Chemical genetics and proteome-wide site mapping reveal cysteine MARylation by PARP-7 on immune-relevant protein targets
Figures
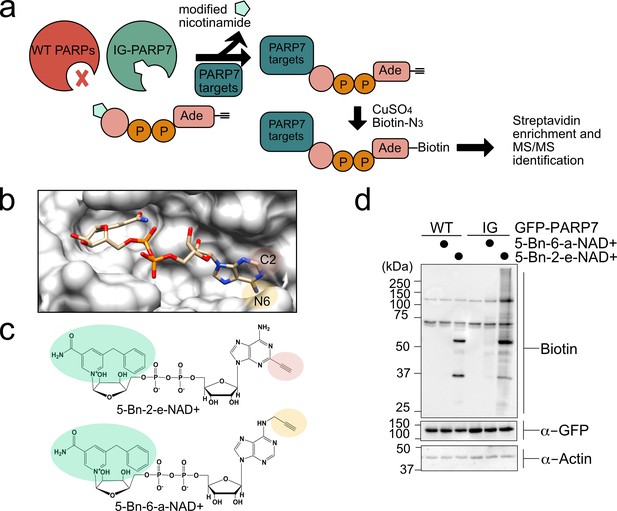
A chemical genetic strategy for identifying the direct MARylation targets of PARP-7.
(a) Schematic of chemical genetic approach for identifying direct targets of PARP-7. (b) Crystal structure of human PARP1 bound to the non-hydrolyzable NAD+ analog benzamide adenine dinucleotide (BAD). PDB: 6bhv. The structure shows that the C-2 position of BAD is pointed out of the ADP-ribose pocket. (c) Chemical structure of the clickable, orthogonal NAD+ analogs. 5-Bn-2-e-NAD+ is the optimized substrate for the floor position engineered PARP-7, IG-PARP-7. 5-Bn-6-a-NAD+ was used in our previous studies by other floor position engineered PARPs (e.g. PARP14). (d) 5-Bn-2-e-NAD+ is an efficient substrate for IG-PARP-7. HEK293T cells were transfected with either GFP-WT-PARP-7 (control) or GFP-IG-PARP-7. Cell lysates were prepared and incubated with either 5-Bn-2-e-NAD+ or 5-Bn-6-a-NAD+ (100 µM). Following copper-catalyzed click chemistry with biotin-azide, proteins were resolved by SDS-PAGE and biotinylated proteins were detected by Western blot detection using Streptavidin-HRP. Biotinylated proteins represent direct MARylation targets of PARP-7.
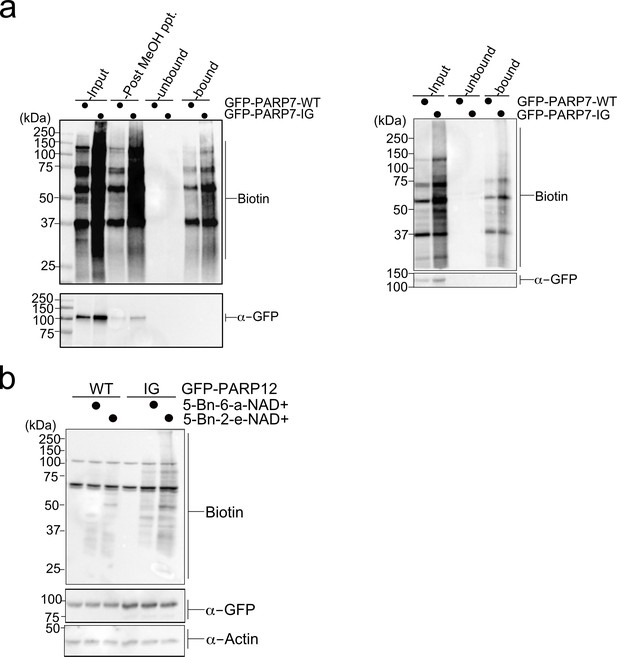
Enrichment of PARP-7 MARylation targets and labeling of PARP-12 IG targets using a chemical genetic approach.
(a) Labeling of PARP-7 MARylation targets was performed as described in Figure 1. Neutravidin agarose was used to enrich biotinylated proteins (i.e. MARylation targets). Proteins were resolved by SDS-PAGE and were detected by Western blot using Strep-HRP (biotin) and an antibody against GFP. The two biological replicates collected in this study are represented here. (b) Labeling of PARP-12 MARylation targets was performed as described in Figure 1. PARP-12 I660G mutant was used. Proteins were resolved by SDS-PAGE and were detected by Western blot using Strep-HRP (biotin) and an antibody against GFP. Representative figure of data collected over two biological replicates.
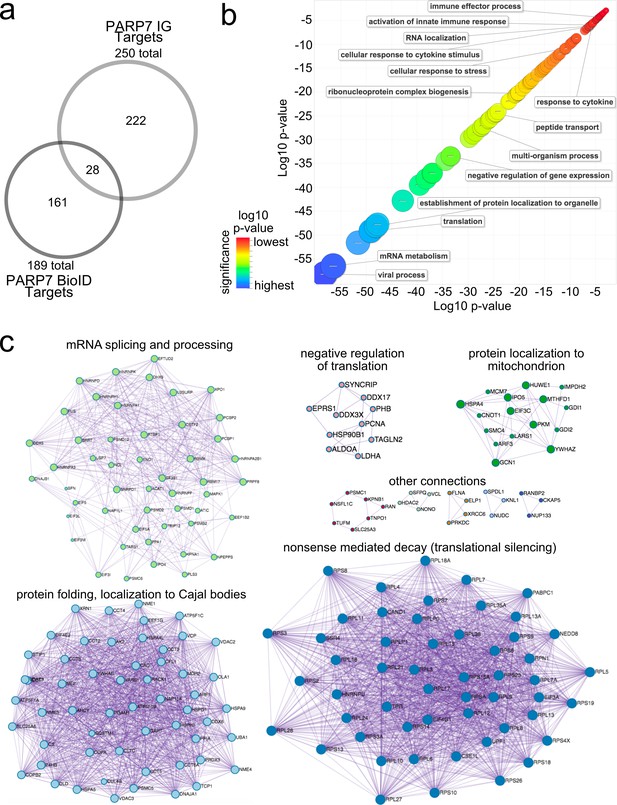
Gene ontology analysis of PARP-7 MARylation targets and interactors reveals roles in innate immune signaling and RNA regulation.
(a) Venn diagram comparing the 250 total PARP-7 targets identified by LC-MS/MS runs in comparison to the 189 PARP-7 interacting proteins identified using Myc-BirA–PARP-7. Data shown are the additive combined targets of two biological replicates for each type of strategy. (b) Scatterplot depicting enriched gene ontology (GO) terms attached to the 439 total PARP-7 MARylation targets (chemical genetics) and PARP-7 interactors (BioID proximity labeling). GO term enrichment was performed using the PANTHER toolkit. Significantly enriched GO terms (p<0.05) were condensed using ReviGO and similar terms were nested based on similarity. Terms are organized by −log10(p-value). Significance ranges from −58.2 (most significant) to −2.8 (least significant). Selected terms are indicated. (c) ‘Protein-protein Interaction Enrichment Analysis’ of the 439 total PARP-7 MARylation targets and PARP-7 interactors. Images were created using Metascape.org. The Molecular Complex Detection (MCODE) algorithm has been applied to identify densely connected network components. The MCODE networks identified for individual gene lists have been gathered and are shown here.
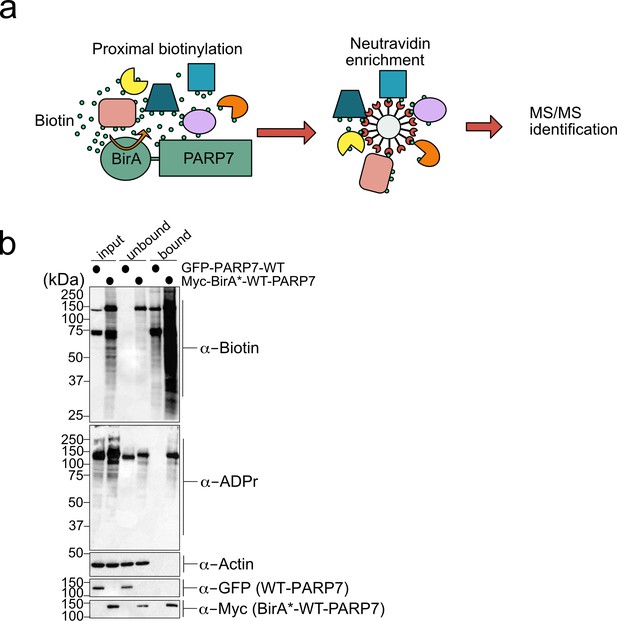
A proximity labeling approach identifies intracellular interactors of PARP-7.
(a) Schematic of proximity labeling approach (referred to as BioID) for identifying interacting proteins of PARP-7 in cells. (b) Proximity labeling of PARP-7 interactors in cells. Myc-BirA*-WT-PARP-7 or GFP-WT-PARP-7 were expressed in HEK 293 T cells (GFP-WT-PARP-7 is used as a negative control). Cells were treated with media supplemented with 50 μM biotin overnight. Neutravidin agarose was used to enrich biotinylated proteins (bound). Proteins were resolved by SDS-PAGE and were detected by Western blot using Strep-HRP (biotin) and an antibody against GFP.
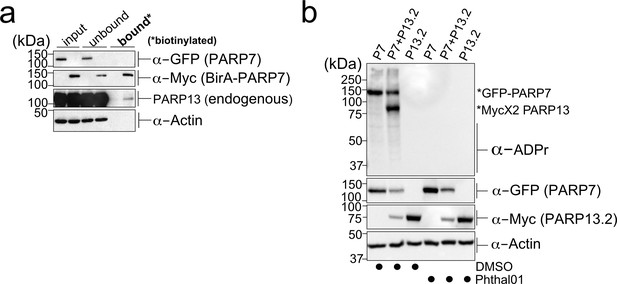
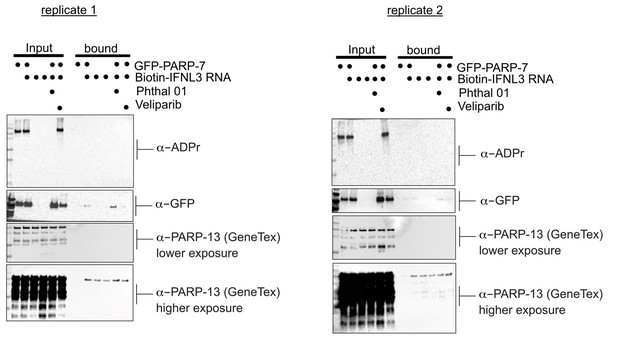
PARP-13 interacts with and is MARylated by PARP-7 in cells.
(a) PARP-7 interacts with endogenous PARP-13 (constitutive isoform PARP-13.1) in cells as shown by proximity labeling. HEK 293 T cells were transfected with Myc-BirA*-PARP-7. Biotin (50 μM) was added to induce biotinylation of Myc-BirA*-PARP-7 interactors. Biotinylated proteins were enriched using Neutravidin agarose. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against GFP, Myc, PARP-13, and actin. Representative figure of data collected over three biological replicates. (b) PARP-7 MARylates PARP-13.2 in cells. GFP-PARP-7 and MycX2-PARP-13.2 were co-expressed in HEK293T cells. Cells were treated overnight either with DMSO or with Phthal01 (1 μM). Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADP-ribose (ADPr), GFP, Myc, and actin. Representative figure of data collected over three biological replicates.
-
Figure 3—source data 1
In vitro IC50 Data for Phthal01 against PARP family members.
Phthal01 (6-(4-(2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)benzoyl)piperazin-1-yl)nicotinonitrile) was tested against all active PARP family members using an ADP-ribosylation plate assay developed in our lab (Kirby et al., 2018). IC50 Data represents ± SEM from at least two biological replicates.
- https://cdn.elifesciences.org/articles/60480/elife-60480-fig3-data1-v3.docx
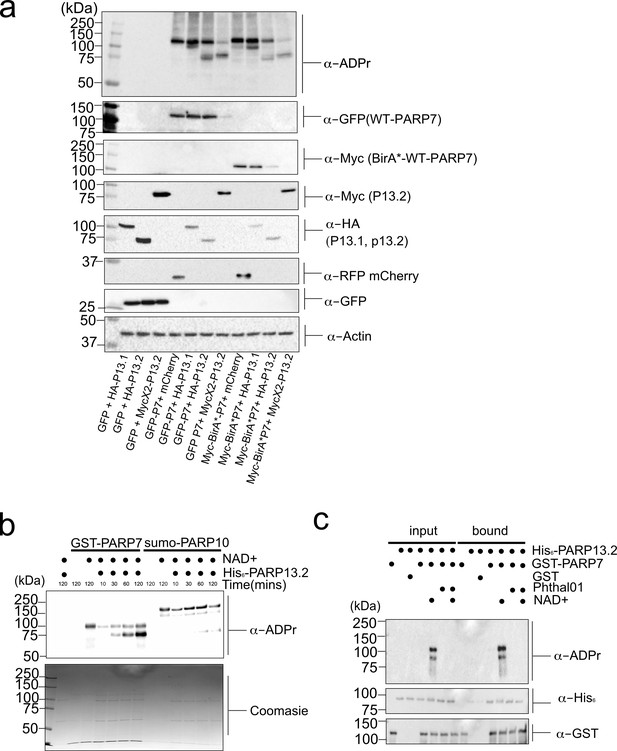
PARP-13 is a MARylation target of PARP-7.
(a) PARP-7 MARylates PARP-13.1 and PARP-13.2 in cells. MARylation of PARP-13.1 and 13.2 by PARP-7. GFP-PARP-7 and PARP-13.1 and PARP-13.2 tagged variants were transfected in HEK 293 T cells. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, HA, Myc, RFP (mCherry), and actin. Representative figure of data collected over three biological replicates. (b) PARP-7 MARylates PARP-13.2 in vitro. GST-PARP-7 (or His6-SUMO-PARP-10) was incubated with His6-SUMO-PARP-13.2 in the presence of NAD+ (100 μM). MARylation reactions were stopped at indicted time points and proteins were resolved by SDS-PAGE and were detected by Coomassie stain and Western blot using the antibody against ADPr. (c) The interaction between PARP-7 and PARP-13.2 is independent of PARP-7 catalytic activity. Recombinant GST-PARP-7 and His6-SUMO-PARP-13.2 were incubated for 1 hr and then subjected to GST pulldown following conditions described in the figure. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, His6, and GST.
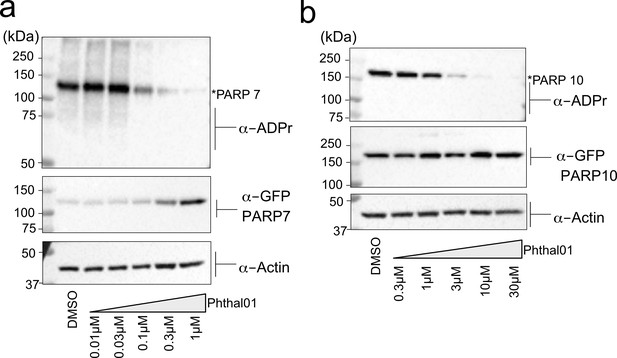
Phthal01 inhibits PARP-7, and at much higher concentrations, PARP-10-mediated MARylation in cells.
(a) HEK 293 T cells expressing GFP-PARP-7 were treated with increasing concentrations of Phthal01 for 16 hr. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, and actin. (b) HEK 293 T cells expressing GFP-PARP-10 were treated with increasing concentrations of Phthal01 for 16 hr. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, and actin.
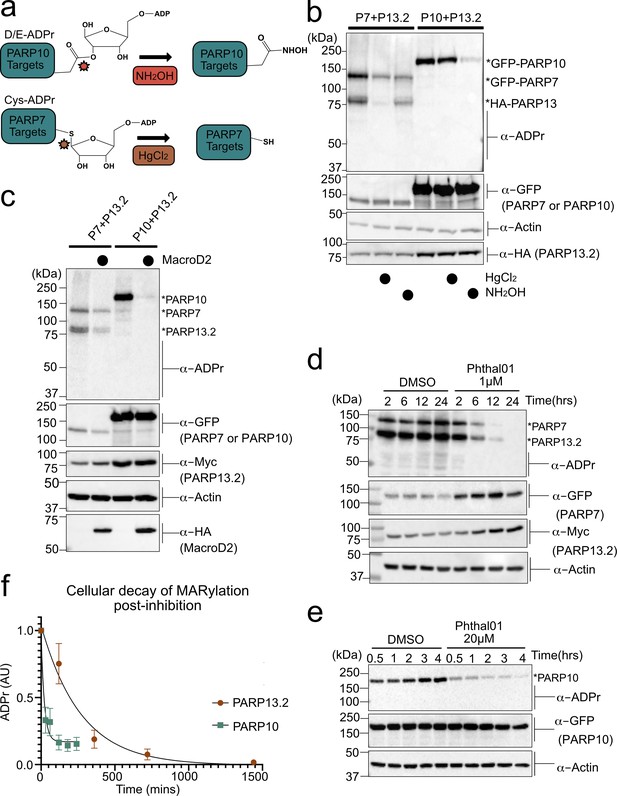
PARP-13 is MARylated by PARP-7 predominately on Cys residues and Cys MARylation is more stable than Asp/Glu MARylation in cells.
(a) Schematic of chemical stability studies for analyzing the amino acid-ADPr linkage. Neutral hydroxylamine (NH2OH) removes ADPr from acid residues to generate a hydroxamic acid, while mercuric chloride (HgCl2) removes ADPr from Cys residues. (b) PARP-7 MARylates predominately on Cys residues whereas PARP-10 MARylates predominately on Glu/Asp residues. GFP-PARP-7/HA-PARP-13.2 or GFP-PARP-10 alone were expressed in HEK 293 T cells. Lysates were prepared and treated with either water control, HgCl2 (2 mM), or NH2OH (0.4 M). Following treatment, proteins were precipitated with ice cold methanol, and resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, HA, and actin. Representative figure of data collected over three biological replicates. (c) The Glu/Asp selective ADPr hydrolase MacroD2 efficiently removes ADPr from PARP-10, but only partially removes ADPr from PARP-7 and PARP-13. GFP-PARP-7/MycX2-PARP-13.2 or GFP-PARP-10 was co-expressed with either HA-MacroD2 or mCherry (negative control) in HEK 293 T cells. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, Myc, and actin. Representative figure of data collected over three biological replicates. (d) Time course of Cys MARylation in cells. GFP-PARP-7 and MycX2-PARP-13.2 were co-expressed in HEK 293 T cells. Cells were treated with either DMSO or Phthal01 (1 µM) to stop PARP-7 MARylation. Cells were harvested at indicated time points and proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, Myc, and actin. Representative figure of data collected over three biological replicates. (e) Time course of Glu/Asp MARylation in cells. GFP-PARP-10 was expressed in HEK 293 T cells. Cells were treated with either DMSO or Phthal01 (20 µM) to stop PARP-10 MARylation. Cells were harvested at indicated time points and proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, and actin. Representative figure of data collected over five biological replicates. (f) Quantification of replicate data from (d) and (e). ADPr signal is normalized to either GFP-PARP-10 or MycX2-PARP-13.2. Data are ± SEM.
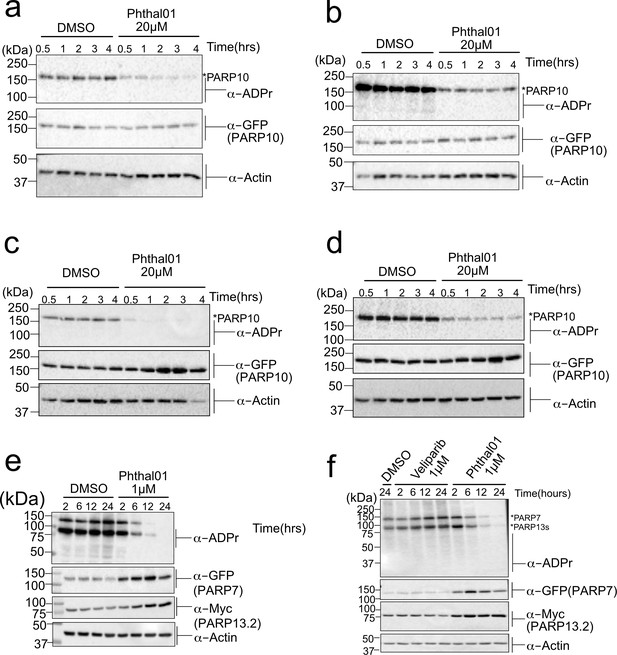
PARP-13.2 Cys MARylation is more stable than PARP-10 Asp/Glu MARylation in cells.
(a–d) Biological replicates of time course inhibition post in-cell (HEK 293T) expression of GFP-PARP-10 using Phthal01 (20 μM). (e–f) Biological replicates of time course inhibition post in-cell (HEK 293T) expression of PARP-7 using Phthal01 (1 μM). See legend of Figure 4D–F.
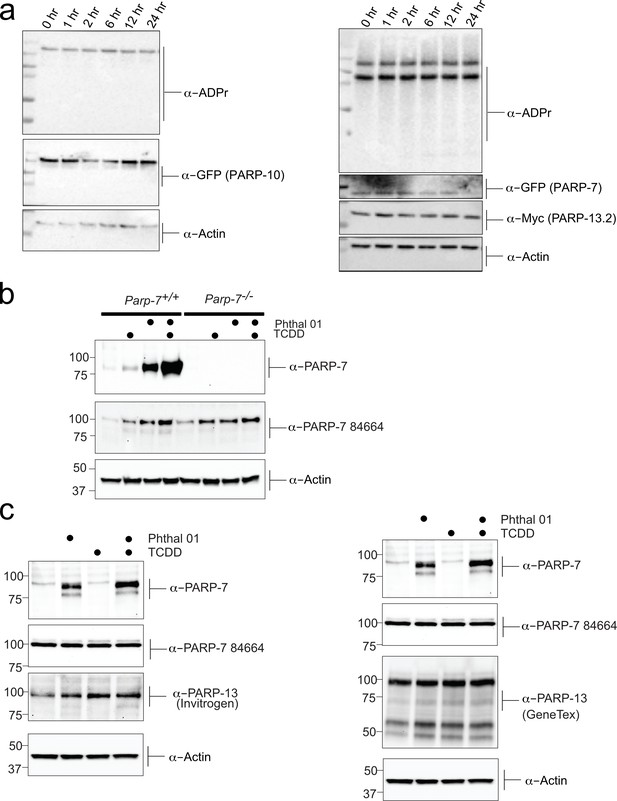
Inhibition of PARP-7 catalytic activity stabilizes PARP-7 protein levels.
(a) Chemical stability of ADPr modification. The chemical stability of PARP-7 and PARP-10 mediated ADPr was assessed by performing a time course post denaturation of HEK293T cellular lysates. Cells we transfected with GFP-PARP-7 and MycX2-PARP-13.2 or GFP-PARP-10, lysed, heat denatured in SDS and allowed to sit at room temperature for the indicated times. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, and actin (b) Validation of PARP-7 (TIPARP) antibody in MEF cells (+/+) and (-/-) PARP-7 cells. Cells were treated with TCDD (2,3,7,8 -Tetrachlorodibenzo-p-dioxin) was used to induce PARP-7 and Phthal01 (24 hr treatment) used to inhibit PARP-7. Proteins from the resulting lysates were resolved by SDS-PAGE and were detected by Western blot using antibodies against novel PARP-7 (TIPARP), commercially available Anti-PARP-7 antibody (ab84664), and Actin. (c) Further investigation of endogenous PARP-7 and PARP-13 response to Phthal 01 PARP-7 inhibitor. Cells treated with TCDD (2,3,7,8 -Tetrachlorodibenzo-p-dioxin) was used to induce PARP-7 and Phthal 01 (24hr treatment) was used to inhibit PARP-7. Proteins from the resulting lysates were resolved by SDS-PAGE and were detected by western blot using antibodies against novel PARP-7 (TIPARP), commercially available Anti-PARP-7 antibody (ab84664), Invitrogen PARP-13 (ZC3HAV1 Antibody PA5-31650), GeneTex PARP-13 antibody (ZC3HAV1 antibody [N3C2]), and Actin.
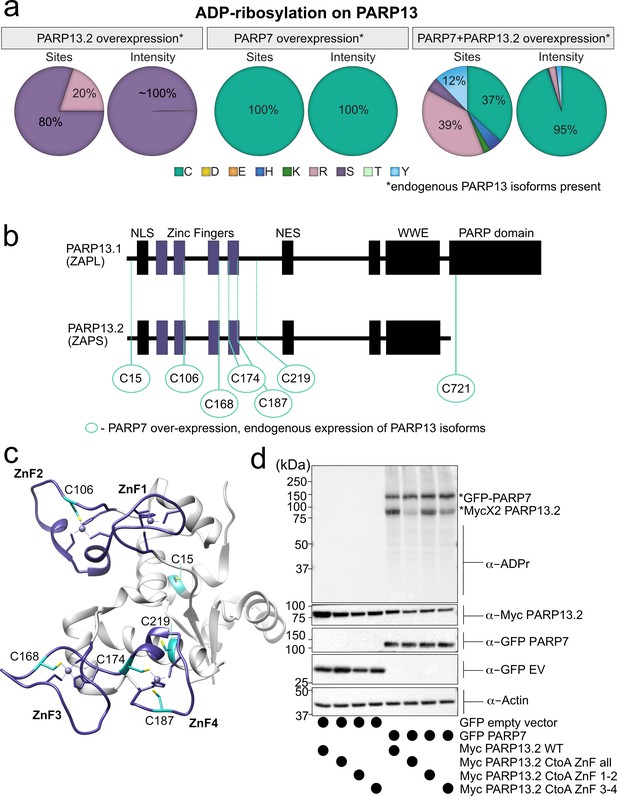
MS-based ADPr site identification and mutagenesis studies demonstrate that PARP-7-mediated MARylation of PARP-13 occurs predominately on Cys residues in the ZnF domains.
(a) Pie-charts showing of the amino acid residue distribution of all ADP-ribosylation sites identified on PARP-13 using EThcD fragmentation. Experiments were performed under three conditions: MycX2-PARP-13.2 overexpressing cells, GFP-PARP-7 overexpressing cells, GFP-PARP-7 and MycX2-PARP-13.2 overexpressing cells. One letter code used for amino acids. (b) Domain architecture of PARP-13.1 and PARP-13.2 showing positions of Cys ADPr sites. (c) Crystal structure of the N-terminal ZnF domains of PARP-13highlighting Cys ADPr sites (cyan). ZnF domains are highlighted (purple). PDB: 6UEI. (d) Mutation of the Cys to Ala in the ZnF domains of PARP-13 decreased PARP-7-mediated PARP-13 MARylation. GFP-PARP-7 was co-expressed with either WT- or CtoA ZnF all, CtoA ZnF1-2, or CtoA ZnF3-4 in HEK 293 T cells. GFP-PARP-7 or GFP-empty vector control co-expressed in HEK293T cells with MycX2-PARP-13.2 WT and mutants. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, Myc, and actin. Representative figure of data collected over three biological replicates.
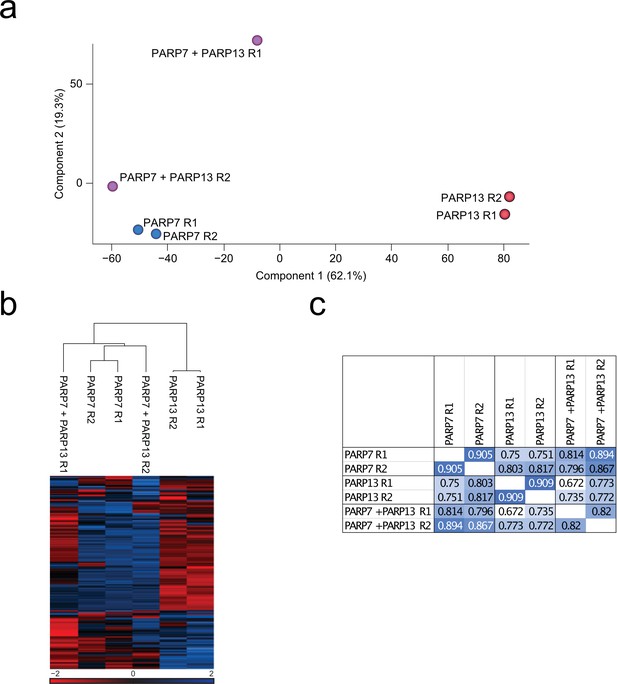
EThcD sample correlation data.
(a) Principal component analysis (PCA) based upon unnormalized data. Blue; PARP-7 overexpressing replicates, red; PARP-13.2 overexpressing replicates, purple; PARP-7 and PARP-13.2 overexpressing replicates. (b) Unsupervised hierarchical clustering analysis of z-scored log2-transformed intensities of ADPr sites. (c) Pearson correlations of identified ADPr sites (localization probability >0.90).
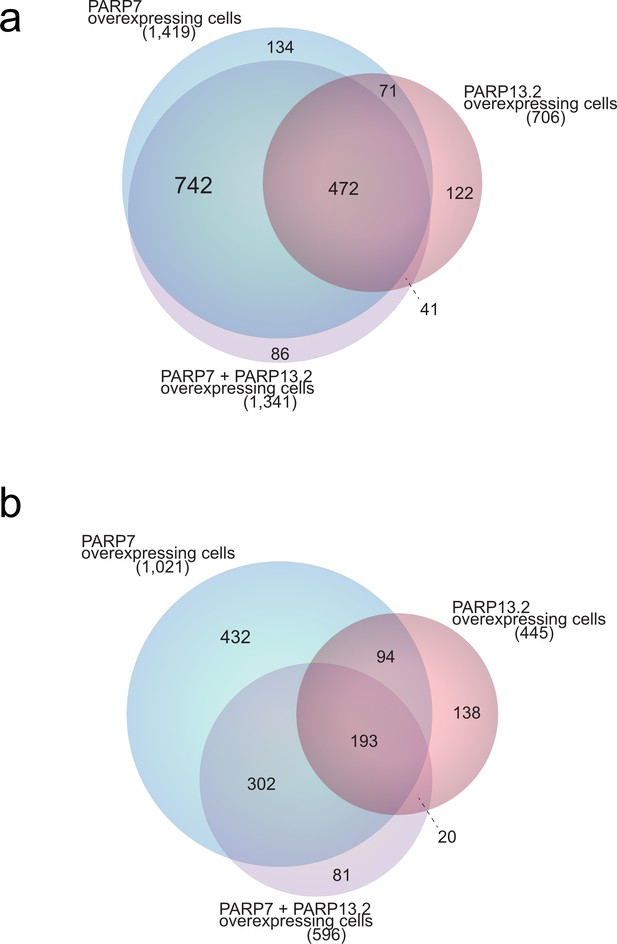
Scaled Venn diagram depicting distributions of ADP-ribosylation sites across samples.
(a) Scaled Venn diagram visualizing distribution of all identified ADP-ribosylation sites (localization probability >0.90) in GFP-PARP-7 overexpressing cells compared to PARP-13.2 overexpressing cells and PARP-7 and PARP-13.2 overexpressing cells. (b) Scaled Venn diagram visualizing distribution of ADP-ribosylation sites (localization probability >0.90) identified in duplicate in PARP-7 overexpressing cells compared to PARP-13.2 overexpressing cells and PARP-7 and PARP-13.2 overexpressing cells.
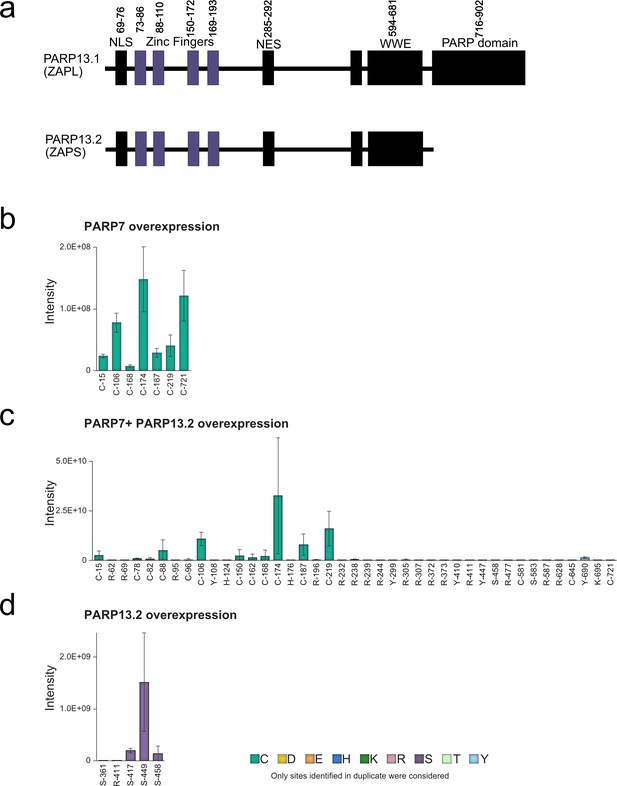
All ADPr sites on PARP-13 identified by EThcD fragmentation.
(a) Annotated domain architecture of PARP-13.1 and PARP-13.2. (b) ADPr sites identified in duplicate on PARP-13in GFP-PARP-7 overexpressing cells. (c) ADPr sites identified in duplicate on PARP-13 in the GFP-PARP-7 and MycX2-PARP-13.2 overexpressing cells. (d) ADPr sites identified in duplicate on PARP-13 in the MycX2-PARP-13.2 overexpressing cells.
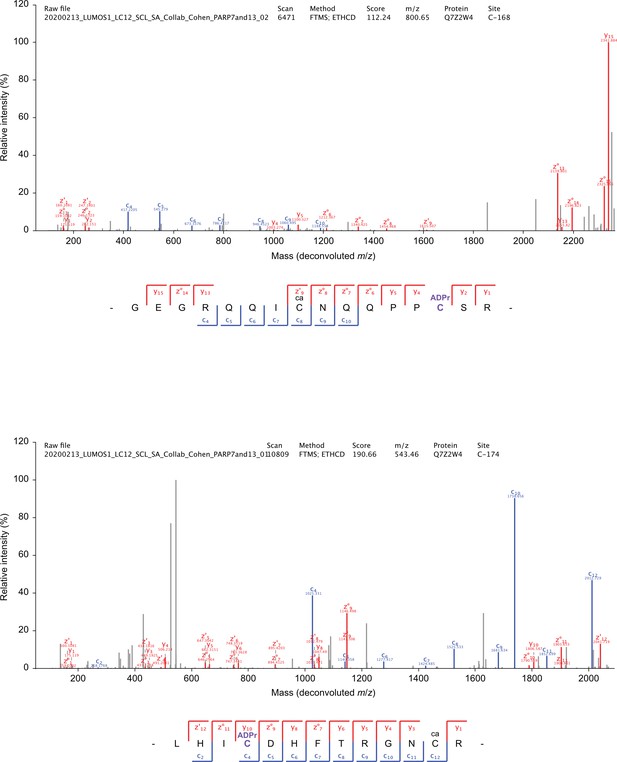
Annotated EThcD MS/MS spectra.
ADP-ribosylation of cysteine residues on PARP-13. Blue; c-ions, red; y-ions and z-ions, grey; unassigned.

Alignment of PARP-13 human with rat and mouse with PARP-7 MARylation sites labeled.
ADPr sites identified on PARP-13 by EThcD indicated by a black box. All sites identified within the ZnF regions are conserved between species. Only one site identified, within the catalytic domain of PARP-13, C721, is not conserved between species.
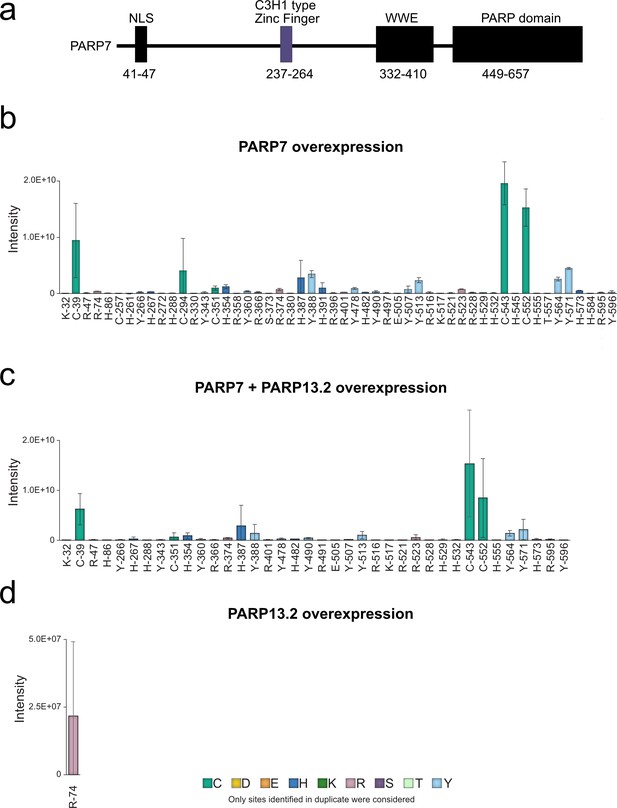
All ADPr sites identified by EThcD MS on PARP-7.
(a) Annotated domain architecture of PARP-7 (b) ADPr sites identified in duplicate on PARP-7 in the GFP-PARP-7 overexpressing cells. (c) ADPr sites identified in duplicate on PARP-7 in the GFP-PARP-7 and MycX2-PARP-13.2 overexpressing cells (d) ADPr sites identified in duplicate on PARP-7 in the MycX2-PARP-13.2 overexpressing cells.
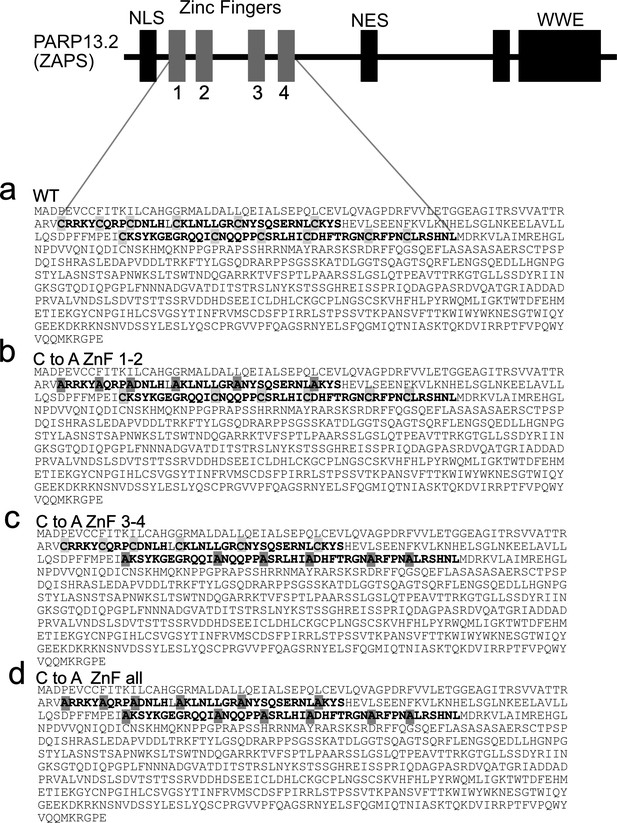
Sequences of CtoA PARP-13.2 mutants.
(a) Wild-type PARP-13.2. (b) PARP-13.2 CtoA ZnF 1–2 mutant, sequence verified. (c) PARP-13.2 CtoA ZnF 3–4 mutant, sequence verified. (d) PARP-13.2 CtoA ZnF all mutant, sequence verified.
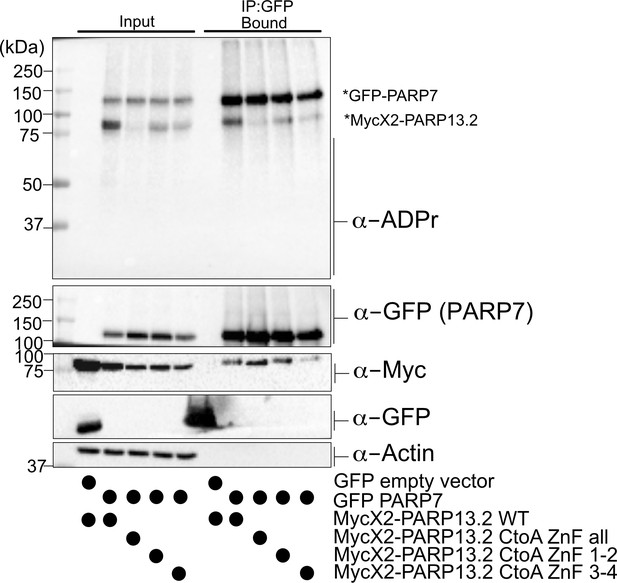
Mutation of Cys residues in the ZnF domain of PARP-13.2 do not disrupt PARP-7 binding.
MycX2 PARP-13.2 WT or ZnF domain mutants were co-expressed with GFP empty vector or GFP PARP-7 in HEK 293 T cells. Proteins were resolved by SDS-PAGE and were detected by Western blot using antibodies against ADPr, GFP, Myc, and actin. GFP-PARP-7 was immunoprecipitated using GFP-nanobody magnetic beads (Chromotek). Representative figure of data collected over two biological replicates.
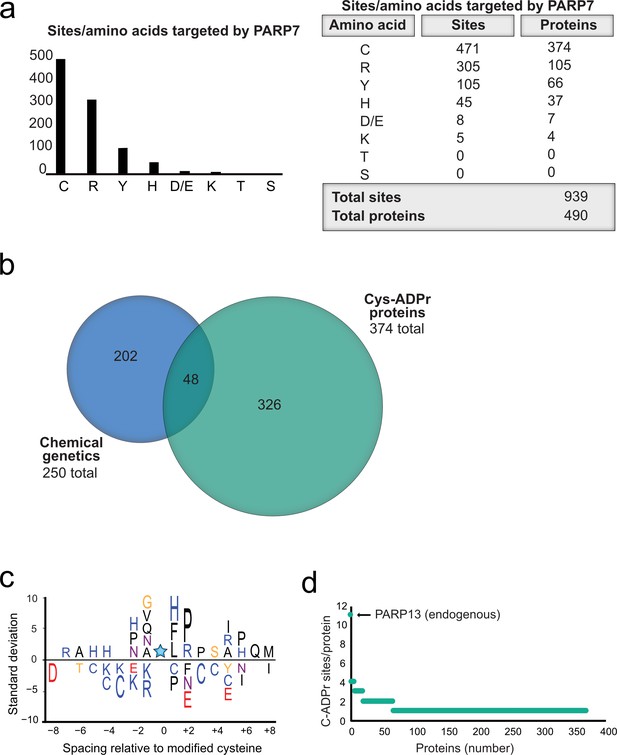
Proteome-wide analysis reveals Cys ADPr as a major MARylation site catalyzed by PARP-7.
(a) Graph and corresponding table showing unique ADPr sites (and proteins) enriched in the PARP-7 overexpression condition (compared to MycX2-PARP-13.2 overexpression condition). (b) Venn diagram depicting the overlap of total proteins identified in our CG strategy compared to total Cys ADPr modified proteins enriched in the PARP-7 condition. (c) IceLogo representation of the sequence context surrounding Cys ADPr sites identified in GFP-PARP-7 overexpressing cells. Amino acids displayed above the line are enriched, and those displayed below are depleted, compared to the sequence context of all cysteine residues derived from the same proteins p=0.05. (d) Graph representing the number of endogenous Cys ADPr sites per protein in the PARP-7 overexpression sample. PARP-13, by far has the highest number of cysteine sites, with 11 sites identified.
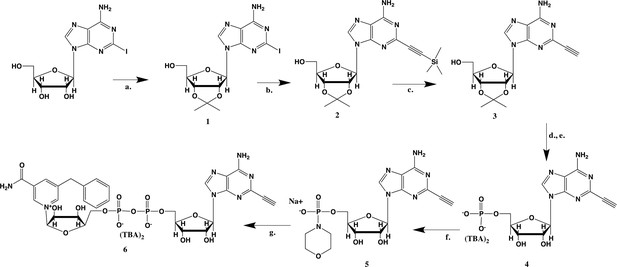
Synthesis of 5-bn-2-ethynyl NAD+.
(a) p-TsOH, acetone, sodium carbonate; (b) (PPh3) PdCl2, CuI, TEA, TMS-acetylene; (c) 7N NH3/MeOH; (d) POCl3,TEA, THF, 0 °C; (e) TFA, 1:1 MeOH H2O; (f) PPh3, Morpholine, 2,2-DPS, 0.2 M NaI in ACN; (g) 5-Bn-NMN, MgSO4, 0.2 M MnCl2 in dry formamide.
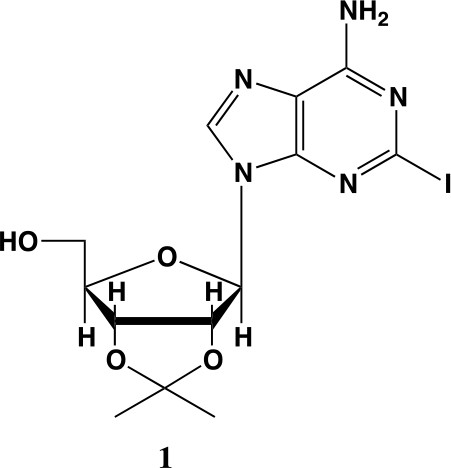
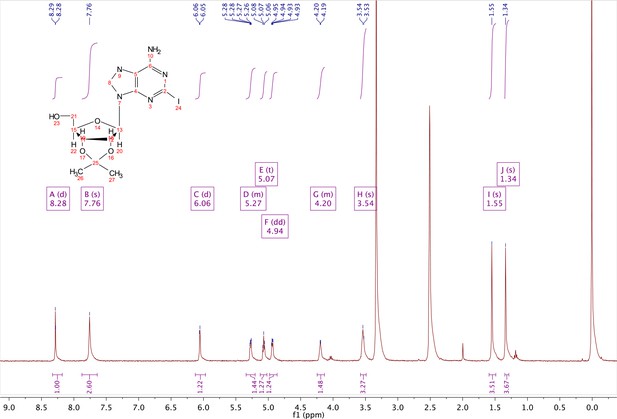
2-iodo-adenosine-acetonide (1).
Commercially available 2-iodo-adenosine (500 mg, 1.3 mmol) was taken up into 13 ml of acetone. To this mixture, p-toluene sulfonic acid (three eq., 3.9 mmol) of was added and stirred for 1 hr. Consumption of the starting material was observed by TLC. The reaction was neutralized with 1 M sodium carbonate until pH = 7.0 was achieved. The product was extracted 3X with chloroform and concentrated in vacuo to yield a fine white crystalline powder. 70% yield. 1H NMR (400 MHz, DMSO-d6) δ 8.28 (d, J = 1.4 Hz, 1H), 7.76 (s, 2H), 6.06 (d, J = 2.7 Hz, 1H), 5.34–5.20 (m, 1H), 5.07 (t, J = 5.4 Hz, 1H), 4.94 (dd, J = 6.2, 2.8 Hz, 1H), 4.25–4.13 (m, 1H), 3.54 (s, 2H), 1.55 (s, 3H), 1.34 (s, 3H).
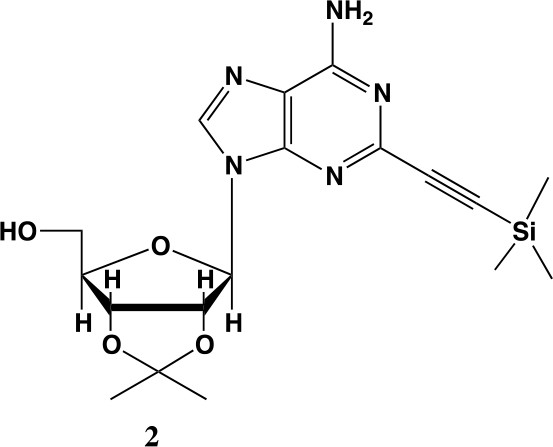
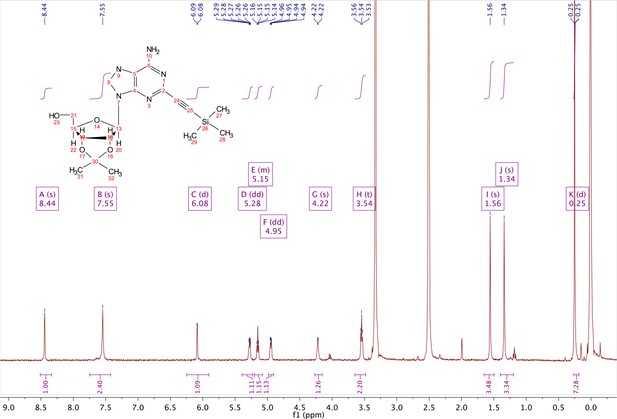
2-ethynyl-TMS-adenosine-acetonide (2).
In a clean, oven dried round bottom flask with stir bar, 1 (342 mg, 0.78 mmol) and copper iodide (0.2 eq., 0.158 mmol) were dissolved in 5 ml dry DMF. The flask was evacuated under high vacuum and refilled with argon several times. Under argon, the flask was capped with a septum and argon was allowed to bubble through a needle through the solution for 20 min. 0.1 equivalents (55 mg, 0.16 mmol) of Bis(triphenylphosphine)palladium(II) Dichloride was added to the solution while still under argon. To the capped solution, trimethylsilylacetylene (1.5, eq., 164 µl, 1.18 mmol) of was added via syringe. To this solution, triethylamine was added (three eq., 330 µl, 2.37 mmol) via syringe. The reaction was covered in aluminum foil and a balloon filled with argon was placed into the septum of the capped reaction and the reaction was allowed to progress, stirring overnight at RT. The reaction turned a deep brown color overnight. DMF was evaporated in vacuo in a water bath set to 40 °C. The residue was taken up into 25 ml ethyl acetate and remaining DMF was extracted from the organic layer with 25 ml brine (3X). The organic layer was dried over sodium sulfate, concentrated and purified via combi flash (MP A: hexanes, MP B: ethyl acetate; 0–3 min: 0–100%B, 3–18 min: 100%B). Fractions containing desired compound were pooled and concentrated to yield 191 mg of product 60% yield. 1H NMR (400 MHz, DMSO-d6) δ 8.44 (s, 1H), 7.55 (s, 2H), 6.08 (d, J = 2.9 Hz, 1H), 5.28 (dd, J = 6.1, 2.9 Hz, 1H), 5.20–5.08 (m, 1H), 4.95 (dd, J = 6.1, 2.7 Hz, 1H), 4.22 (s, 1H), 3.54 (t, J = 5.1 Hz, 2H), 1.56 (s, 3H), 1.34 (s, 3H), 0.25 (d, J = 1.0 Hz, 9H).
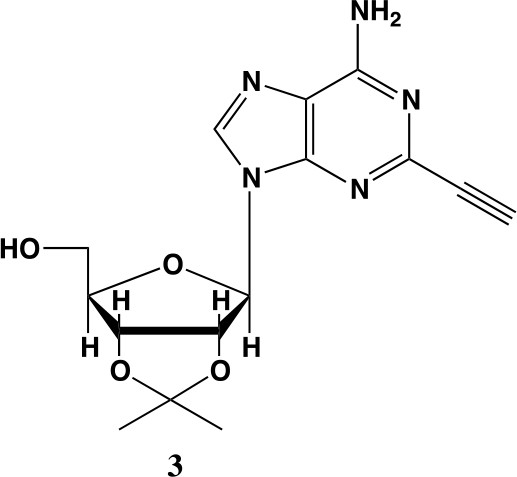
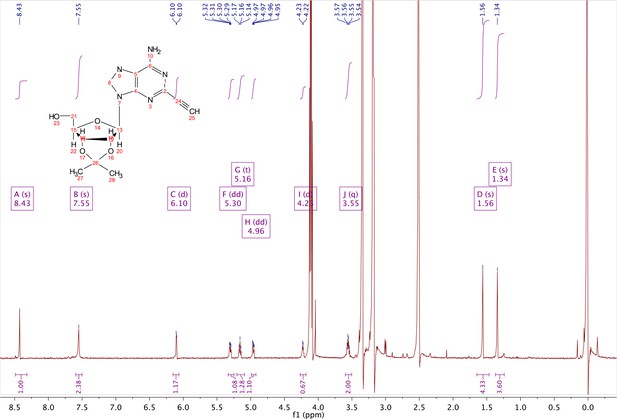
2-ethynyl-adenosine-acetonide (3).
In a clean, dry round bottom flask, 20 ml of saturated 7N ammonia in methanol was added to 2 (200 mg, 0.5 mmol) and stirred for 1.5 hr until complete trimethylsilyl deprotection was observed by TLC. The reaction was concentrated in vacuo. The crude residue was purified via combi flash (MP A: hexanes, MP B: ethyl acetate; 0–1 min: 0–100% B, 1–18 min: 100% B). Fractions containing desired compound were pooled and concentrated to yield 122 mg of off-white powder (74% yield). 1H NMR (400 MHz, DMSO-d6) δ 8.43 (s, 1H), 7.55 (s, 2H), 6.10 (d, J = 2.9 Hz, 1H), 5.30 (dd, J = 6.1, 3.1 Hz, 1H), 5.16 (t, J = 5.5 Hz, 1H), 4.96 (dd, J = 6.2, 2.5 Hz, 1H), 4.23 (d, J = 3.0 Hz, 1H), 3.55 (q, J = 5.0 Hz, 2H), 1.56 (s, 3H), 1.34 (s, 3H).
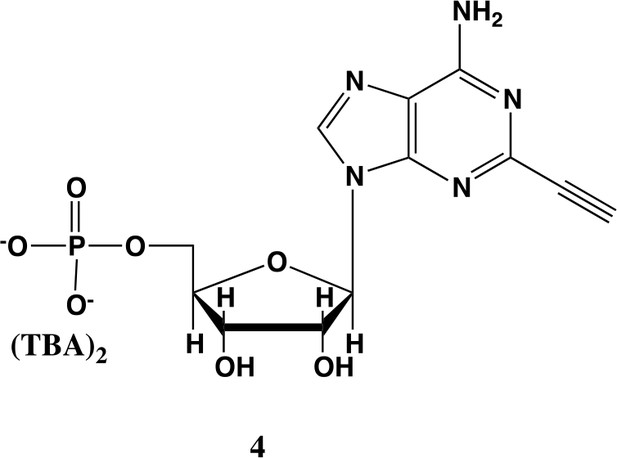
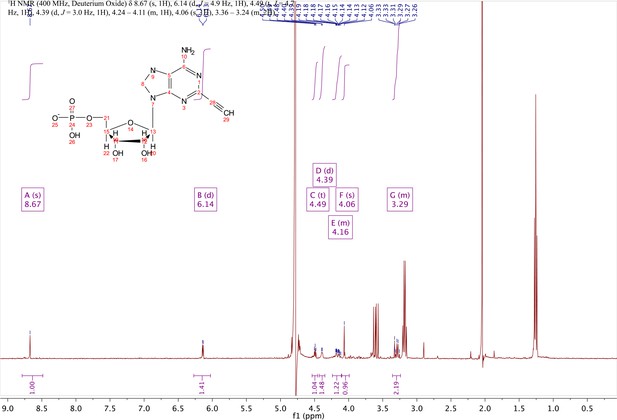
2-ethynyl-AMP (4).
In a clean, dry round bottom flask, 5.1 ml dry THF was added to 3 (0.51 mmol, 170 mg). Triethylamine (12 eq., 858 µl, 6.1 mmol) were added to the solution and cooled to 0 °C in an ice bath. Once cooled, Phosphorous (V) oxychloride (POCl3) (two eq., 95 µl, 1.02 mmol,) was added dropwise to the solution. Consumption of the starting material was monitored by TLC (100% ethyl acetate). To quench the reaction, 5.1 ml of H2O was added to the solution. The solution was concentrated to dryness and co-evaporated with acetonitrile and methanol. The residue was rinsed with ether. The resulting residue was resuspended in chloroform. The desired compound, insoluble in chloroform, crashed out and the resulting TEA salt was filtered out from the mixture. Without further purification the solid was dissolved in 8.2 ml of a 1:1 mixture of H2O and methanol. Trifluoracetic acid (65 eq., 2.0 ml, 27.0 mmol,) was added dropwise to the solution at room temperature and stirred until complete deprotection of the acetonide moiety was observed by analytical HPLC. The mixture was concentrated to a small volume (less than 2 ml) in vacuo and the liquid was purified on reverse phase chromatography using a Combiflash Companion system (C18Aq 5.5 g Redisep Rf; MP A: 10 mM tributylamine/30 mM acetic acid pH 4.4 (aq.), MP B: methanol; 0–1 min: 0% B, 1–12 min: 0–50% B, 12–16 min: 100% B). Fractions containing desired product were pooled and concentrated in vacuo to yield the tributylammonium (TBA) salt of the product (80 mg, 35% yield over two steps).1H NMR (400 MHz, D2O) δ 8.67 (s, 1H), 6.14 (d, J = 4.9 Hz, 1H), 4.49 (t, J = 4.7 Hz, 1H), 4.39 (d, J = 3.0 Hz, 1H), 4.24–4.11 (m, 1H), 4.06 (s, 1H), 3.36–3.24 (m, 2H).
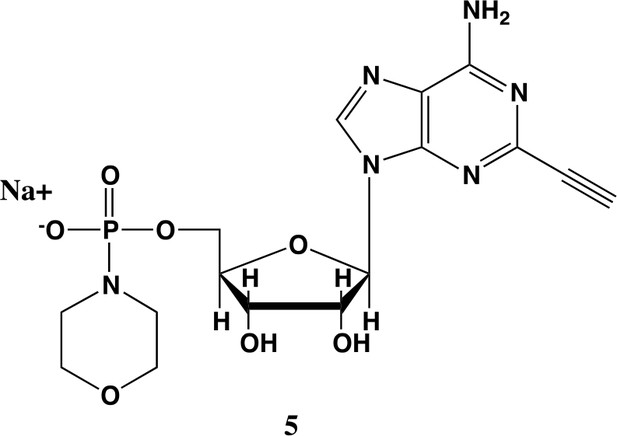
2-ethynyl-AMP-morpholidate (5).
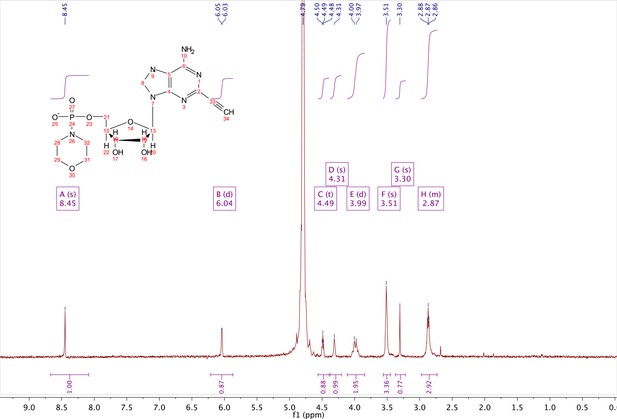
In a clean oven-dried round bottom flask with stir bar, 4 (80 mg, 0.144 mmol) was taken up into 2.5 mL of DMSO (dried over 3 Å sieves overnight). 5 ml of dry DMF was co-evaporated three times in vacuo with the solution in a water bath set to 35 °C. After the third co-evaporation, the solution was allowed to sit under high vacuum pressure for 30 min. The solution was taken off of vacuum pressure under argon gas and capped with a septum and argon balloon. Under argon pressure and in the order listed, triphenyl phosphine (202 mg, 0.77 mmol), morpholine (108 µl, 1.24 mmol), and 2,2'-dipyridyl disulfide (170 mg, 0.77 mmol) were added to the solution. The reaction was allowed to take place for 1.5 hr until consumption of the starting material was observed by HPLC. The desired compound was precipitated out in 0.2 M NaI in acetonitrile (dry) and filtered using a fine filter glass funnel. The result was an off-white powder (36 mg, 57% yield). 1H NMR (400 MHz, D2O) δ 8.45 (s, 1H), 6.04 (d, J = 4.8 Hz, 1H), 4.49 (t, J = 4.7 Hz, 1H), 4.31 (s, 1H), 3.99 (d, J = 12.0 Hz, 2H), 3.51 (s, 4H), 3.30 (s, 1H), 2.98–2.73 (m, 4H).
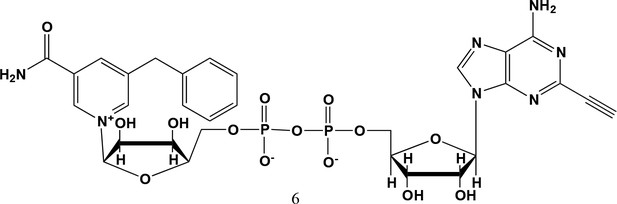
5-benzyl-2-ethynyl-NAD+ (6).
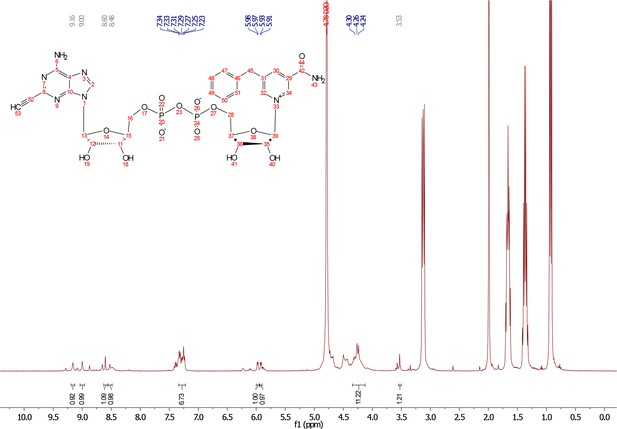
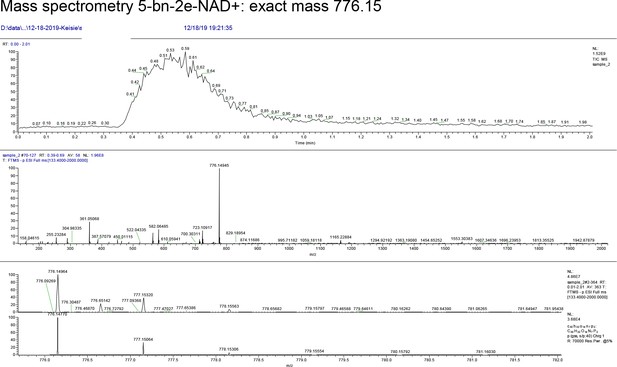
5-benzyl-nicotinamide mononucleotide (two eq., 29 mg, 0.068 mmol), synthesized as previously reported (Carter-O'Connell et al., 2016), was taken up into 2 ml dry toluene in a scintillation vile and concentrated in vacuo (3X) to remove excess water. 30 min under high vacuum, the solid was capped with a septum under argon pressure. 5 (15 mg, 0.034 mmol), after overnight desiccation in a chamber containing P2O5, was added along with MgSO4 (8 mg, 0.068 mmol) to the scintillation vile containing 5-Bn-NMN under argon pressure. 500 µl of 0.2 M MnCl2 in dry formamide was added to the solids via syringe and the reaction was stirred overnight under Argon balloon at room temperature. The reaction was checked by HPLC. After complete consumption of 5 was observed overnight, the reaction was diluted with 1.5 mL 10 mM tributylamine/30 mM acetic acid [pH = 4.4] and purified using HPLC. Retention time 8.5 min. The fractions containing desired compound were pooled and co-evaporated with acetonitrile (to remove excess water) and methanol to remove excess tributylamine. The resulting compound was a white powder (13 mg, 33% yield). 1H NMR (400 MHz, D2O) δ 9.16 (s, 1H), 9.00 (s, 1H), 8.87 (s, 1H), 8.60 (s, 1H), 8.52 (s, 1H), 7.29 (m, 5H), 5.98 (d, J = 5.0 Hz, 1H), 5.92 (d, J = 5.2 Hz, 1H), 4.30–4.24 (m, 11H), 3.53 (s, 1H).

2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)benzamide.
2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)benzoic acid (50 mg, 0.17 mmol) was added to a flame dried flask and dissolved in SOCl2 (20 ml) and refluxed at 80 °C for 45 min. The reaction mixture was cooled to 0 °C and then added dropwise to ice cold anhydrous NH3/MeOH (7 N, 100 ml). White solids quickly precipitated from an orange solution accompanied by rapid heating of the reaction vessel and smoke. The reaction was left to stir over night and the product collected as a solid by vacuum filtration: 50 mg (100%).
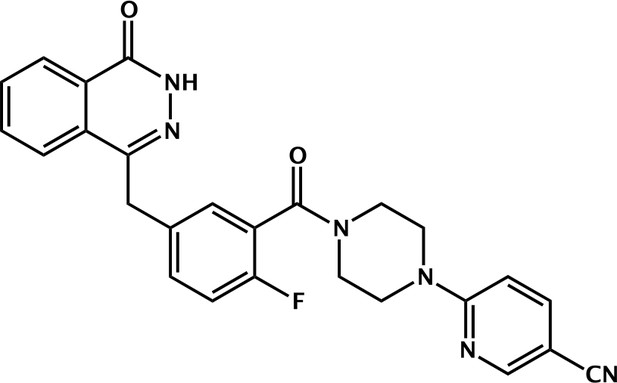
6-(4-(2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)benzoyl)piperazin-1-yl)nicotinonitrile (Phthal 01).
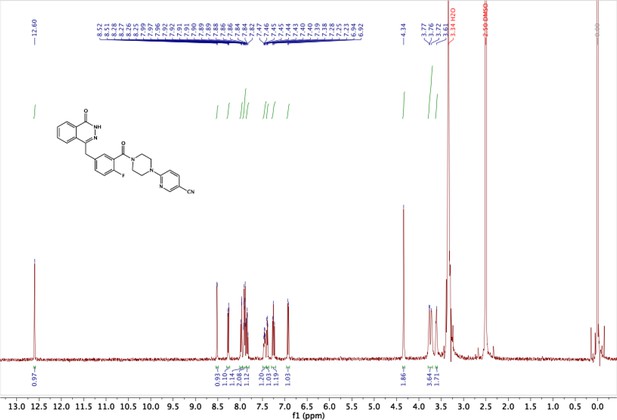
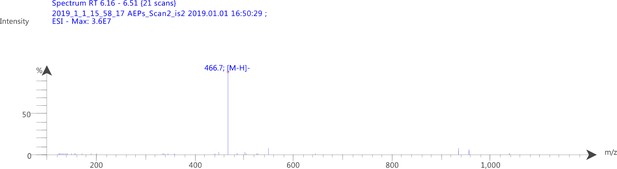
2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)benzoic acid (50 mg, 0.17 mmol), TBTU (60 mg, 0.19 mmol) and 6-(piperazin-1-yl)nicotinonitrile (36 mg, 0.19 mmol) were added to a flame dried flask and dissolved in anhydrous DMF/DCM (1:10) (5 mL). DIPEA (0.06 mL, 0.34 mmol) was added under argon and the reaction stirred at RT for 18 h. The product was then precipitated in water (100 mL) and collected by vacuum filtration as a cream solid: 70 mg (89%). 1H NMR (400 MHz, DMSO-d6) δ 12.60 (s, 1H), 8.51 (d, J = 2.3 Hz, 1H), 8.27 (dd, J = 7.7, 1.5 Hz, 1H), 8.01 – 7.95 (m, 1H), 7.95 – 7.87 (m, 2H), 7.87 – 7.80 (m, 1H), 7.45 (td, J = 5.5, 5.0, 2.7 Hz, 1H), 7.39 (dd, J = 6.5, 2.3 Hz, 1H), 7.25 (t, J = 9.0 Hz, 1H), 6.93 (d, J = 9.1 Hz, 1H), 4.34 (s, 2H), 3.79 – 3.69 (m, 4H), 3.61 (s, 2H). MS m/z [M-H]- for C26H21FN6O2: 466.7. Rt = 8.95 min.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-ADP-ribose, E6F6A (Rabbit monoclonal) | Cell signaling technology | 83732: RRID:AB_2749858 | WB(1:1000) |
| Antibody | Anti-GFP, PABG1 (Rabbit polyclonal) | Chromotek | PABG1-100; RRID:AB_2749857 | WB(1:1000) |
| Antibody | Anti-HA.11, 16B12 (Mouse monoclonal) | BioLegend | 901502; RRID:AB_2565007 | WB(1: 2000) |
| Antibody | Anti-Myc, 9B11 (Mouse monoclonal) | Cell signaling technology | 2276S; RRID:AB_331783 | WB(1:1000) |
| Antibody | Anti-β-Actin, C-4 (Mouse monoclonal) | Santa Cruz Biotechnology | sc-47778; RRID:AB_2714189 | WB(1:1000) |
| Antibody | Anti-ZC3HAV1/PARP-13 (Rabbit polyclonal) | Proteintech | 16820–1-AP; RRID:AB_2728733 | WB(1:1000) |
| Antibody | Goat anti-rabbit IgG, HRP linked | Jackson Immuno Research Lab | 20402–1 mL | WB(1:10000) |
| Antibody | Anti rabbit IgG, HRP linked | Cell signaling technology | 7074S | WB(1:1000) |
| Antibody | Goat anti-mouse IgG (H+L), HRP linked | Invitrogen | 62–6520 | WB(1:5000) |
| Antibody | Anti-ZC3HAV1/PARP-13 (Rabbit polyclonal) | Invitrogen | PA5-106389; RRID:AB_2854063 | WB(1:1000) |
| Antibody | Anti-ZC3HAV1, N3C2 (Mouse monoclonal) | GeneTex | GTX120134; AB_10721153 | WB(1:2,000) |
| Antibody | Anti- TIPARP/PARP-7 (mouse monoclonal) | This paper | WB(1:1000) | |
| Peptide, recombinant protein | Streptavidin HRP (Peroxidase Streptavidin 1.0 mg) | Jackson Immuno Research Lab | NC9705430 | WB(1:2500) |
| Other | GFP-Trap Magnetic Agarose | Chromotek | gtma-20 | |
| Other | Myc-trap magnetic agarose | Chromotek | ytma-10 | |
| Other | Pierce NeutrAvidin Agarose | Thermo Scientific | 29201 | |
| Other | Cytiva (Formerly GE Healthcare Life Sciences) Glutathione Sepharose 4B Media | Fisher Scientific | 45-000-139 | |
| Chemical compound, drug | Biotin-Peg3-Azide | Click Chemistry Tools | AZ104-5 | |
| Chemical compound, drug | Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) | Sigma-Aldrich | 762342–100 MG | |
| Chemical compound, drug | cOmplete EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | 11873580001 | |
| Chemical compound, drug | Hydroxylamine solution | Sigma-Aldrich | 438227–50 ML | |
| Chemical compound, drug | Fetal bovine serum | Sigma-Aldrich | TMS-013-B | |
| Chemical compound, drug | Gibco DMEM- high glucose | Fisher Scientific | 11965092 | |
| Chemical compound, drug | Gibco GlutaMAX supplement | Fisher Scientific | 35050061 | |
| Chemical compound, drug | 5bn-6a-NAD+ | JACS, 136, 5201 (2014) | ||
| Chemical compound, drug | 5bn-2e-NAD+ | This paper | See Methods Supplement | |
| Chemical compound, drug | Phthal01 | This paper | See Methods Supplement | |
| Recombinant DNA reagent | Myc-BirA*-PARP-7 | This paper | Cloned form Myc-BirA*-PARP-14 described in ACS Chem. Biol., 13, 2841 (2018) | |
| Recombinant DNA reagent | GFP-WT-PARP-7 | This paper | WT-PARP-7 gene (gBlock gene fragment) was cloned into pEGFP-C1 | |
| Recombinant DNA reagent | GFP-IG-PARP-7 | This paper | IG-PARP-7 gene (gBlock gene fragment) was cloned into pEGFP-C1 | |
| Recombinant DNA reagent | HA-PARP-13.1 | ACS Chem. Biol.,13, 2841 (2018) | ||
| Recombinant DNA reagent | HA-PARP-13.2 | This paper | WT-HA-PARP-13.2 gene (gBlock gene fragment) was cloned into CMV vector using standard restriction enzyme methods | |
| Recombinant DNA reagent | MycX2-PARP-13.2 | This paper | WT-MycX2-PARP-13.2 gene (gBlock gene fragment) was cloned into CMV vector using standard restriction enzyme methods | |
| Recombinant DNA reagent | MycX2-PARP-13.2 CtoA ZnF all mutant | This paper | Mutant MycX2-PARP-13.2 gene (gBlock gene fragment) was cloned into CMV vector using standard restriction enzyme methods | |
| Recombinant DNA reagent | MycX2-PARP-13.2 CtoA ZnF 1–2 mutant | This paper | Mutant MycX2-PARP-13.2 gene (gBlock gene fragment) was cloned into CMV vector using standard restriction enzyme methods | |
| Recombinant DNA reagent | MycX2-PARP-13.2 CtoA ZnF 3–4 mutant | This paper | mutant MycX2-PARP-13.2 gene (gBlock gene fragment) was cloned into CMV vector using standard restriction enzyme methods | |
| Recombinant DNA reagent | HA-MacroD2 | This paper | HA-MacroD2 gene (gBlock gene fragment) was cloned into CMV vector using standard restriction enzyme methods | |
| Recombinant DNA reagent | GFP-PARP-10 | Cell Reports, 25, 4770 (2015) | ||
| Recombinant DNA reagent | mCherry | gift plasmid from RY Tsien | ||
| Recombinant DNA reagent | GST-PARP-7 | Gomez et al., Biochemical Journal,475, 3827 (2018) | ||
| Recombinant DNA reagent | His6-SUMO-PARP-13.2 | gift plasmid from I. Carter-O'Connell |
Additional files
-
Supplementary file 1
See Excel file.
Tab 1: PARP-7 MARylation targets identified from HEK 293 T cell lysates using a chemical genetic approach. Tab 2: PARP-7 interactors identified from HEK 293 T cells using proximity labeling (BioID approach) Tab 3: reviGO analysis, related to Figure 1a. Tab 4: Metascape GO analysis, related to Figure 1b. Tab 5: ADP-ribosylation targets and sites identified from HEK 293 T cells expressing PARP-13.2 alone, PARP-7 alone, or PARP-13.2 and PARP-7. Tab 6: Number of cysteine sites per protein target, related to Figure 4d.
- https://cdn.elifesciences.org/articles/60480/elife-60480-supp1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60480/elife-60480-transrepform-v3.docx