Identification of PARP-7 substrates reveals a role for MARylation in microtubule control in ovarian cancer cells
Figures
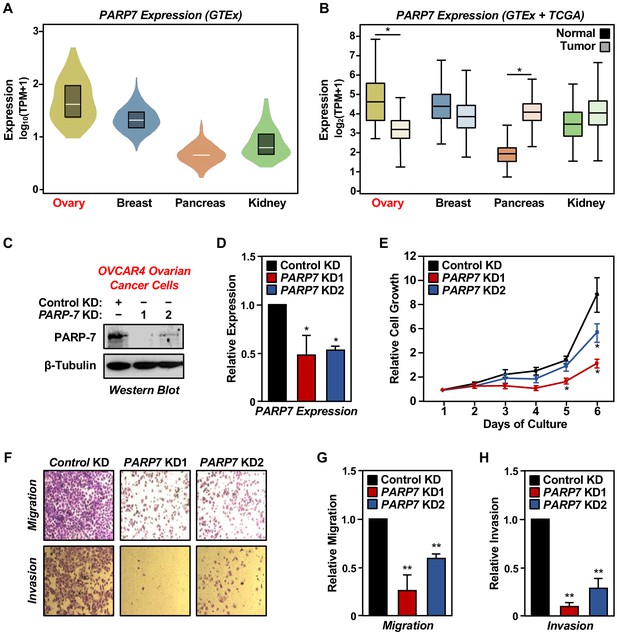
PARP-7 expression in cancers and role in ovarian cancer cell phenotypes.
(A) Violin plots showing normalized expression of PARP7 mRNA from GTEx data presented as log10(TPM+1) (Transcripts Per Million), calculated from a gene model with all isoforms collapsed to a single gene. The data were obtained from the GTEx portal and expressed as normalized TPM scores as described (Li et al., 2010). Expression profiles of PARP7 mRNA in the following cancers: breast (n = 459, median TPM = 21.1), ovary (n = 180, median TPM = 37.6), pancreas (n = 328, median TPM = 4.1), and kidney cortex (n = 85, median TPM = 5.6). (B) Box plots showing normalized expression of PARP7 mRNA across four tissues (ovary, breast, pancreas, and kidney) and their corresponding cancer types (matched TCGA normal and GTEx) analyzed using GEPIA and presented as log2(TPM+1). Ovary and pancreas show significantly different expression levels for PARP7 between normal and tumor tissues. Asterisks indicate significant differences between normal and tumor samples (Student’s t-test, *p<0.05). (C) Western blots showing PARP-7 protein levels with or without siRNA-mediated knockdown (KD) of PARP7 in OVCAR4 human ovarian cancer cells. Two different siRNAs were used. β-tubulin was used as loading control. (D) Expression of PARP7 mRNA with or without siRNA-mediated knockdown (KD) of PARP7 in OVCAR4 cells as determined by RT-qPCR. Each bar represents the mean ± SEM, n = 3. Asterisks indicate significant differences from the control (Student’s t-test; *p<0.05). (E) Growth of OVCAR4 cells with or without siRNA-mediated knockdown (KD) of PARP7 was assayed by crystal violet staining. Each point represents the mean ± SEM, n = 3. Asterisks indicate significant differences from the corresponding control (Student’s t-test; *p<0.05). (F) Cell migration assays (top) and cell invasion assays (bottom) for OVCAR4 cells with or without siRNA-mediated knockdown (KD) of PARP7. (G) Quantification of cell migration assays like those shown in (F), top. Each bar represents the mean ± SEM, n = 3. Asterisks indicate significant differences from the control (Student’s t-test; **p<0.01). (H) Quantification of cell invasion assays like those shown in (F), bottom. Each bar represents the mean ± SEM, n = 3. Asterisks indicate significant differences from the control KD (Student’s t-test; **p<0.01).
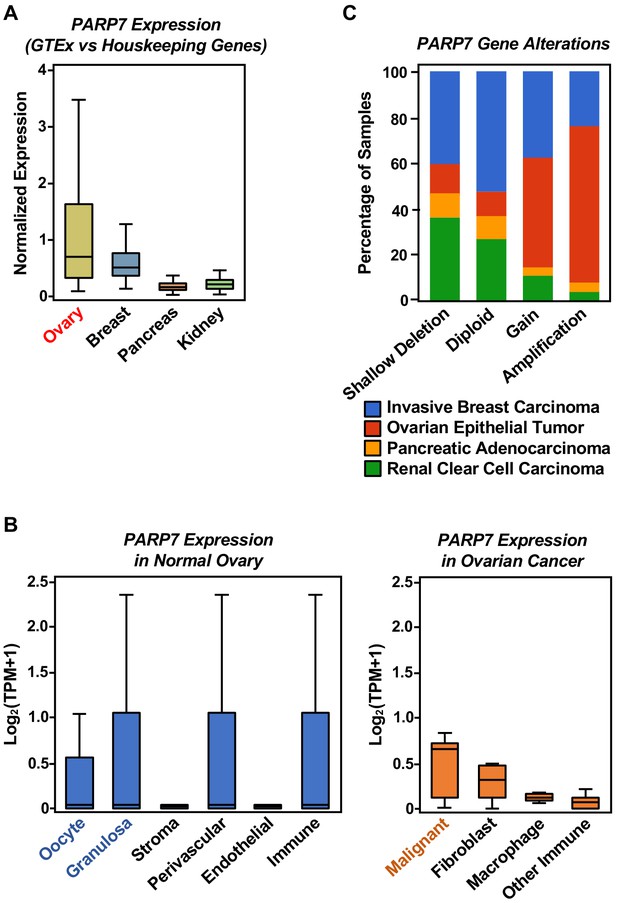
Expression of PARP7 mRNA in normal and cancerous tissues, and alterations in the PARP7 gene across four cancer types.
(A) The average TPMs for mRNAs expressed from 50 ‘housekeeping’ genes were obtained from GTEx data for breast, ovary, pancreas, and kidney, as described previously (Eisenberg and Levanon, 2013). The expression of PARP7 mRNA was normalized to the average TPMs for mRNAs expressed from the 50 ‘housekeeping’ genes to derive a normalized expression value. (B) Analysis of PARP7 expression in individual cell populations from normal ovary tissue (left) and ovarian cancer tissue (right) using published single-cell RNA-seq (scRNA-seq) data. Each bar represents: (1) The average expression (log2[TPM+1]) of PARP7 in clusters of similar cell types revealed by uniform manifold approximation and projection (UMAP) analysis of scRNA-seq from sorted ovarian DDX4 Ab+ and Ab- cortex cells (Wagner et al., 2020) (left) and (2) the average expression (log2[TPM + 1]) of PARP7 in clusters of similar cell types from ovarian cancer ascites by scRNA-seq (Izar et al., 2020) (right). (C) PARP7 gene alterations across four cancer types (ovary, breast, pancreas, and kidney). Alterations are classified in four types: Shallow deletion, diploid (unaltered), gain, and amplification. Data are from the Pan-Cancer Atlas in the TCGA database (Hoadley et al., 2018).
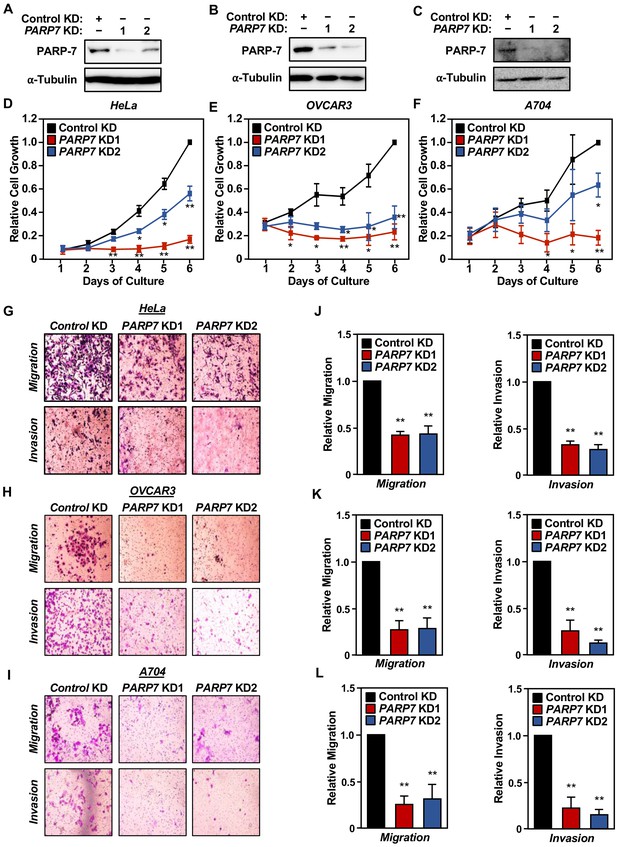
Effects of PARP7 knockdown on cancer-related phenotypes in HeLa, OVCAR3, and A704 cells.
(A–C) Western blots showing PARP-7 protein levels after siRNA-mediated knockdown (KD) of PARP7 in HeLa (A), OVCAR3 (B) and A704 (C) cells. Two different siRNAs were used. α-tubulin was used as loading control. (D–F) The growth of HeLa (D), OVCAR3 (E), and A704 (F) cells with or without siRNA-mediated knockdown (KD) of PARP7 was assayed by crystal violet staining. Each point represents the mean ± SEM, n = 3. Asterisks indicate significant differences from the corresponding control (Student’s t-test; *p<0.05). (G–I) Cell migration assays (top) and cell invasion assays (bottom) for HeLa (G), OVCAR3 (H), and A704 (I) cells with or without siRNA-mediated knockdown (KD) of PARP7. (J–L) Quantification of cell migration (left) and cell invasion (right) assays like those shown in (G–I). Asterisks indicate significant differences from the control (n = 3, Student’s t-test; **p<0.01).
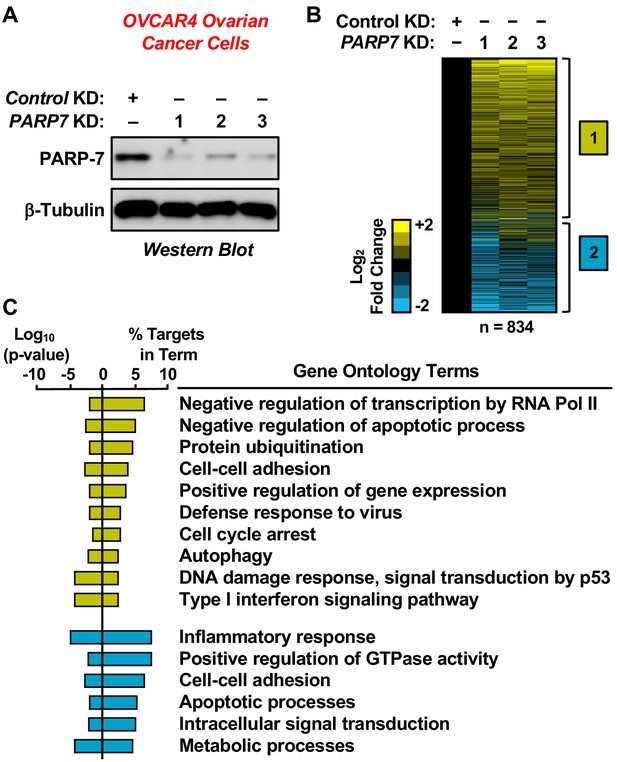
RNA-seq analysis of gene expression in ovarian cancer cells following PARP-7 depletion.
(A) Western blots showing PARP-7 protein levels after siRNA-mediated knockdown (KD) of PARP7 in OVCAR4 cells. Three different siRNAs were used. β-tubulin was used as loading control. (B) Heat maps showing the results of RNA-seq assays from OVCAR4 cells upon siRNA-mediated knockdown (KD) of PARP7. Three different siRNAs were used. Results represent fold changes in FPKM values for genes significantly regulated versus the control KD, expressed as log2 fold change. A fold change ≥1.5 was classified as upregulated, while a fold change ≤−0.5 was classified as downregulated. (C) Gene ontology terms enriched for the significantly upregulated genes (yellow) and significantly downregulated genes (blue) shown in (B). The percent of targets from each GO term identified in the analysis is shown, along with the log10 p-value for the enrichment.
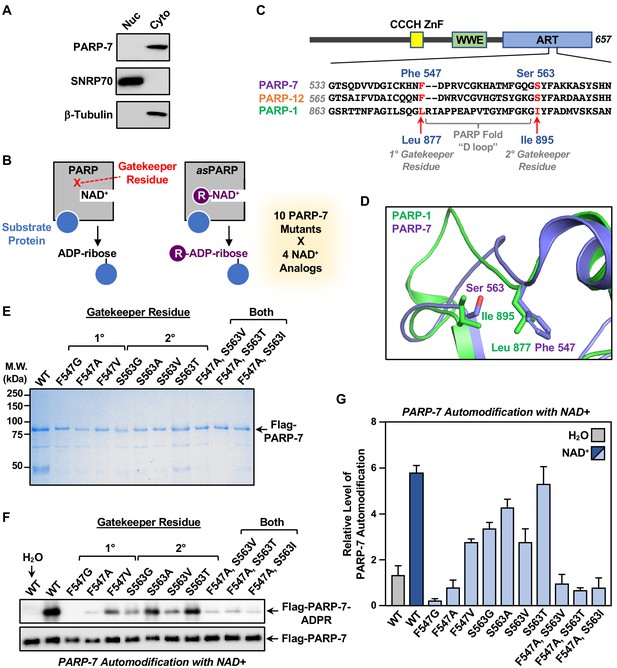
Generation of NAD+ analog-sensitive PARP-7 (asPARP-7) mutants.
(A) PARP-7 localizes to the cytosolic compartment in OVCAR4 cells. Western blots of nuclear and cytosolic fractions of OVCAR4 ovarian cancer cells. SNRP70 and β-tubulin were used as loading controls for the nuclear and cytoplasmic fractions, respectively. (B) Schematic diagram illustrating the NAD+ analog-sensitive PARP approach. R, unnatural chemical moieties added to NAD+. In this study, 10 PARP-7 mutants targeting two potential gatekeeper residues (shown in red) were screened with 14 different NAD+ analogs. (C) (Top) Schematic diagram of PARP-7 showing the functional domains, including the Cys3His1 zinc finger (CCCH ZnF), the WWE PAR-binding domain (WWE), and the ADP-ribosyl transferase domain (ART). (Bottom) Multiple sequence alignment of PARP-7 (Q7Z3E1), PARP-1 (P09874) and PARP-12 (Q9H0J9). The two potential gatekeeper residues in PARP-7 selected for mutation are highly conserved among PARP-7, PARP-1, and PARP-12. Residues F547 and S563 in PARP-7, as well as the corresponding residues in PARP-1 (residues L877 and I895) and PARP-12 (residues F579 and S595), are indicated in red. (D) Close-up view of residues F547 and S563 in PARP-7. The structure was generated by comparison of available structures of PARP-1 (PDB: 3PAX) and PARP-12 (PDB: 2PQF). The molecular graphic was generated with Pymol. (E) Recombinant PARP-7 proteins used for asPARP-7 activity screening. SDS-PAGE analysis with subsequent Coomassie blue staining of purified FLAG-tagged wild-type (WT) PARP-7 and 10 PARP-7 site-specific mutants expressed in Sf9 insect cells. Molecular weight markers in kilodaltons (kDa) are shown. (F) Western blots showing the automodification (MARylation) activity of the PARP-7 mutants with NAD+ detected using an MAR-binding reagent. PARP-7 was used as loading control for the reactions. (G) Quantification of the automodification activity of the PARP-7 mutants shown in (F).
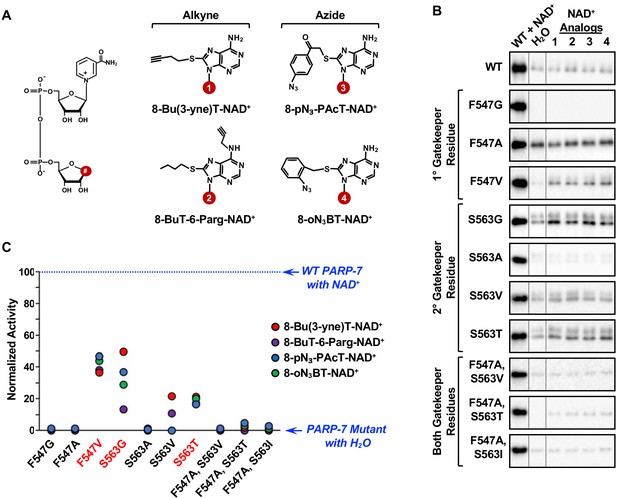
Screening the activity of potential analog-sensitive PARP-7 mutants with clickable NAD+ analogs.
(A) Chemical structures of four clickable NAD+ analogs used for the asPARP-7 screen. (Left) The R groups are linked at position 8 (#) of the adenine ring of NAD+. (Right) 8-BuT-6-Parg-NAD+ and 8-Bu(3-yne)T-NAD+ are clickable through their alkyne groups, whereas 8-oN₃-BT-NAD+ and 8-pN3-PAcT-NAD+ are clickable through their azide groups. (B) Western blots showing the wild-type and mutant PARP-7 automodification reactions performed with NAD+ or the clickable NAD+ analogs shown in (A). The MAR signals were detected using a MAR binding reagent. (C) Normalized automodification activity of wild-type and mutant PARP-7 proteins with NAD+ or the clickable NAD+ analogs shown in (A). PARP-7 automodification assays like those shown in (B) were used to determine the activity of the mutants with the NAD+ analogs. This screen was performed twice and yielded similar results each time; the results from one replicate are shown.
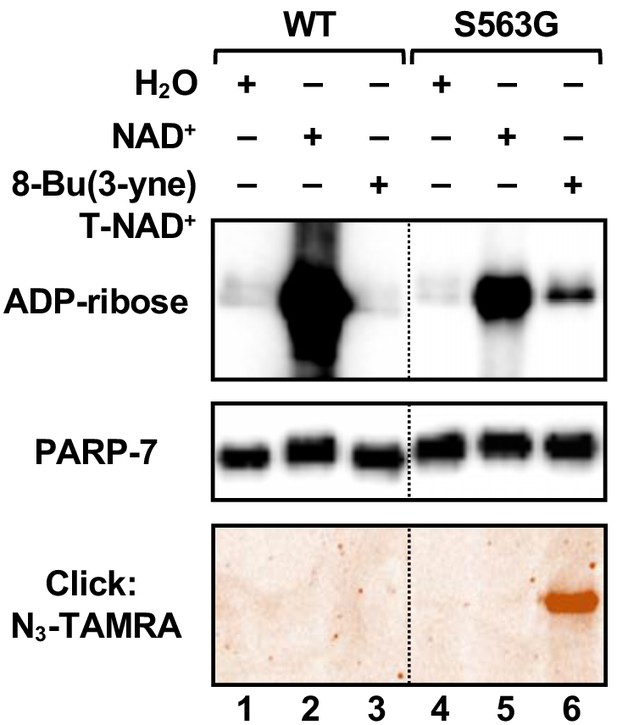
Activity of the S563G analog-sensitive PARP-7 mutant with 8-Bu(3-yne)T-NAD+.
Automodification reactions with wild-type and S563G mutant PARP-7 performed with NAD+ or 8-Bu(3-yne)T-NAD+. The autoMARylation signals were detected by (top) western blotting using a MAR binding reagent or (bottom) click chemistry-based in-gel fluorescence. PARP-7, detected by western blotting (middle), was used as loading control for the reactions. TAMRA, tetramethylrhodamine.
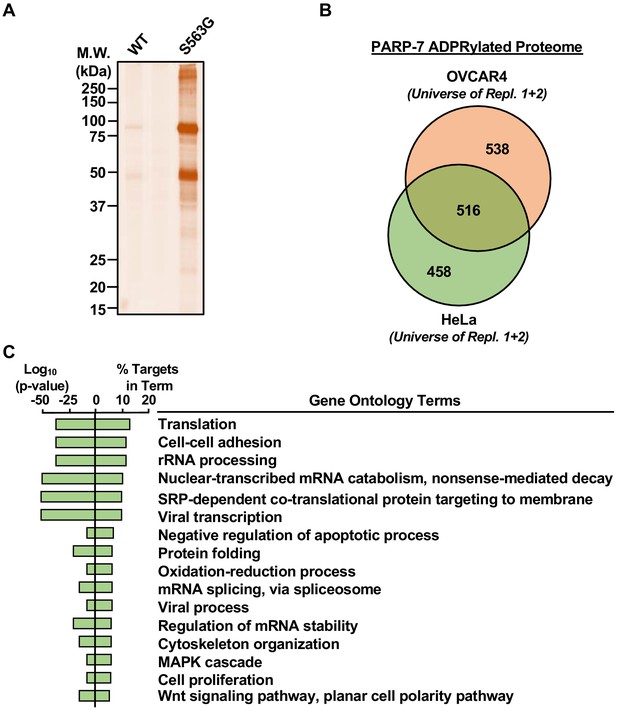
Identification of PARP-7-specific substrates using an NAD+ analog-sensitive PARP-7 approach.
(A) In-gel fluorescence of cytoplasmic extract from OVCAR4 cells conjugated to azido-TAMRA after labeling reactions with 8-Bu(3-yne)T-NAD+ in the presence of wild-type or S563G mutant PARP-7. Molecular weight markers in kDa are shown. Similar assays were performed with cytoplasmic extract from HeLa cells. The assay was performed twice in each cell line. (B) Venn diagram depicting the overlap of the protein substrates of PARP-7-mediated MARylation between OVCAR4 and HeLa cells identified using asPARP-7 and mass spectrometry. (C) Gene ontology terms enriched for the protein targets of PARP-7-mediated MARylation common between OVCAR4 and HeLa cells.
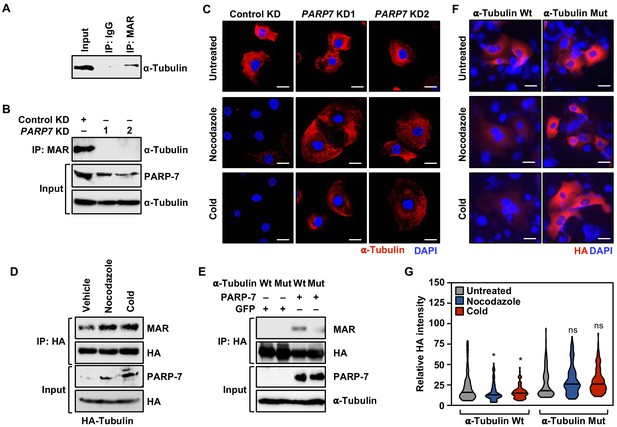
PARP-7 links MARylation of α-tubulin to the regulation of microtubule stability in ovarian cancer cells.
(A) MARylation of α-tubulin in OVCAR4 cells. MARylated proteins were immunoprecipitated from OVCAR4 cells using a MAR detection reagent and subjected to western blotting for α-tubulin. The experiment was performed three times (n = 3) to ensure reproducibility. (B) Knockdown of PARP-7 reduces the MARylation of α-tubulin. MARylated proteins were immunoprecipitated from OVCAR4 cells after siRNA-mediated knockdown (KD) of PARP7 and the subjected to western blotting for α-tubulin. The experiment was performed three times (n = 3) to ensure reproducibility. (C) Knockdown of PARP-7 promotes microtubule stability in OVCAR4 cells. Immunofluorescent staining of α-tubulin in OVCAR4 cells after siRNA-mediated knockdown (KD) of PARP7 and treatment with cold or nocodazole. Scale bars = 25 µm. The experiment was performed three times (n = 3) to ensure reproducibility. (D) Cold or nocodazole treatment increases MARylation of α-tubulin. HA-tagged α-tubulin was immunoprecipitated from OVCAR4 cells subjected to cold or nocodazole treatment using an HA antibody. The IP material was subjected to western blotting for MAR and HA, and the input material was subjected to western blotting for PARP-7 and HA. (E) Ectopic expression of PARP-7 enhances the MARylation of α-tubulin. Wild-type (Wt) or MAR-deficient mutant α-tubulin was immunoprecipitated from HEK 293 T cells after ectopic expression of PARP-7. The IP material was subjected to western blotting for MAR and HA, and the input material was subjected to western blotting for PARP-7 and α-tubulin. (F) Inhibition of α-tubulin MARylation promotes microtubule stability in OVCAR4 cells. Immunofluorescent staining of HA-epitope tagged wild-type or MARylation site mutant α-tubulin in OVCAR4 cells after treatment with cold or nocodazole. Scale bars = 100 µm. The experiment was performed three times (n = 3) to ensure reproducibility. (G) Quantification of α-tubulin staining from experiments like those shown in panel (F). Asterisks indicate significant differences from the control (n = 3; ANOVA, *p<0.05).
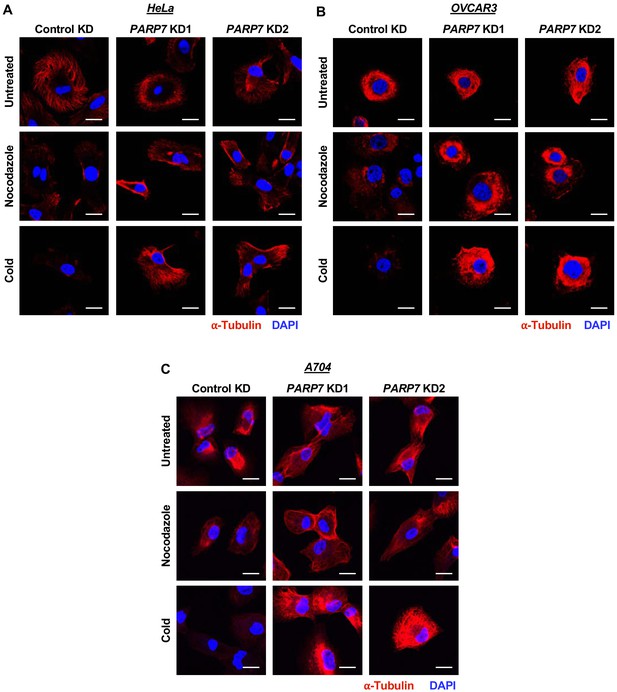
Effects of PARP7 knockdown on microtubule stability in HeLa, OVCAR3, and A704 cells.
Knockdown of PARP7 promotes microtubule stability. Immunofluorescent staining of α-Tubulin in HeLa (A), OVCAR3 (B), and A704 (C) cells after siRNA-mediated knockdown (KD) of PARP7 and treatment with cold or nocodazole. Scale bars = 25 µm. Each experiment was performed three times (n = 3) to ensure reproducibility.
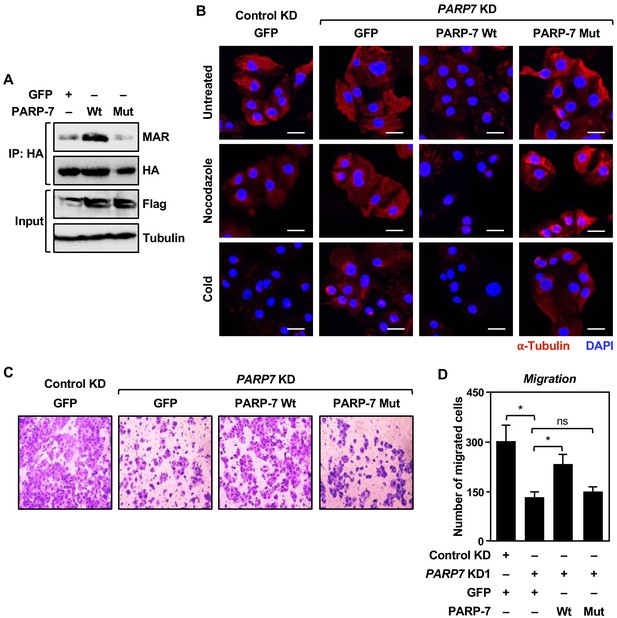
PARP-7 catalytic activity is required for MARylation of α-tubulin and functional outcomes.
(A) PARP-7 catalytic activity is required for MARylation of α-tubulin. HA-tagged α-tubulin was immunoprecipitated from OVCAR4 cells expressing wildtype or catalytic dead mutant PARP-7 using HA-antibody and subjected to western blotting for MAR. (B) PARP-7 catalytic activity is required for the control of microtubule stability in OVCAR4 cells. Immunofluorescent staining of α-tubulin in OVCAR4 cells subjected to ectopic expression of PARP-7 followed by siRNA-mediated knockdown (KD) of PARP7. Scale bars = 25 µm. The experiment was performed three times (n = 3) to ensure reproducibility. (C) PARP-7 catalytic activity is required for controlling cell motility. Cell migration assays for OVCAR4 cells subjected to ectopic expression of PARP-7 followed by siRNA-mediated knockdown (KD) of PARP7. (D) Quantification of cell migration assays. Asterisks indicate significant differences from the control (n = 3, Student’s t-test; **p<0.01).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | TIPARP (PARP7) | Genbank | Gene ID: 25976 | |
| Gene (Mus musculus) | Tiparp (Parp7) | Genbank | GeneID: 99929 | |
| Gene (Homo sapiens) | TUBA1A | Genbank | GeneID: 7846 | |
| Strain, strain background (Escherichia coli) | DH10BAC | ThermoFisher | Cat#10361012 | |
| Cell line (Homo sapiens) | OVCAR4 (female adult ovarian cancer) | ATCC | RRID:CVCL_1627 | |
| Cell line (Homo sapiens) | OVCAR3 (female adult ovarian cancer) | ATCC | RRID:CVCL_0465 | |
| Cell line (Homo sapiens) | HeLa (female adult cervical cancer) | ATCC | RRID:CVCL_0030 | |
| Cell line (Homo sapiens) | A704 (male adult kidney cancer) | ATCC | RRID:CVCL_1065 | Maintained and obtained from Dr. Laura Banaszynski, UT Southwestern |
| Cell line (Homo sapiens) | HEK 293T (normal embryonic kidney) | ATCC | RRID:CVCL_0063 | |
| Cell line (Spodoptera frugiperda) | Sf9 | ATCC | RRID:CVCL_0549 | |
| Transfected construct (Homo sapiens) | PARP7 siRNA #1 | Sigma-Aldrich | Cat# D-013948–08 | |
| Transfected construct (Homo sapiens) | PARP7 siRNA #2 | Sigma-Aldrich | Cat# D-013948–09 | |
| Transfected construct (Homo sapiens) | PARP7 siRNA #4 | Sigma-Aldrich | Cat# D-013948–07 | Our PARP7 siRNA #3 |
| Antibody | Anti-mono-ADP-ribose binding reagent (rabbit monoclonal; IgG Fc) | Millipore, MABE1076 | RRID:AB_2665469 | Gibson and Kraus, 2017 WB (8 µg/mL) IP (3 µg) |
| Antibody | PARP-7 (rabbit polyclonal) | Invitrogen, PA5-40774 | RRID:AB_2607074 | WB (1:500) |
| Antibody | α-tubulin (mouse monoclonal) | Santa Cruz, sc-8035 | RRID:AB_628408 | WB (1:1000) IF (1:1000) |
| Antibody | β-tubulin (rabbit polyclonal) | Abcam, ab6046 | RRID:AB_2210370 | WB (1:1000) |
| Antibody | SNRP70 (rabbit polyclonal) | Abcam, ab51266 | RRID:AB_10673827 | WB (1:1000) |
| Antibody | Flag (mouse monoclonal) | Sigma-Aldrich, F3165 | RRID:AB_259529 | WB (1:2000) |
| Antibody | HA (mouse monoclonal) | Sigma-Aldrich, H3663 | RRID:AB_262051 | WB (1:1000) IF (1:200) IP (1.5 µg) |
| Antibody | Rabbit IgG fraction (polyclonal) | Invitrogen, 10500C | RRID:AB_2532981 | IP (3 µg) |
| Antibody | HRP-conjugated anti-rabbit IgG (goat polyclonal) | Pierce, 31460 | RRID:AB_228341 | WB (1:5000) |
| Antibody | HRP-conjugated anti-mouse IgG (goat polyclonal) | Pierce, 31430 | RRID:AB_228307 | WB (1:5000) |
| Antibody | Alexa Fluor 488-conjugated anti-mouse IgG (goat polyclonal) | ThermoFisher, A-11001 | RRID:AB_2534069 | IF (1:500) |
| Recombinant DNA reagent | pFastBac1 | ThermoFisher | Cat# 10360014 | |
| Recombinant DNA reagent | pFastBac1-human wild-type PARP-7 | This study | ||
| Recombinant DNA reagent | pFastBac1-human analog sensitive PARP-7 | This study | ||
| Recombinant DNA reagent | pcDNA3.1 | ThermoFisher | Cat# V79020 | |
| Recombinant DNA reagent | pcDNA3.1-mouse wild-type PARP-7 | This study | ||
| Recombinant DNA reagent | pcDNA3.1-mouse catalytic mutant PARP-7 | This study | ||
| Recombinant DNA reagent | pINDUCER20 | Addgene | Plasmid no. 44012 | |
| Recombinant DNA reagent | pINDUCER20-mouse wild-type PARP-7 | This study | ||
| Recombinant DNA reagent | pINDUCER20-mouse catalytic mutant PARP-7 | This study | ||
| Recombinant DNA reagent | pCMV-VSV-G | Addgene | Plasmid no. 8454 | |
| Recombinant DNA reagent | pAd-VAntage | Promega | Cat# TB207 | |
| Recombinant DNA reagent | psPAX2 | Addgene | Plasmid no. 12260 | |
| Recombinant DNA reagent | pCMV3-C-HA-human wild-type-α-tubulin | Sino Biologicals | Cat# HG14201-CY | |
| Recombinant DNA reagent | pCMV3-C-HA-human MARylation site mutant-α-tubulin | This study | ||
| Commercial assay or kit | Bac-to-Bac Baculovirus Expression System | ThermoFisher | Cat# 10359016 | |
| Commercial assay or kit | RNAeasy Plus Mini kit | Qiagen | Cat# 74136 | |
| Chemical compound, drug | 8-Bu (3-yne)T-NAD+ | BIOLOG Life Science Institute | Cat# N 055 | |
| Software, algorithm | FastQC | Babraham Bioinformatics | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | |
| Software, algorithm | Tophat | Trapnell et al., 2010 | https://ccb.jhu.edu/software/tophat/index.shtml | |
| Software, algorithm | Cufflinks | Trapnell et al., 2010; Trapnell et al., 2012 | http://cole-trapnell-lab.github.io/cufflinks/ | |
| Software, algorithm | DAVID Bioinform-atics Resources | LHRI Huang et al., 2009a; Huang et al., 2009b | https://david.ncifcrf.gov/home.jsp | |
| Software, algorithm | Java TreeView | Saldanha, 2004 | http://jtreeview.sourceforge.net/ | |
| Software, algorithm | GTEx Analysis Release v. 8 | The Genotype-Tissue Expression (GTEx) project | https://gtexportal.org/home/ |
Additional files
-
Supplementary file 1
RNA-seq analysis of gene expression in ovarian cancer cells following PARP-7 depletion.
RNA-seq was performed after siRNA-mediated knockdown (KD) of PARP7 in OVCAR4 cells. Three different siRNAs were used versus a control siRNA. The table lists the expression values in FPKM obtained using the Cufflinks pipeline described in the Methods for all the genes across all the samples for the RNA-seq data.
- https://cdn.elifesciences.org/articles/60481/elife-60481-supp1-v2.xlsx
-
Supplementary file 2
NAD+ analogs used in this study.
The synthesis of many of these NAD+ analogs was reported in Gibson et al., 2016. Most of the compounds can be purchased from the BIOLOG Life Science Institute (https://www.biolog.de). a The number assigned to the NAD+ analog for this study. b Abbreviation assigned by BIOLOG Life Science Institute and used herein. For the clickable analogs: alkyne = contains an alkyne group, or azide = contains an azide group for use in copper-catalyzed alkyne-azide cycloaddition (‘click’) reactions. c BIOLOG Life Science Institute catalog number.
- https://cdn.elifesciences.org/articles/60481/elife-60481-supp2-v2.docx
-
Supplementary file 3
PARP-7 protein substrates from OVCAR4 and HeLa cells identified using an asPARP-7 approach with 8-Bu(3-yne)T-NAD+.
Cytosolic extracts prepared from OVCAR4 and HeLa cells were incubated with purified asPARP-7 S563G and 8-Bu(3-yne)T-NAD+. The 8-Bu(3-yne)T-ADP-ribosylated proteins from the reactions were covalently linked to azido-agarose beads via copper-catalyzed cycloaddition. The conjugated azido-agarose beads were washed extensive and trypsinized to release peptides for protein identification by LC-MS/MS analysis. The ‘Column Heading’ key provides annotation information describing the metrics of each of the LC-MS/MS-identified peptides. The ‘Tab’ key provides details about all of the other worksheets contained within this spreadsheet.
- https://cdn.elifesciences.org/articles/60481/elife-60481-supp3-v2.xlsx
-
Supplementary file 4
Sequences of tryptic peptides from the asPARP-7 protein identifications.
Sequences of the trypsinized peptides from the asPARP-7 protein identifications shown in Supplementary file 3.
- https://cdn.elifesciences.org/articles/60481/elife-60481-supp4-v2.xlsx
-
Supplementary file 5
Identification of PARP-7-mediated sites of ADPRylation on α-tubulin using the NAD+ analog-sensitive approach.
OVCAR4 and HeLa cell cytosolic extracts were incubated with recombinant analog sensitive PARP-7 (asPARP-7) in the presence of 8-Bu(3-yne)T-NAD+. Following in vitro modification, the extract proteins were covalently linked to azido-agarose beads via copper-catalyzed cycloaddition. The conjugated beads were washed, trypsinized to release peptides for protein identification (Supplementary files 3 and 4), and then washed again. The remaining peptides containing ADP-ribosylation sites were eluted from the resin using hydroxylamine (NH2OH). The cleaved modification produces a 15.019 m/z shift identifying the specific site of glutamate or aspartate modification. Both the tryptic digest (PeptideID) and hydroxylamine eluate (SiteID) were subjected to LC-MS/MS analysis. The ‘Summary’ worksheet in this spreadsheet provides a summary of all data identifying sites of PARP-7-mediated ADPRylation on α-tubulin. The ‘Data’ worksheet provides mass spec data and annotation information describing the metrics of each of the LC-MS/MS-identified peptides. All analyses and numbering of amino acids are for human α-tubulin.
- https://cdn.elifesciences.org/articles/60481/elife-60481-supp5-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60481/elife-60481-transrepform-v2.pdf





