Identification of distinct pH- and zeaxanthin-dependent quenching in LHCSR3 from Chlamydomonas reinhardtii
Figures
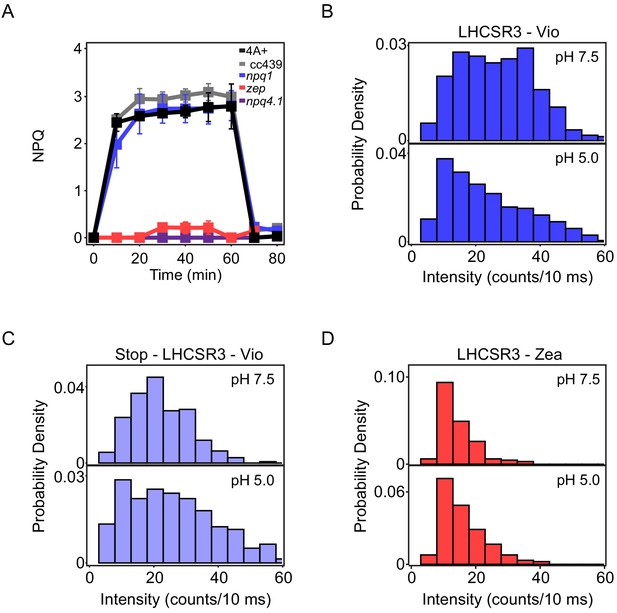
Fluorescence measurements of quenching in vivo and in vitro.
(A) In vivo nonphotochemical quenching (NPQ) induction kinetics for high-light acclimated samples measured upon 60 min of high light (1500 μmol m -2 s -1 ) in vivo. The results are reported as the mean of three independent biological replicates (N=3). Error bars are reported as standard deviation. Kinetics for low-light acclimated samples are shown in Figure 1—figure supplement 4. In vitro single-molecule fluorescence spectroscopy was performed on LHCSR3 (Figure 1—figure supplement 5). The fluorescence intensities measured from ~100 single complexes were used to construct the histograms shown for (B) LHCSR3- Vio, (C) stop-LHCSR3-Vio, and (D) LHCSR3-Zea at pH 7.5 (top) and pH 5.0 (bottom). Statistical parameters are provided in Figure 1—source data 1.
-
Figure 1—source data 1
Table of statistical parameters for the histograms of the single-molecule fluorescence intensity shown in Figure 1.
The median (med) and standard deviation (std) are given for each sample. The 95% confidence intervals for each value were determined by bootstrapping the samples 10,000 times.
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig1-data1-v2.docx
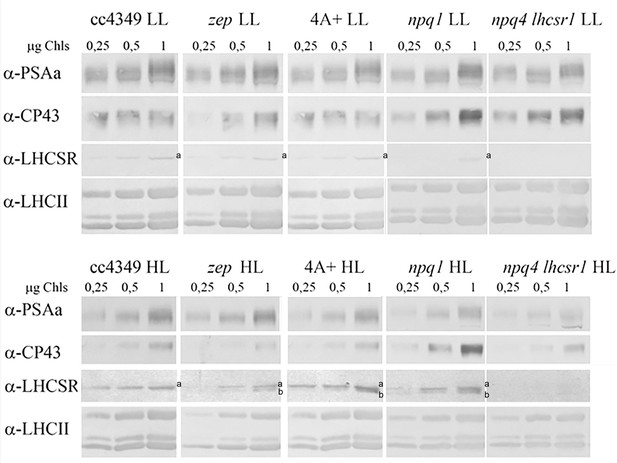
Immunoblot analysis of LHCSR accumulation in vivo.
Total protein extracts from low-light (LL) or high-light (HL) acclimated cells were loaded on a chlorophyll basis (μg of chlorophylls loaded are reported in the figure). Immunoblot analysis was performed using specific antibodies recognizing PsaA, CP43, LHCSR3 (a), and LHCII subunits. The filter used for LHCSR3 detection was then used for LHCSR1 (b) detection using specific antibody. The results reported are representative of two independent experiments with different biological replicates and three technical replicates for each genotype in LL or HL.
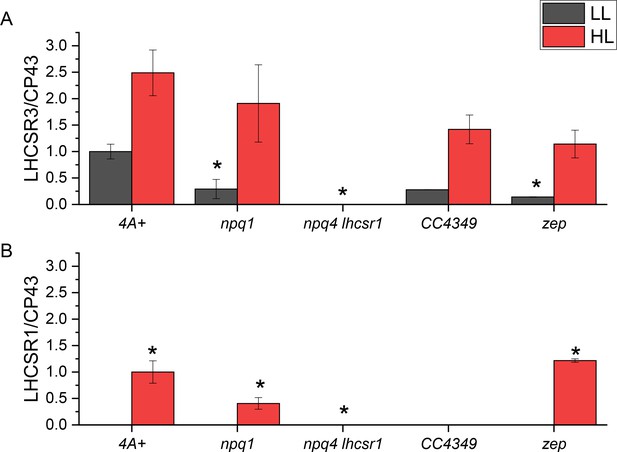
Quantification of LHCSR3 and LHCSR1 accumulation per PSII.
Immunoblotting results were analyzed by densitometry. LHCSR3 (A) and LHCSR1 (B) content was then normalized to the amount of CP43 as a reference for PSII. The results obtained were then normalized to the 4A+ low light (LL) case in the case of LHCSR3 and 4A+ high light (HL) case in the case of LHCSR1. The results reported are representative of two independent experiments with different biological replicates and three technical replicates for each genotype in LL or HL. Error bars are reported as standard deviation. The statistical significance of differences compared to WT (4A+ for npq1 and npq1 lhcsr1 mutants, CC4349 for zep mutant) is indicated as * (p<0.05), as determined by unpaired two sample t-test (N = 3).
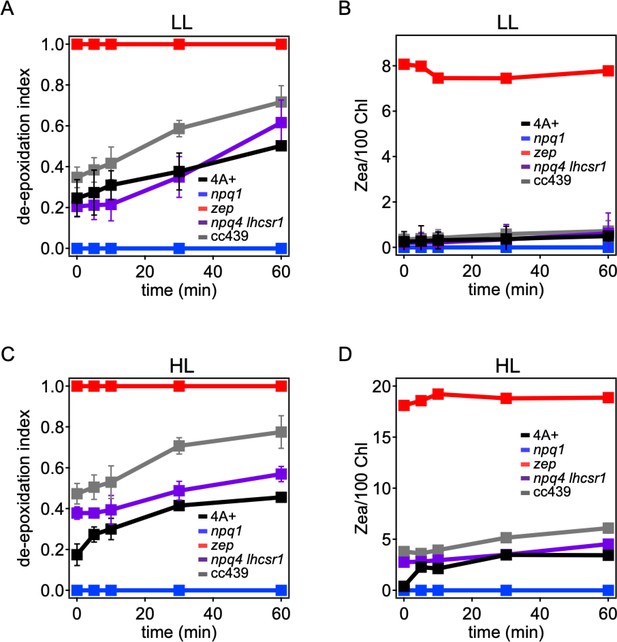
Violaxanthin de-epoxidation kinetics in Chlamydomonas reinhardtii WT and mutant strains.
Violaxanthin de-epoxidation kinetics were measured upon 60 min of strong light treatment (red light 1500 μmol m−2s−1) of low-light (LL) or high-light (HL) acclimated cells. Panel A, C: de-epoxidation indexes at different time points. Panel B, D: zeaxanthin content per 100 chlorophylls. De-epoxidation index was calculated from the molar concentration of zeaxanthin, anteraxanthin, and violaxanthin as ([zeaxanthin]+ [anteraxanthin]/2)/([zeaxanthin]+ [anteraxanthin]+ [violaxanthin]). The results reported are representative of three independent biological replicates for each genotype in LL or HL. Error bars are reported as standard deviation (N = 3).
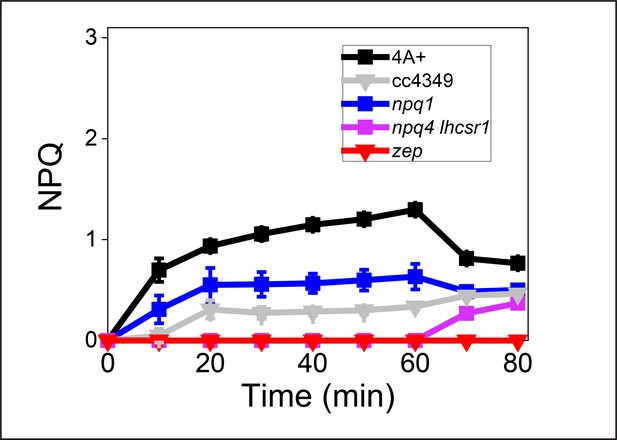
Nonphotochemical quenching (NPQ) induction in low-light (LL) acclimated Chlamydomonas reinhardtii cells.
NPQ induction kinetics measured upon 60 min of high light (HL; 1500 μmol m−2s−1) followed by 20 min of dark recovery. The results reported are representative of three independent biological replicates for each genotype in LL or HL. Error bars are reported as standard deviation (n = 3).
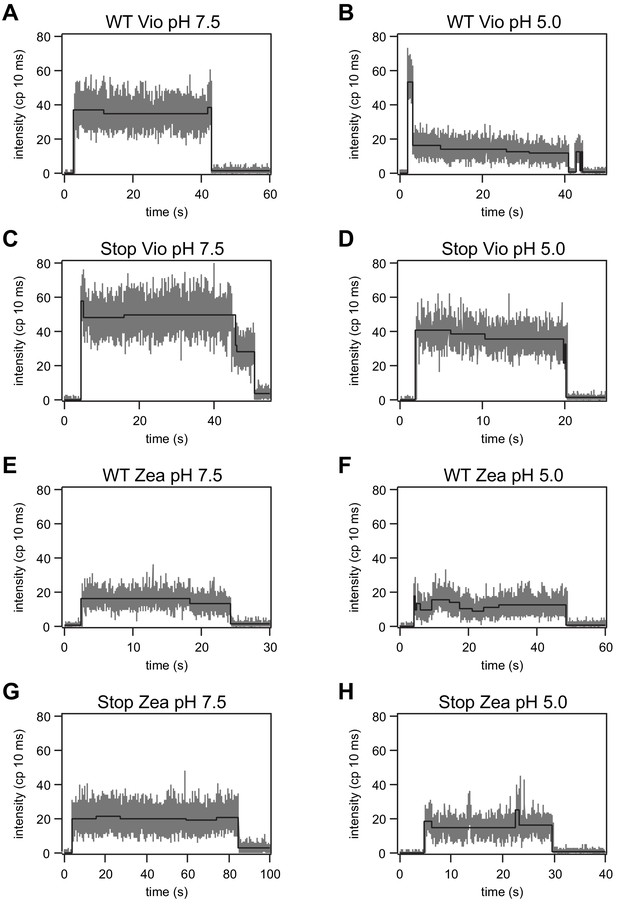
Representative fluorescence intensity traces of single LHCSR3 complexes at pH 7.5 and 5.0.
The intensity levels determined by the change-point-finding algorithm are shown in black.
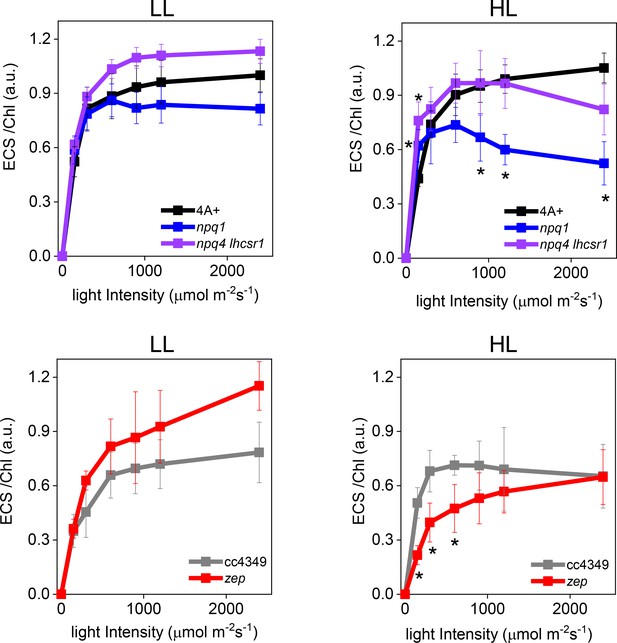
Electrochromic shift measurements at different light intensities in low-light (LL) and high-light (HL) acclimated Chlamydomonas reinhardtii cells.
Electrochromic shift (ECS) measurements were performed in WT (4A+ and cc4349) and mutant strains (npq4 lhcsr1, npq1 and zep) acclimated to LL (Panels A and B) or HL (Panels C, D). Genotypes having the same background are shown in the same Panel. The results reported are representative of three independent biological replicates for each genotype in LL or HL. Error bars are reported as standard deviation. The statistical significance of differences compared to WT (4A+ for npq1 and npq1 lhcsr1 mutants, CC4349 for zep mutant) is indicated as * (p<0.05), as determined by unpaired two sample t-test (N = 3).
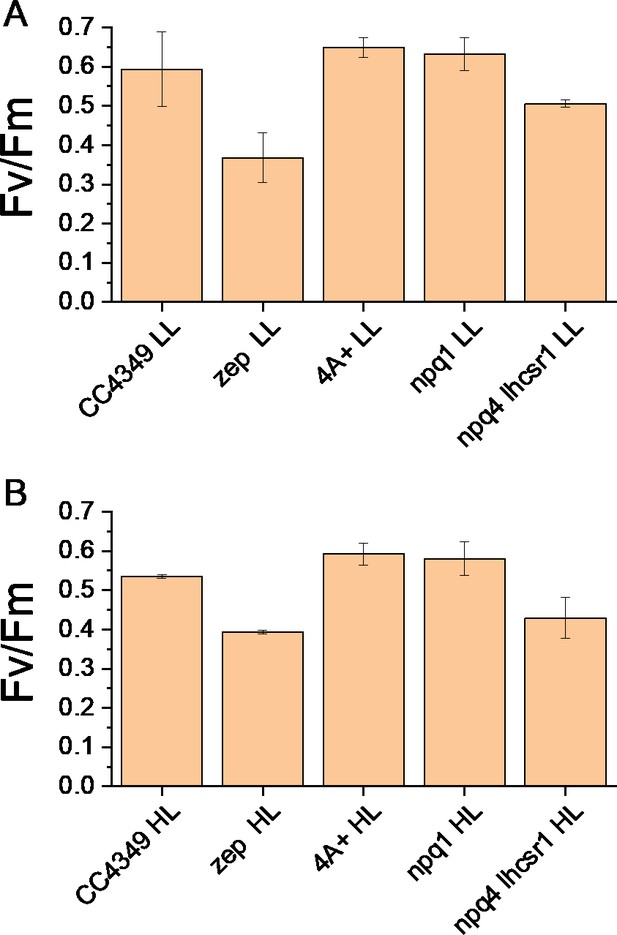
Maximum quantum yield of Photosystem II in WT (CC4349 and 4A+) and mutant (zep, npq1, npq4 lhcsr1) strains acclimated to low light (LL, Panel A) and high light (HL, Panel B).

Alignment of LHCSR-like proteins: protonatable residues are red written while insertion site for TAA mutation to generate the stop-LHCSR3 mutant is indicated by black arrow.
Protein number for LHCSR-like proteins used for alignment are listed below: XP_001696064.1 Chlamydomonas reinhardtii LHCSR3, XP_001696125.1 Chlamydomonas reinhardtii LHCSR1, XP_002948670.1 Volvox carteri f. nagariensis, ADP89594.1 Chlamydomonas sp. ICE-L LHCSR2, XP_001768071.1 Physcomitrella patens LHCSR2, ABD58893.1 Acutodesmus obliquus, ADY38581.1 Ulva linza, ADU04518.1 Ulva prolifera, XP_005848576.1 Chlorella variabilis, XP_002178699.1 Phaeodactylum tricornutum, AHH80644.1 Durinskia baltica, AA05890.1 Bigelowiella natans, CCO66741.1 Bathycoccus prasinos.
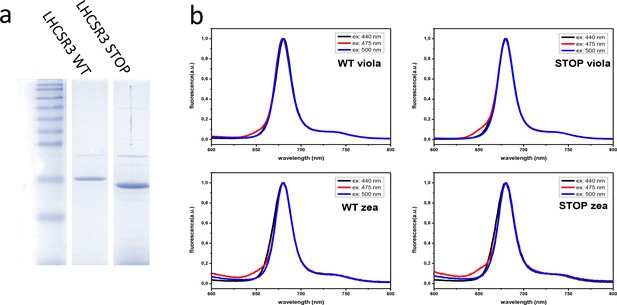
Recombinant LHCSR protein purification and in vitro refolding.
(a) Coomassie brilliant blue stained SDS-page of LHCSR3 apo-protein separated on Tris-Glycine 12%. LHCSR3 STOP protein shows high mobility conferred by its shorter C-terminal with respect to LHCSR3 WT; (b) Fluorescence spectra of LHCSR3 complexes measured at room temperature at different excitation wavelengths (reported in each panel). The results reported are representative of two independent experiments.
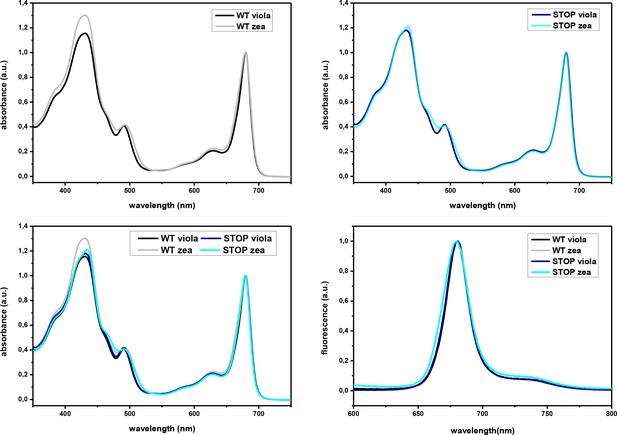
Absorption and fluorescence emission spectra of LHCSR3 WT and STOP.
(a–c) Absorption spectra of WT (a) or STOP (b) refolded with violaxanthin or zeaxanthin. (d) Fluorescence emission spectra of LHCSR3 WT and STOP mutant upon excitation at 440 nm with both violaxanthin and zeaxanthin pigments composition. The results reported are representative of two independent experiments.
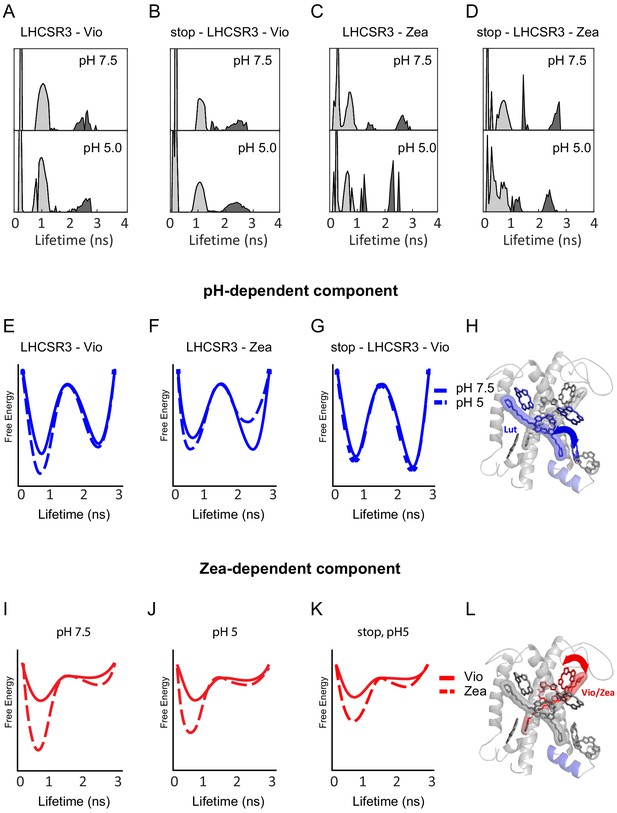
pH- and Zea-dependent quenching in LHCSR3.
(A-D) Fluorescence lifetime distributions from the global fit in the 2D-FLC analysis for each LHCSR3 sample at pH 7.5 and 5.0 estimated using the maximum entropy method (MEM) to perform an inverse Laplace Transform on the single-molecule emission times. States 1 and 2 are shown in light gray and dark gray, respectively. Free-energy diagrams constructed from the populations and transition rates (Figure 2—source data 1) extracted from the 2D-FLC analysis (E-G and I-K). Structural model with likely quenching sites (H and L) for the effects of pH (top) and Zea (bottom) on protein dynamics.
-
Figure 2—source data 1
Table of parameters estimated through the global fit to the correlation functions using the 2D-FLC analysis.
The number of dynamic components, fluorescence lifetime states, intensity of each lifetime state, population of each state, and transition rates between states were estimated by global fitting of the correlation functions as shown in Figure 2—figure supplement 1 using the model function described in the Materials and methods and the fluorescence lifetime distributions shown in Figure 2A–D. The fluorescence intensity is a relative intensity that is normalized by the total measurement time for each sample and scaled to set the maximum intensity to 1. The free-energy differences were calculated as described in the Materials and methods.
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig2-data1-v2.docx
-
Figure 2—source data 2
Pigment binding properties of recombinant LHCSR3 WT and stop-LHCSR3 refolded in vitro.
Binding pigments are reported referred to 7 Chlorophyll. The results reported are representative of two independent experiments. Errors are reported as standard deviation.
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig2-data2-v2.xlsx
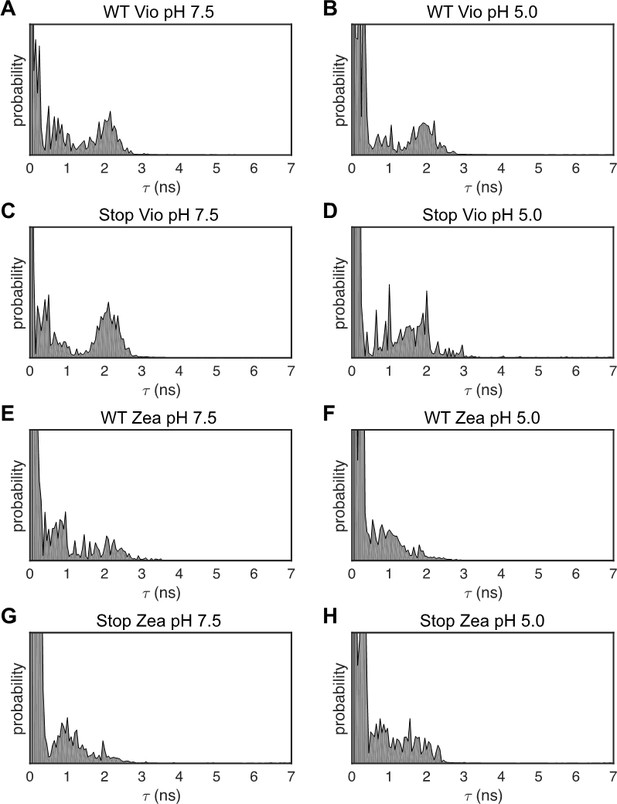
1D Lifetime distributions.
Lifetime distributions from the single-molecule fluorescence from all LHCSR3 samples determined using a one-dimensional inverse Laplace transform (1D-ILT) of the 1D fluorescence lifetime decay. Lifetime states identified from the 1D distribution were used as initial parameters in the fit to the 2D distributions in the 2D-FLC analysis.
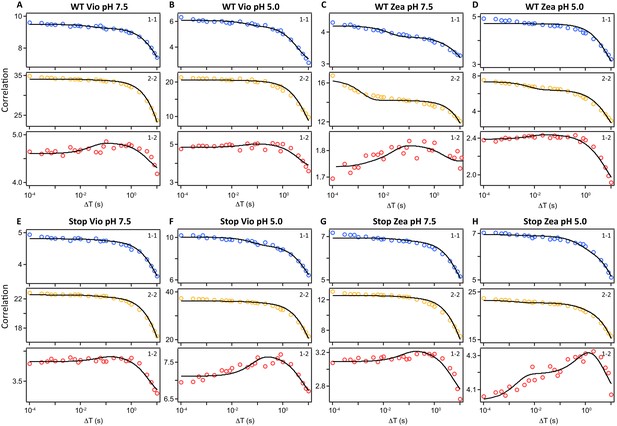
Correlation functions used in the 2D-FLC analysis of LHCSR3 complexes.
The auto-correlation and cross-correlation functions of the lifetime states were determined from the single-molecule photon emission for each sample. The auto-correlation is shown in blue for the quenched state and in yellow for the unquenched state. The cross-correlation of the two states is shown in red. The black lines shows the global fitting curves calculated using the model function given by the 2D-FLC analysis using Equation 1 and the lifetime distribution as described in the Materials and methods. The correlation functions are shown for single LHCSR3 complexes with Vio at pH 7.5 (A) and pH 5.0 (B), Zea at pH 7.5 (C) and 5.0 (D), stop-LHCSR3 with Vio at pH 7.5 (E) and pH 5.0 (F) and stop-LHCSR3 with Zea at pH 7.5 (G) and pH 5.0 (H).
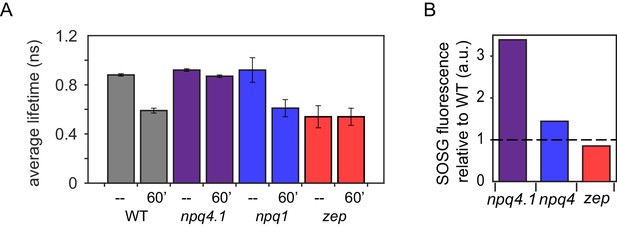
Fluorescence lifetime decay of Chlamydomonas reinhardtii whole cellsat 77K and singlet oxygen formation.
(A) Fluorescence lifetimes were measured on high light (HL; 500 μmol m -2 s -1 ) acclimated samples. Each genotype was measured at a dark-adapted state (--) or after 60 min of HL treatment (1500 μmol m -2 s -1 , 60’). WT samples shown here are 4A + strain. Similar results were obtained in the case of CC4349 strain. The npq4 lhcsr1 mutant is indicated here as npq4.1. Fluorescence lifetime values for all genotypes and light conditions are shown in Figure 3—figure supplements 1 and 2. The fluorescence data are provided in Figure 3—source data 1 with fit values in Figure 3—source data 2. (B) Singlet Oxygen Sensor Green (SOSG) fluorescence emission measured for HL acclimated samples relative to WT (4A+ for npq1 and npq4 lhcsr1, CC4349 for zep). Dotted line represents WT level at 1. The results reported are representative of three independent biological replicates for each genotype in low light (LL) or HL. SOSG kinetics are shown in Figure 3—figure supplement 3. LL acclimated samples are shown in Figure 3—figure supplement 4. SOSG emission data is provided in Figure 3—figure supplement 5—source data 1.
-
Figure 3—source data 1
In vivo 77K fluorescence lifetime decays from TCSPC where individual sheets demarcate different genotypes acclimated to either low light (LL) or high light (HL) with the individual data from two independent biological and technical replicates after exposure to 60’ of HL or in the dark.
For each measurement, the first column is the time (ns) and the second column is photon counts.
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig3-data1-v2.xlsx
-
Figure 3—source data 2
77K time resolved fluorescence analysis and average fluorescence decay lifetimes of whole cells.
Kinetics of Figure 3—source data 1 were fitted with a three-exponential decay function using Vinci two software from ISS. Fractions (fi) and time constants (τi) are reported. Average fluorescence lifetimes were calculated as Σfiτi. The results reported are two independent experiments with two independent biologic replicates.
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Time resolved fluorescence analysis and average fluorescence decay lifetimes of isolated monomeric (b2) and trimeric (b3) light-harvesting complexes.
Kinetics of Figure 3—figure supplement 5—source data 1 were fitted with a two-exponential decay function using Vinci two software from ISS. Fractions (fi) and time constants (τi) are reported. Average fluorescence lifetimes were calculated as Σfiτi. Errors are reported as standard deviation (n = 2).
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig3-data3-v2.docx
-
Figure 3—source data 4
In vivo SOSG fluorescence emission kinetics where individual sheets demarcate different genotypes acclimated to either low light (LL) or high light (HL).
For each measurement, the first column is the time at which the fluorescence (arbitrary units) was measured. The following columns are the measured fluorescence of replicate experiments as well as the standard deviation for the specific sample.
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig3-data4-v2.xlsx
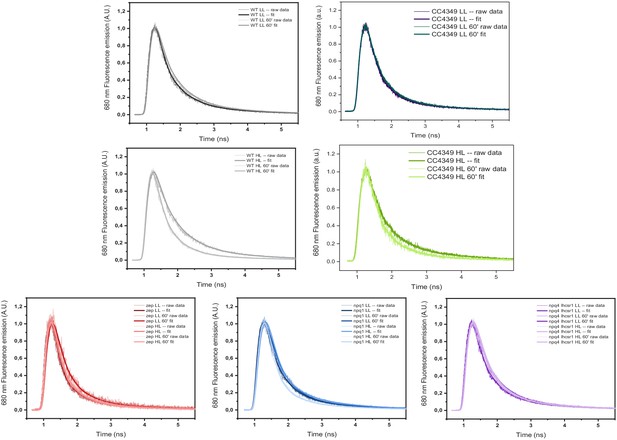
77K raw and fitted traces acquired by TCSPC of Chlamydomonas reinhardtii WT (4a+) and mutant strains.
The results reported are representative of two independent experiments with two independent biologic replicates.
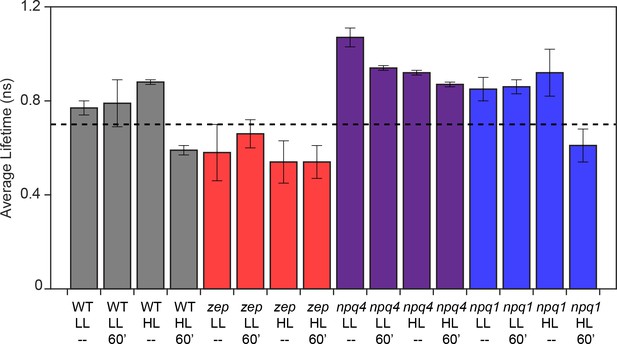
Average fluorescence lifetime for WT (4A+) and mutant strains under all light conditions.
The other WT strain (cc4349) has similar values. Above dotted line at 0.75 ns is referred to as unquenched and below the dotted line is referred to as quenched. The results reported are representative of two independent experiments with two independent biologic replicates.
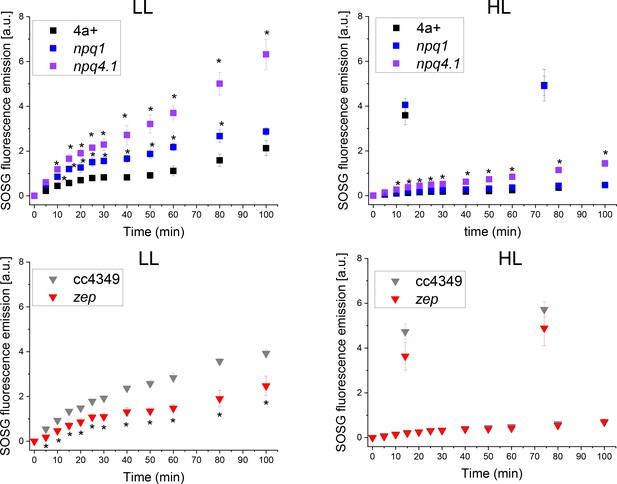
SOSG fluorescence emission kinetics in WT and mutant strains.
Singlet oxygen production was measured via the singlet oxygen sensor green (SOSG) fluorescence probe. Low-light (LL) or high-light (HL) acclimated samples were exposed to strong red light (2000 μmol m−2s−1) and singlet oxygen production rate was probed at the different time points by following SOSG fluorescence at 530 nm. Genotypes having the same background are shown in the same Panel. The results reported are representative of three independent biological replicates for each genotype in LL or HL. Error bars are reported as standard deviation (N = 3). The statistical significance of differences compared to WT (4A+ for npq1 and npq1 lhcsr1 mutants, CC4349 for zep mutant) is indicated as * (p<0.05), as determined by unpaired two sample t-test (N = 3).
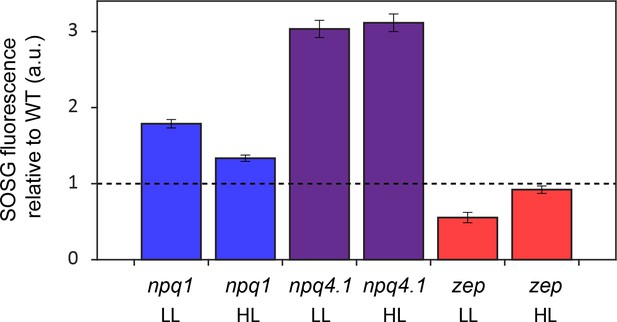
Singlet oxygen sensor green (SOSG) fluorescence emission in WT and mutant strains.
Singlet oxygen production rate was measured by SOSG as a fluorescence probe. Low light (LL), left, or high light (HL), right, acclimated samples were exposed to strong red light (2000 μmol m−2s−1), right. SOSG emission relative to WT (4A+ for npq1 and npq4 lhcsr1, cc4349 for zep). The results reported are representative of three independent biological replicates for each genotype in LL or HL. Unnormalized data are reported in Figure 3—figure supplement 3.
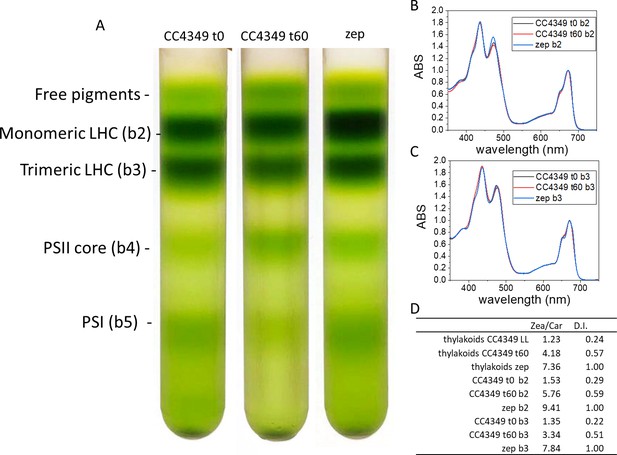
Isolation and characterization of monomeric and trimeric light-harvesting complexes (LHC).
(A) CC4349 cells acclimated to low light (LL) after 60 min of dark adaptation (t0) were exposed for 60 min to high light at 1500 μmol m−2s−1 (t60). Thylakoid membranes were purified, solubilized in 0.8% n-dodecyl-α-D-maltoside and the different pigment binding complexes were separated by ultracentrifugation in 0.1–1M sucrose gradient. A similar procedure was conducted in the case of dark-adapted zep cells acclimated to LL. Absorption spectra of fractions corresponding to monomeric (b2) and trimeric (b3) complexes are reported in (B) and (C), respectively. (D) The Zea fraction of total carotenoids (Zea/Car) and the de-epoxidation index (D.I.) where standard deviation is below 15% (n = 2).
-
Figure 3—figure supplement 5—source data 1
Fluorescence lifetime decays from TCSPC of monomeric (b2) and trimeric (b3) light-harvesting complexes isolated after exposure to 60’ of high light or in the dark as described in Figure 3—figure supplement 5.
For each measurement, the first column is the time (ns), the second is photon counts for the emission decay, and the third is photon counts for the instrument response function (IRF). The values from each biological and technical replicate are shown.
- https://cdn.elifesciences.org/articles/60383/elife-60383-fig3-figsupp5-data1-v2.xlsx
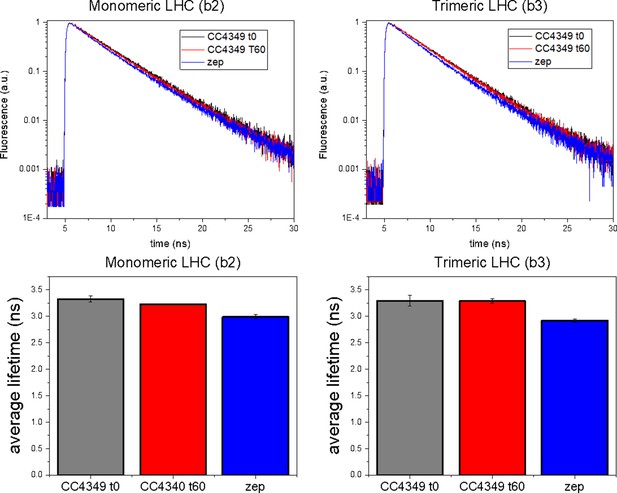
Time-resolved fluorescence emission decays were measured by time-correlated single-photon counting on isolated monomeric (b2) and trimeric (b3) light-harvesting complexes (LHC) as reported in Figure 3—figure supplement 5.
Average fluorescence lifetimes for each sample are reported as an average of two independent experiments.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) | Sigma-Aldrich | CMC0016 | Electrocompetent cells |
| Strain, strain background (Chlamydomonas reinhardtii) | 4A+ | https://www.chlamycollection.org/ | CC-4051 4A+ mt+ | Wild type strain |
| Strain, strain background (Chlamydomonas reinhardtii) | CC4349 | https://www.chlamycollection.org/ | Cell wall deficient strain | |
| Strain, strain background (Chlamydomonas reinhardtii) | npq1 | Niyogi, K. K., Bjorkman, O. and Grossman, A. R. Chlamydomonas Xanthophyll Cycle Mutants Identified by Video Imaging of Chlorophyll Fluorescence Quenching. Plant Cell 9, 1369–1380, doi:10.1105/tpc.9.8.1369 (1997). | Strain mutated on cvde gene | |
| Strain, strain background (Chlamydomonas reinhardtii) | npq4 lhcsr1 | Ballottari, M. et al. Identification of pH-sensing Sites in the Light Harvesting Complex Stress-related 3 Protein Essential for Triggering Nonphotochemical Quenching in Chlamydomonas reinhardtii. J Biol Chem 291, 7334–7346, doi:10.1074/jbc.M115.704601 (2016). | Strain mutated on lhcr1, lhcsr3.1 and lhcsr3.2 genes | |
| Strain, strain background (Chlamydomonas reinhardtii) | zep | Baek, K. et al. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep 6, 30620, doi:10.1038/srep30620 (2016). | Strain obtained by CRISPR CAS9 being mutated on zep gene | |
| Antibody | αCP43 (Rabbit polyclonal) | Agrisera (Sweden) | AS11 1787 | Dilution used (1:3000) |
| Antibody | αPSAA (Rabbit polyclonal) | Agrisera (Sweden) | AS06 172 | Dilution used (1:5000) |
| Antibody | αLHCBM5 (Rabbit polyclonal) | Agrisera (Sweden) | AS09 408 | Dilution used (1:5000) |
| Antibody | αLHCSR3 (Rabbit polyclonal) | Agrisera (Sweden) | AS14 2766 | Dilution used (1:3000) |
| Recombinant DNA reagent | pETmHis containing LHCSR3 CDS | Ballottari, M. et al. Identification of pH-sensing Sites in the Light Harvesting Complex Stress-related 3 Protein Essential for Triggering Nonphotochemical Quenching in Chlamydomonas reinhardtii. J Biol Chem 291, 7334–7346, doi:10.1074/jbc.M115.704601 (2016). | ||
| Recombinant protein | LHCSR3- Vio | This paper | Recombinant protein expressed in E. coli Purified as inclusion bodies and refolded by adding pigments, including violaxanthin but not zeaxanthin | |
| Recombinant protein | LHCSR3- Zea | This paper | Recombinant protein expressed in E. coli Purified as inclusion bodies and refolded by adding pigments, including zeaxanthin but not violaxanthin | |
| Recombinant protein | stop-LHCSR3- Vio | This paper | Recombinant protein expressed in E. coli with the deletion of the last 13 residues at the C-terminus. Purified as inclusion bodies and refolded by adding pigments, including violaxanthin but not zeaxanthin | |
| Recombinant protein | stop-LHCSR3- Zea | This paper | Recombinant protein expressed in E. coli with the deletion of the last 13 residues at the C-terminus. Purified as inclusion bodies and refolded by adding pigments, including zeaxanthin but not violaxanthin | |
| Commercial assay or kit | Agilent QuikChange Lightning Site-Directed Mutagenesis Kit. | Agilent | 210519 | |
| Chemical compound, drug | n-dodecyl-α-D-maltoside | Anatrace | D310HA | |
| Chemical compound, drug | Singlet Oxygen Sensor Green (SOSG) | Thermo Fisher | S36002 | Fluorescent probe for singlet oxygen |
| Software, algorithm | OriginPro 2018 | https://www.originlab.com/2018 | ||
| Software, algorithm | MATLAB | Mathworks, Inc | ||
| Software, algorithm | 2D-FLC | https://github.com/PremashisManna/2D-FLC-code; Kondo et al., 2020 Kondo, T. et al. Microsecond and millisecond dynamics in the photosynthetic protein LHCSR1 observed by single-molecule correlation spectroscopy. PNAS 116, 11247–11252, doi: 10.1073/pnas.1821207116 (2019). |





