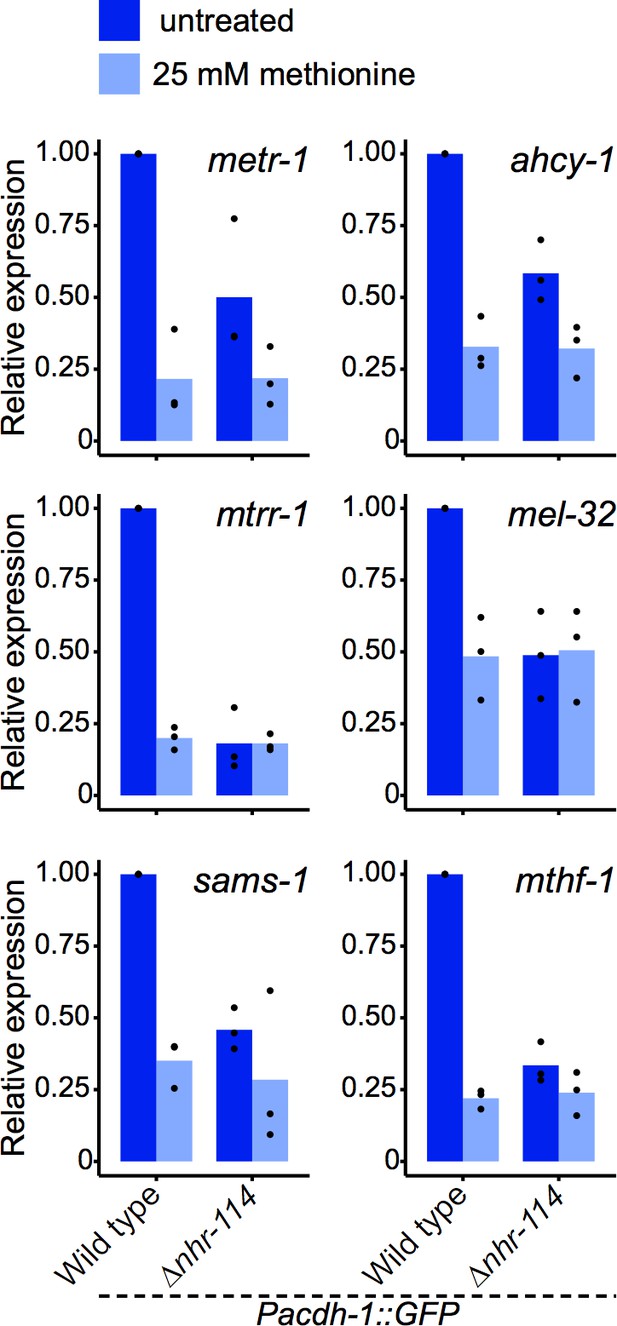
Relative mRNA level expression (fold change) as determined by qRT-PCR of Met/SAM cycle genes whose p adjusted value was high in RNA-seq shown in Figure 5H.Datapoints show each biological replicate and bar represents the mean.
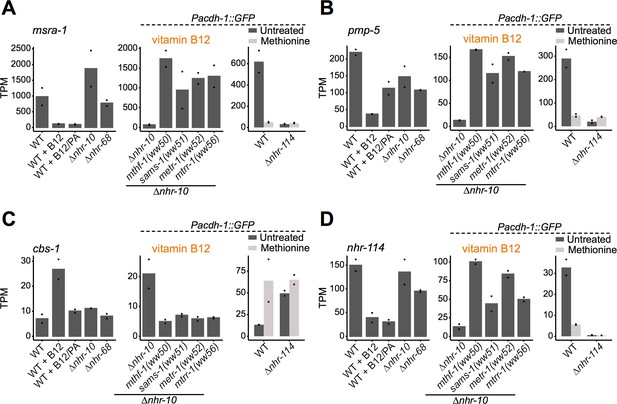
nhr-114 transcriptionally regulates Met/SAM cycle influx and efflux.
(A, B, C) RNA-seq data from each of the experiments for msra-1 (A), pmp-5 (B), cbs-1 (C) and nhr-114 (D). Datapoints show each biological replicate and the bar represents the mean. TPM – transcripts per million. p adjusted values are provided in Supplementary files 1, 2 and 4.
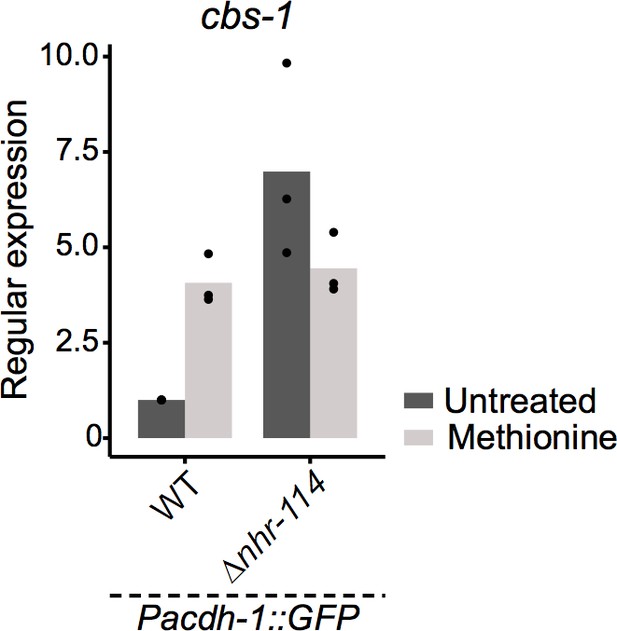
Relative mRNA level expression (fold change) as determined by qRT-PCR of cbs-1 whose p adjusted value was high in RNA-seq shown in Figure 6C.
Datapoints show each biological replicate and bar represents the mean.
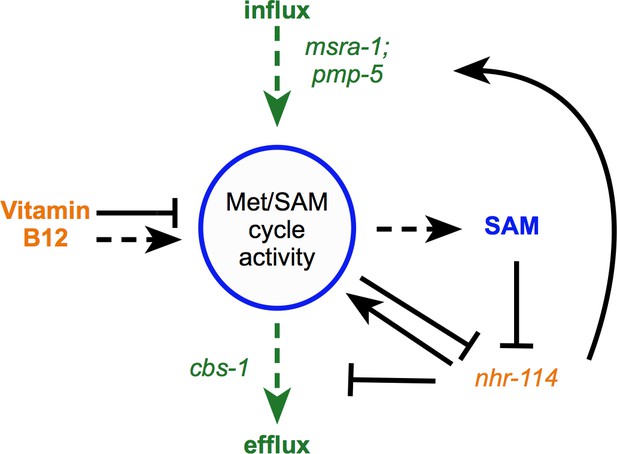
Model of B12-mechanism-II.
Vitamin B12 modulates the transcription of Met/SAM cycle genes and controls in/efflux through the sensing of SAM by nhr-114. Dashed arrows indicate regulation of metabolic activity. Solid arrows indicate transcriptional regulation. Vitamin B12 increases Met/SAM cycle metabolic activity producing SAM, which represses nhr-114 transcription thereby reducing the expression of Met/SAM cycle related genes.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Caenorhabditis elegans) | N2; wild type | Wormbase | RRID:WB-STRAIN: WBStrain00000001 | Laboratory reference strain/wild type |
| Strain, strain background (C. elegans) | TM4695; ∆nhr-10 | • Wormbase | Wormbase ID: WBVar00253059 | Genotype: nhr-10(tm4695) III |
| Strain, strain background (C. elegans) | VC1527; ∆nhr-68 | Wormbase | RRID:WB-STRAIN: WBStrain00036665 | Genotype: nhr-68(gk708) V |
| Strain, strain background (C. elegans) | VL749; Pacdh-1::GFP | Wormbase | RRID:WB-STRAIN: WBStrain00040155 | Genotype: wwIs24[Pacdh-1::GFP;unc-119(+)] |
| Strain, strain background (C. elegans) | VL868; ‘Pacdh-1::GFP;∆nhr-10’ | PMID:23540701 | Genotype: wwIs24[Pacdh-1::GFP; unc-119(+)];nhr-10(tm4695) | |
| Strain, strain background (C. elegans) | VL1127; ‘Pacdh-1::GFP;∆nhr-114’ | PMID:26430702 | Genotype: wwIs24[Pacdh-1::GFP;unc-119(+)];nhr-114(gk849) | |
| Strain, strain background (C. elegans) | VL1102; ‘Pacdh-1::GFP;∆metr-1’ | PMID:23540702 | Genotype: wwIs24[Pacdh-1::GFP;unc-119(+)];metr-1(ok521) | |
| Strain, strain background (C. elegans) | VL1115; ‘Pacdh-1::GFP;∆sams-1’ | PMID:23540702 | Genotype: wwIs24[Pacdh-1::GFP;unc-119(+)];sams-1(ok2946) | |
| Strain, strain background (C. elegans) | CB4856 | Wormbase | RRID:WB-STRAIN: WBStrain00004602 | Wild isolate |
| Strain, strain background (C. elegans) | VL1199; ‘Pacdh-1::GFP;∆nhr-10;mthf-1(ww50)’ | This paper | See Materials and methods, Genotype: wwIs24[Pacdh-1::GFP;unc-119(+)];nhr-10(tm4695);mthf-1(ww50) | |
| Strain, strain background (C. elegans) | VL1200; ‘Pacdh-1::GFP;∆nhr-10;sams-1(ww51)’ | This paper | See Materials and methods, Genotype: wwIs24[Pacdh-1:: GFP;unc-119(+)];nhr-10(tm4695);sams-1(ww51) | |
| Strain, strain background (C. elegans) | VL1201; ‘Pacdh-1::GFP;∆nhr-10;metr-1(ww52)’ | This paper | See Materials and methods, Genotype: wwIs24[Pacdh-1::GFP;unc-119(+)];nhr-10(tm4695);metr-1(ww52) | |
| Strain, strain background (C. elegans) | VL1205; ‘Pacdh-1::GFP;∆nhr-10;mtrr-1(ww56)’ | This paper | See Materials and methods, Genotype:wwIs24[Pacdh-1::GFP;unc-119(+)];nhr-10(tm4695);mtrr-1(ww56) | |
| Sequence-based reagent | q_act-1_F | PMID:23540702 | qPCR primers | ctcttgccccatcaaccatg |
| Sequence-based reagent | q_act-1_R | PMID:23540702 | qPCR primers | cttgcttggagatccacatc |
| Sequence-based reagent | q_ama-1_F | PMID:23540702 | qPCR primers | agtgccgagattgaaggaga |
| Sequence-based reagent | q_ama-1_R | PMID:23540702 | qPCR primers | gtattgcatgttacctttttcaacg |
| Sequence-based reagent | q_metr-1_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, GGAGCAGCTACTGGTAGAC |
| Sequence-based reagent | q_metr-1_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, CACAGATGGCGAAATTGAGAG |
| Sequence-based reagent | q_mtrr-1_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, TACGTTCTTCTCGGTCTCG |
| Sequence-based reagent | q_mtrr-1_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, AGAGCTGTCAGTTGTTTGTC |
| Sequence-based reagent | q_sams-1_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, ATTATCAAGGAGCTCGACCT |
| Sequence-based reagent | q_sams-1_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, ATGGGAACTCAGAGTGACC |
| Sequence-based reagent | q_ahcy-1_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, CGATTGCGAGATTGACGTC |
| Sequence-based reagent | q_ahcy-1_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, GTGTAACGGTCAACCTGTG |
| Sequence-based reagent | q_mel-32_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, TGACTCATGGATTCTTCACCC |
| Sequence-based reagent | q_mel-32_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, GATCAACCTTGTATGGAAGAGAC |
| Sequence-based reagent | q_mthf-1_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, GTTGAGACCGATGAGAATGC |
| Sequence-based reagent | q_mthf-1_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, TTCATAATGCTTTGGTGACCAG |
| Sequence-based reagent | q_pmt-2_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, TTCATGTCGAAGTTTACCCA |
| Sequence-based reagent | q_pmt-2_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, GTCCTTCTCGATGTATCCG |
| Sequence-based reagent | q_cbs-1_F | This paper | qPCR primers | Used for qRT-PCR as described in methods, GAAGCTAGAGTATCTCAATATTGCG |
| Sequence-based reagent | q_cbs-1_R | This paper | qPCR primers | Used for qRT-PCR as described in methods, CCAATCTCTTCAGCAAACTGG |
| Software, algorithm | CloudMap | PMID:23051646 | ||
| Software, algorithm | DolphinNext | PMID:32306927 | ||
| Software, algorithm | DESeq2 | PMID:25516281 | RRID:SCR_01568 | |
| Software, algorithm | WormFinder.m | This paper | See methods, Available on github: https://github.com/shiaway/wormFinder/blob/master/wormFinder.m | |
| Software, algorithm | GETprime | PMID:21917859 | ||
| Software, algorithm | fiji/imageJ | PMID:22743772 | RRID:SCR_002285 |
Additional files
-
Supplementary file 1
mRNA fold change and p adjusted values of genes involved in the propionate shunt, Met/SAM cycle and phospholipid metabolism in the following conditions.
Wild type on vitamin B12 vs.wild type on untreated; wild type on vitamin B12 and propionate vs. wild type on vitamin B12; ∆nhr-10 untreated vs. wild type untreated; ∆nhr-10 on vitamin B12 vs. ∆nhr-10 untreated; ∆nhr-68 untreated vs. wild type untreated; ∆nhr-68 on vitamin B12 vs. ∆nhr-68 untreated. This table is a subset of data published in Bulcha et al., 2019 (GSE123507). Related to Figures 1, 3, 5 and 6.
- https://cdn.elifesciences.org/articles/60259/elife-60259-supp1-v2.xlsx
-
Supplementary file 2
Genes with an mRNA fold change greater or equal to 1.5 and a p adjusted value less than 0.01 in Met/SAM cycle mutants vs.the parental strain Pacdh-1::GFP;∆nhr-10.
- https://cdn.elifesciences.org/articles/60259/elife-60259-supp2-v2.xlsx
-
Supplementary file 3
mRNA fold change and p adjusted values of all detected genes in the following conditions.
Pacdh-1::GFP on nhr-114 RNAi vs. vector; Pacdh-1::GFP;∆metr-1 on vector vs. Pacdh-1::GFP on vector; Pacdh-1::GFP;∆metr-1 on nhr-114 RNAi vs. Pacdh-1::GFP on vector; Pacdh-1::GFP;∆metr-1 on nhr-114 RNAi vs. Pacdh-1::GFP;∆metr-1 on vector; Pacdh-1::GFP;∆nhr-10;metr-1(ww52) on vector vs. Pacdh-1::GFP on vector; Pacdh-1::GFP;∆nhr-10;metr-1(ww52) on nhr-114 RNAi vs. Pacdh-1::GFP on vector; Pacdh-1::GFP;∆nhr-10;metr-1(ww52) on nhr-114 RNAi vs. Pacdh-1::GFP;∆nhr-10;metr-1(ww52) on vector. Related to Figure 4.
- https://cdn.elifesciences.org/articles/60259/elife-60259-supp3-v2.xlsx
-
Supplementary file 4
mRNA fold change and p adjusted values of all detected genes in the following conditions.
- https://cdn.elifesciences.org/articles/60259/elife-60259-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60259/elife-60259-transrepform-v2.pdf





