The Arabidopsis V-ATPase is localized to the TGN/EE via a seed plant-specific motif
Figures
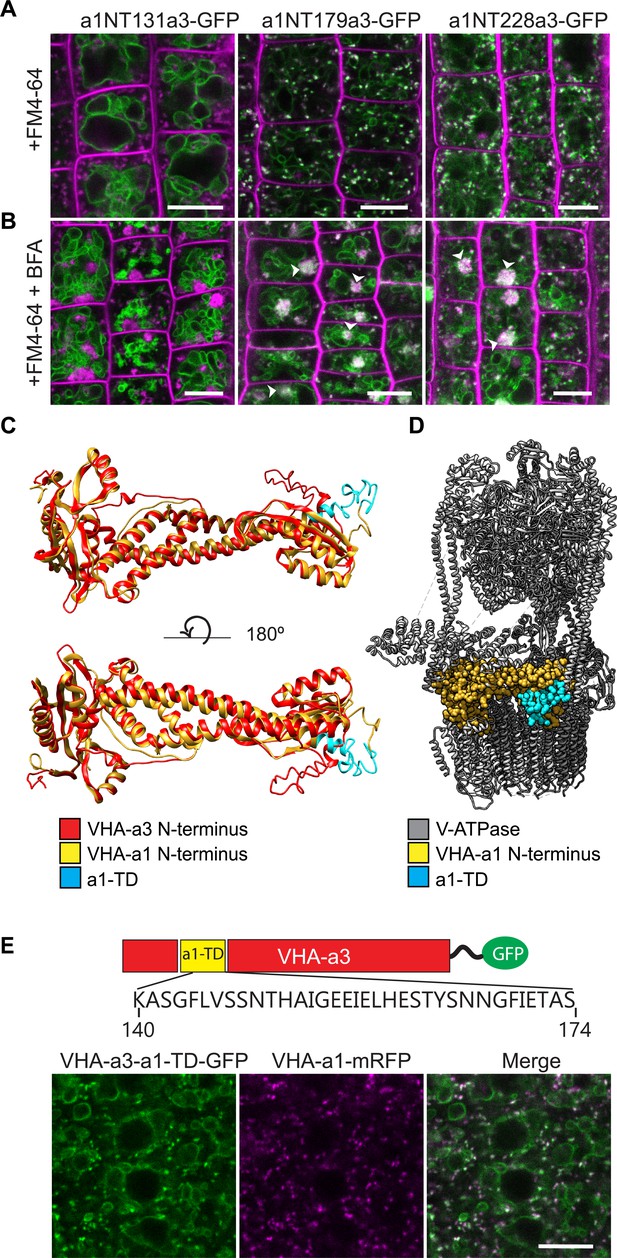
The targeting signal of VHA‐a1 is located between L132 and E179 and the a1-TD is sufficient for targeting of VHA‐a3 to the TGN/EE.
Root tips of 6-day-old Arabidopsis seedlings were analyzed by confocal laser scanning microscopy (CLSM) (A) The chimeric constructs a1NT179a3-GFP and a1NT228a3-GFP show dual localization at the TGN/EE and tonoplast. (B) TGN/EE localization of a1NT179a3-GFP and a1NT228a3-GFP was confirmed by treatment of Arabidopsis root cells with 50 µM BFA for 3 hr followed by staining with FM4-64 for 20 min. Scale bars = 10 µm. Green and magenta pseudo colors indicate fluorescence from GFP and FM4-64 respectively. (C) One of the structural differences between the VHA-a1 NT and VHA-a3 NT corresponds to the location of the a1-TD (blue). (D) An alignment of the VHA-a1 NT model with a model of one of the rotational states of the yeast V-ATPase (PDB = 6O7V) revealed that the VHA‐a1 targeting domain is exposed and accessible for recognition. (E) VHA-a3 with the a1-TD (VHA‐a3-a1TD‐GFP) partially colocalizes with VHA‐a1‐mRFP. Scale bar = 10 µm.
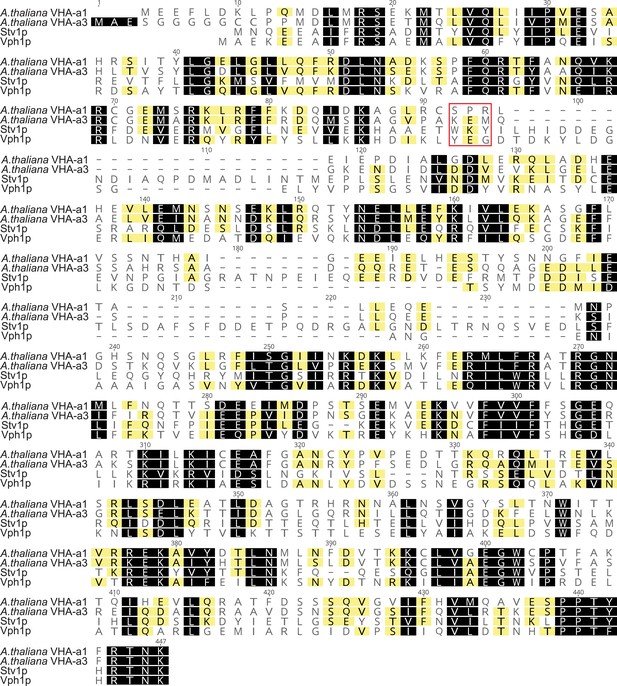
The tri-peptide motif that is responsible for the targeting of Stv1p is absent in VHA-a1.
Amino acid sequence alignment of the yeast subunit a isoforms Vph1p and Stv1p with the Arabidopsis isoforms VHA-a1 and VHA-a3. Residues similar between all proteins at the same position are shown against a black background. Residues similar between only three of the proteins at the same position are shown against a yellow background. Un-highlighted residues have no similarity in all four sequences at the same position. The position of the tri-peptide motif that is responsible for the targeting of Stv1p is indicated with a red box.
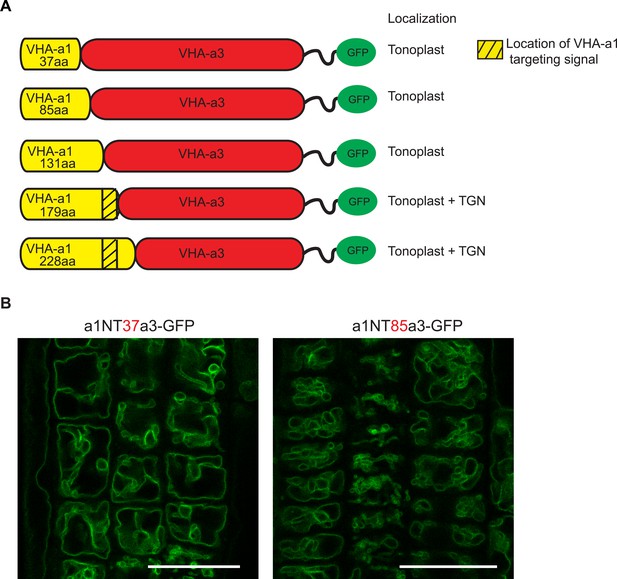
The targeting signal of VHA‐a1 is not located in the first 85 amino acids.
(A) Chimeric proteins were made which consisted of increasing lengths of the VHA‐a1 N‐terminus fused to decreasing lengths of the C‐terminal domain of VHA‐a3. All constructs were fused to GFP. (B) Root tips of 6-day-old Arabidopsis seedlings were analyzed by CLSM. The first two chimeric constructs consisting of 37 aa and 85 aa of the VHA-a1 N-terminal domain localized at the tonoplast. Scale bars = 25 µm.
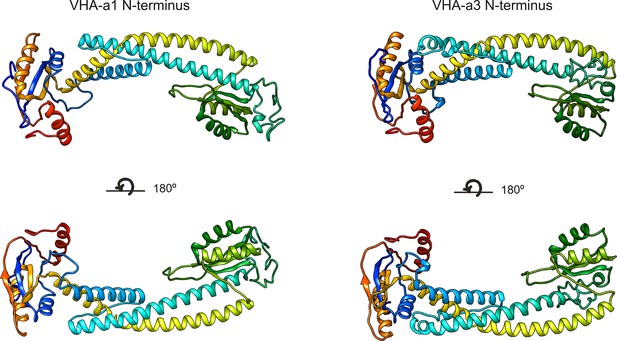
Three-dimensional models of the VHA-a1 and VHA-a3 N-termini.
Homology modeling of the N- termini of VHA-a1 and VHA-a3 was done using cryo-EM models of Stv1p (PDB607U) and Vph1p (PDB607T) respectively as templates. The models are color ramped.
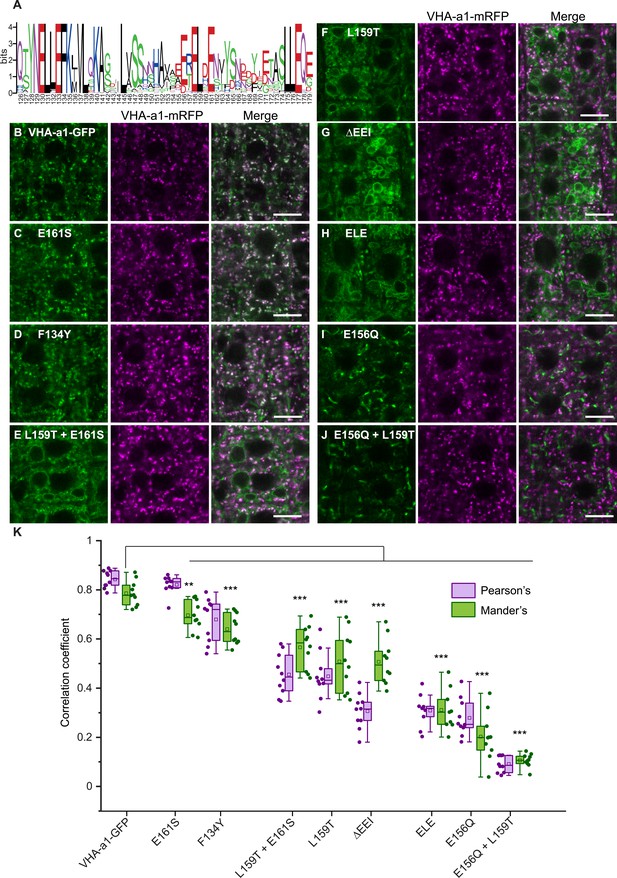
Site-directed mutagenesis reveals the importance of conserved amino acids in the targeting of VHA‐a1 to the TGN/EE.
(A) The VHA-a1-clade consensus sequence for the a1-TD region was made on the weblogo platform (Crooks et al., 2004). Sequence numbers are based on Arabidopsis VHA-a1. Conserved amino acids were mutated and their effect on the localization of GFP tagged VHA-a1 was analyzed in the VHA-a1-mRFP background. Root tips of 6-day-old Arabidopsis seedlings were analyzed by CLSM. (B) VHA-a1-GFP was co-expressed with VHA-a1-mRFP as a control. The mutations in VHA-a1 produced three classes of punctate patterns that could be classified as TGN/EE only (C and D), TGN/EE and tonoplast (E, F, G and H) and different from VHA-a1-mRFP (I and J) Scale bars = 10 µm. Green and magenta pseudo colors indicate fluorescence from GFP and VHA-a1-mRFP respectively. (K) Mutated VHA-a1-GFP proteins colocalization with VHA-a1-mRFP as shown by Pearson’s and Mander’s coefficients. The Mander’s coefficient indicates the fraction of mutated VHA-a-GFP in the VHA-a1-mRFP location. Box plot center lines, medians; center boxes, means with n = 9–10 ratios calculated from 9 to 10 images; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 interquartile range. Asterisks indicate significant differences of the mean Mander’s correlation coefficients between mutated VHA-a1-GFP and VHA-a1-GFP (Two-sample t-Test, p<0.05) (*p<0.05, **p<0.01 and ***p<0.001).
-
Figure 2—source data 1
Source data for Figure 2K.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig2-data1-v2.xlsx
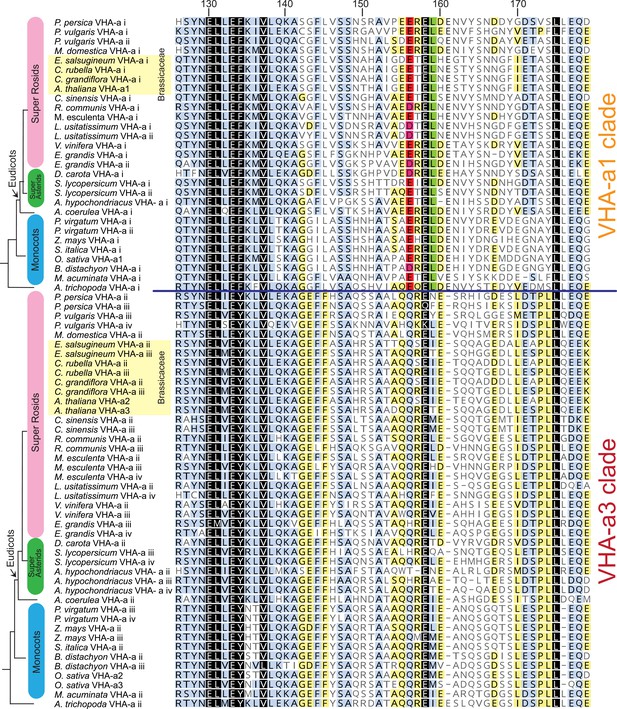
The VHA-a1-TD is conserved in Angiosperms.
Amino acid sequence alignment of representative sequences from the VHA-a1 and VHA-a3 clades. The sequence numbers are in reference to the A. thaliana VHA-a1 sequence. Residues highlighted in black, blue and yellow are 100%, 80–100% and 60–80% similar, respectively. Un-highlighted residues are less than 60% similar. The alignment is restricted to the VHA‐a1 targeting domain (His126 to Glu 179 in A. thaliana VHA-a1). The alignment reveals that there is an acidic cluster that is only present in the VHA-a1 clade.
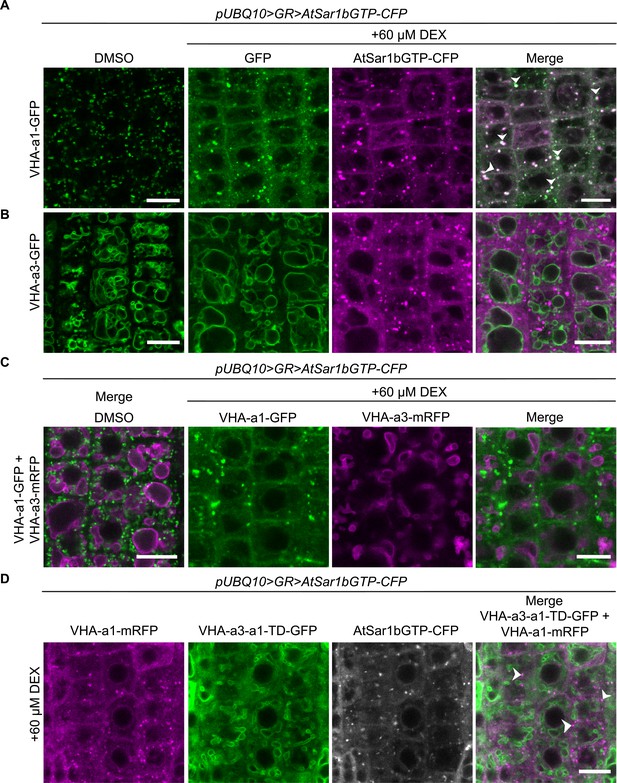
VHA-a1 is retained at the ER after AtSar1b-GTP-CFP expression.
After 6 hr of induction with 60 µM DEX, AtSar1b-GTP-CFP is expressed in 6-day-old Arabidopsis root tip cells. (A) VHA-a1-GFP is retained in the ER and also agglomerates to produce bright punctae which colocalize with AtSar1b-GTP-CFP (white arrows). For the DMSO control, only the GFP channel is shown. (B) VHA-a3-GFP does not accumulate when exit from the ER via COPII vesicles is blocked by expression of AtSar1b-GTP-CFP. For the DMSO control, only the GFP channel is shown. (C) When VHA-a1 and VHA-a3 are co-expressed in the same cell only VHA-a1 accumulates in the ER upon induction of AtSar1b-GTP-CFP. (D) VHA-a3-a1-TD-GFP is partially retained in the ER after induction of AtSar1b-GTP-CFP. Arrows point to VHA-a1-mRFP aggregates. Scale bars = 10 µm.
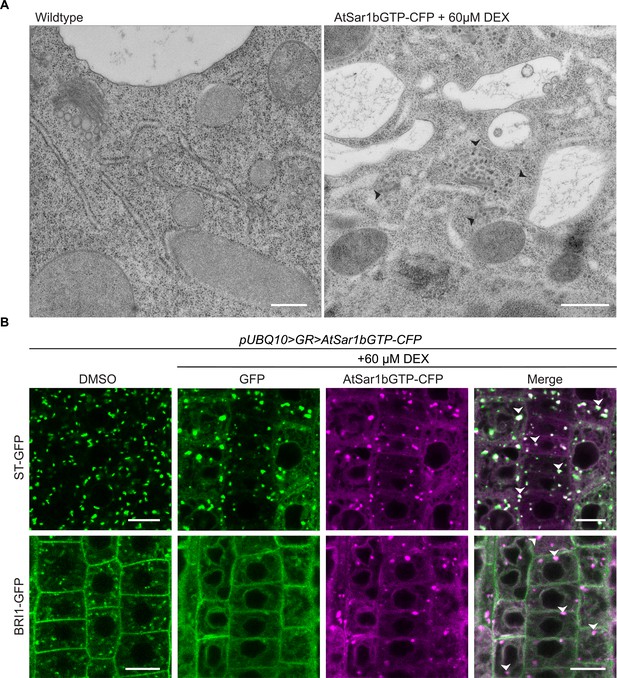
AtSar1b-GTP-CFP expression blocks the ER exit of secretory pathway proteins.
(A) EM of high-pressure frozen Arabidopsis root tips. After 6 hr of induction with 60 µM DEX, AtSar1b-GTP-CFP expression causes bloating of the ER, aggregation of Golgi stacks and leads to an accumulation of vesicles in the cell (black arrows). Scale bars = 500 nm. (B) After 6 hr of induction with 60 µM DEX, AtSar1b-GTP-CFP is expressed. Both the Golgi targeted protein; ST-GFP and plasma membrane destined BRI1-GFP are retained at the ER when exit from the ER via COPII vesicles is blocked by expression of AtSar1b-GTP-CFP. Green and magenta pseudo colors indicate fluorescence from GFP and CFP respectively. Scale bars = 10 µm.
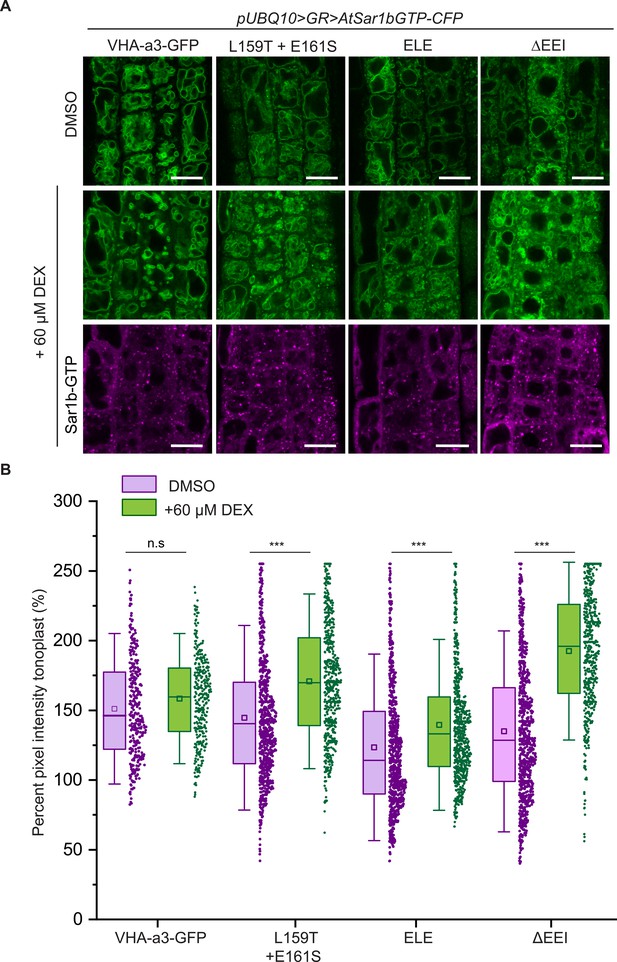
The mutations in the a1-TD affect an ER-exit motif.
The localization and fluorescence intensity of mutated VHA-a1-GFP proteins was analyzed in the presence (+DEX) and absence (DMSO) of AtSar1b-GTP-CFP. Root tips of 6-day-old Arabidopsis seedlings were used for the analysis. (A) The mutated VHA-a1 proteins still localized at the tonoplast when exit from the ER via COPII vesicles was blocked by expression of AtSar1b-GTP-CFP. Green and magenta pseudo colors indicate fluorescence from GFP and CFP respectively Scale bars = 15 µm. (B) The tonoplast fluorescence intensity was measured in the presence and absence of DEX. There is a significant increase in the GFP fluorescence intensity at the tonoplast of mutated VHA-a1 proteins when exit from the ER via COPII vesicles is blocked by expression of AtSar1b-GTP-CFP. Box plot center lines, medians; center boxes, means with n ≥ 319 measurements; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 SD. Asterisks indicate significant differences between the uninduced and induced conditions (Mann-Whitney test, p<0.001). (n.s: not significant,*p<0.05, **p<0.01 and ***p<0.001).
-
Figure 4—source data 1
Source data for Figure 4B.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig4-data1-v2.xlsx
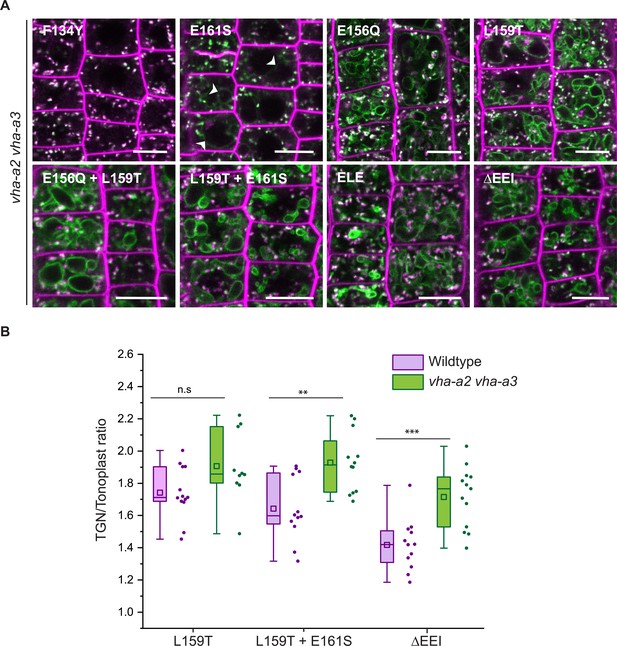
Tonoplast localization of mutated VHA-a1 increases in the absence of VHA-a2 and VHA-a3.
(A) The localization of mutated VHA-a1 proteins tagged to GFP were analyzed in the vha-a2 vha-a3 double mutant background after 20 min staining with FM4-64. Root tips of 6-day-old Arabidopsis seedlings were used for the analysis. Scale bars = 10 µm. Green and magenta pseudo colors indicate fluorescence from GFP and FM4-64 respectively. (B) TGN/EE-to-tonoplast fluorescence intensity ratios were calculated for selected mutations in the wildtype background and in the vha-a2 vha-a3 double mutant background. The ratio of TGN/EE-to-tonoplast fluorescence intensity of the mutated VHA-a1 proteins (L159T + E161S and ΔEEI) is significantly higher in the vha-a2 vha-a3 double mutant as compared to the wildtype background. Box plot center lines, medians; center boxes, means with n ≥ 10 ratios calculated from individual images; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 interquartile range. Asterisks indicate significant differences between the wildtype and vha-a2 vha-a3 double mutant mean ratios (Two-sample t-Test, p<0.05). (n.s: not significant, *p<0.05, **p<0.01 and ***p<0.001).
-
Figure 5—source data 1
Source data for Figure 5B.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig5-data1-v2.xlsx
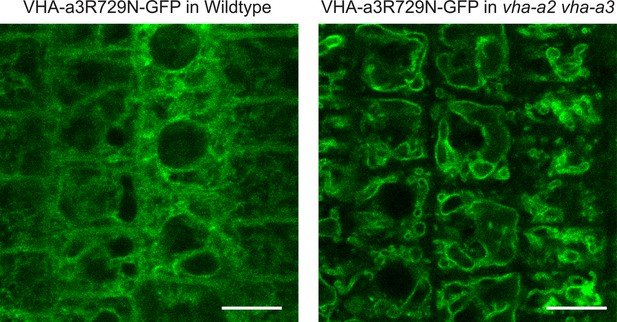
A competition exists to enter provacuoles in wild type Arabidopsis root tip cells.
VHA-a3-R729N-GFP is retained in the wild type background and localizes to the tonoplast in the vha-a2 vha-a3 double mutant background. Scale bars = 10 µm.
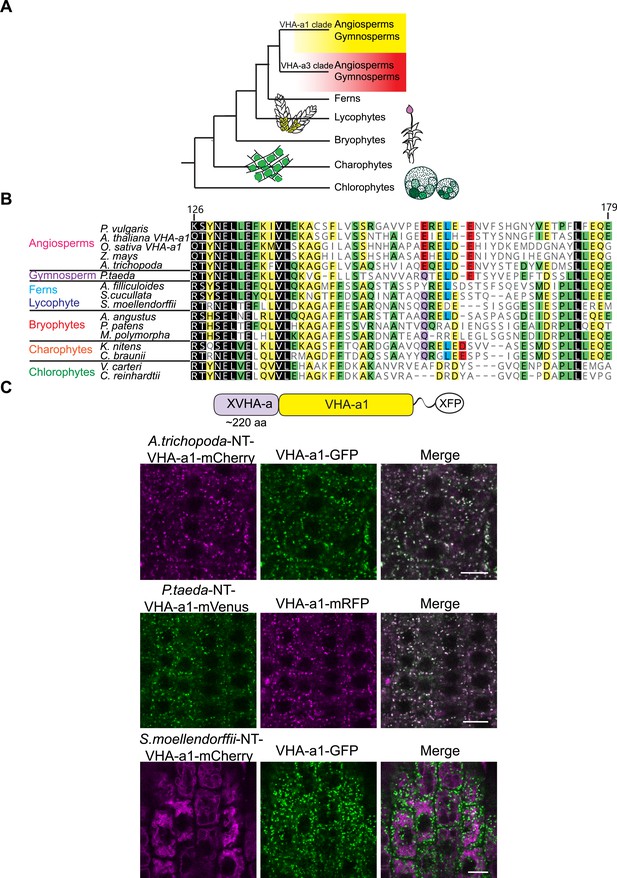
Spermatophyte VHA-a isoforms cluster into two distinct clades, the a1-TD is conserved in the VHA-a1 clade and originates in the gymnosperm sequences.
(A) Phylogenetic analysis of the N-terminal sequences of VHA-a proteins was done. A graphical summary of the tree is depicted. VHA-a isoforms from seed plants including the gymnosperm Pinus taeda cluster into two distinct clades. (B) The a1-TD originates in the gymnosperms and it is absent from the chlorophytes, bryophytes, lycophytes and pteridophytes. The sequence of the gymnosperm, P. taeda and that of the charophytes contain all the amino acids thought to comprise the ER-exit signal with the exception of one flanking glutamic acid residue. The sequence numbers are in reference to the A. thaliana VHA-a1 sequence. (C) The a1-TD is functionally conserved in gymnosperms and angiosperms. Root tips of 6-day-old Arabidopsis seedlings were analyzed by CLSM. Pine and Amborella chimeric proteins colocalized with VHA-a1 at the TGN/EE. The Selaginella chimeric protein localized to the tonoplast only. Scale bars = 10 µm.
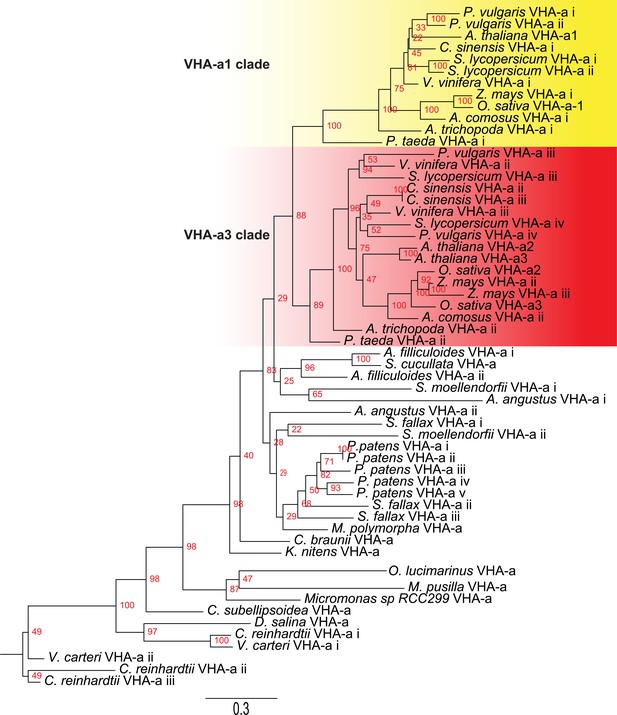
Phylogenetic analysis of the N-terminal sequences of VHA-a related proteins.
VHA-a related protein sequences for selected species are shown. Branch support is calculated on the basis of 500 bootstraps.
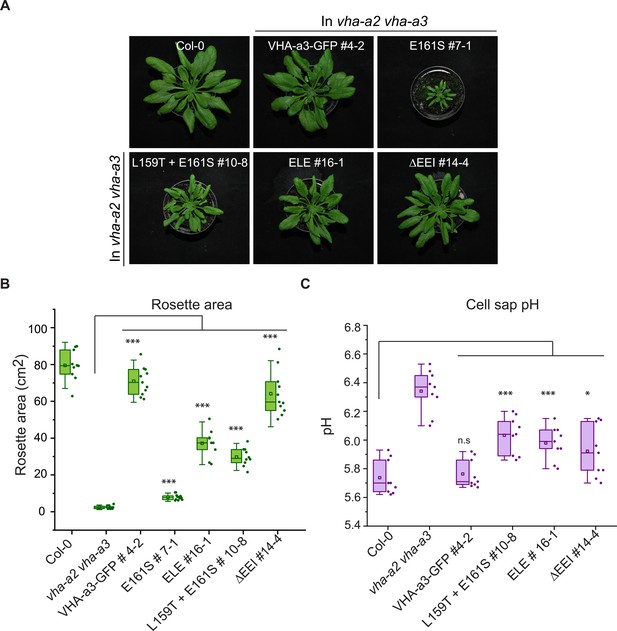
The mutated VHA-a1 subunits complement the vha-a2 vha-a3 double mutant to varying degrees.
Plants were grown in short day conditions (22 °C and 10 hr light) for 6 weeks. (A) All mutant variants of VHA-a1-GFP displayed bigger rosette size than the vha-a2 vha-a3 double mutant. E161S which had a faint signal at the tonoplast in the vha-a2 vha-a3 background also partially complemented the dwarf phenotype of the vha-a2 vha-a3 double mutant. (B) Rosette area of 6-week-old plants grown under short day conditions. VHA-a1-GFP with ΔEEI mutation complements the vha-a2 vha-a3 double mutant the best. (C) Mutated VHA-a1 containing V-ATPases have more alkaline cell sap pH values. Box plots center lines, medians; center boxes, means with n ≥ 9 measurements; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 interquartile range. Asterisks indicate significant differences in the mean rosette area and cell sap pH. (Two-sample t-Test, p<0.05). (n.s: not significant, *p<0.05, **p<0.01 and ***p<0.001).
-
Figure 7—source data 1
Source data for Figure 7B and C.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig7-data1-v2.xlsx
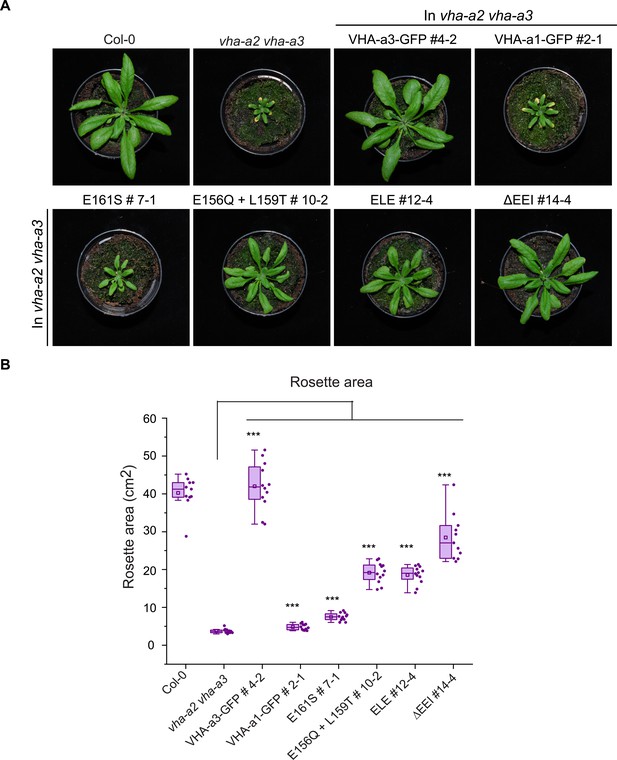
The mutated VHA-a1-GFP proteins complement the vha-a2 vha-a3 double mutant to varying degrees in long day conditions.
Plants were grown in long day conditions (22 °C and 16 hr of light) for 4 weeks. All mutant variants of VHA-a1-GFP displayed bigger rosette size than the vha-a2 vha-a3 double mutant. VHA-a1 with the ΔEEI mutation (E155+ E156 + I157 deletion) complements the vha-a2 vha-a3 double mutant the best. Box plot center lines, medians; center boxes, means with n ≥ 11; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 interquartile range. Asterisks indicate significant differences in the mean rosette area from the vha-a2 vha-a3 double mutant mean rosette area. (Two-sample t-Test, p<0.05). (*p<0.05, **p<0.01 and ***p<0.001).
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1B.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig7-figsupp1-data1-v2.xlsx
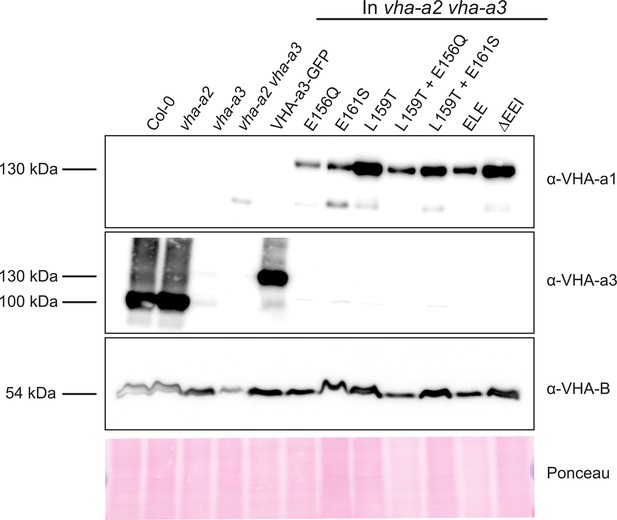
Protein levels of the mutated VHA-a proteins at the tonoplast.
Abundance of the GFP tagged proteins was determined via western blot. Tonoplast membrane proteins were separated by SDS-PAGE and subsequently immunoblotted with an anti-VHA-a1, VHA-a3 and VHA-B antibodies. Protein loading is indicated by Ponceau staining of the membrane.
-
Figure 7—figure supplement 2—source data 1
Source data for Figure 7—figure supplement 2.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig7-figsupp2-data1-v2.zip
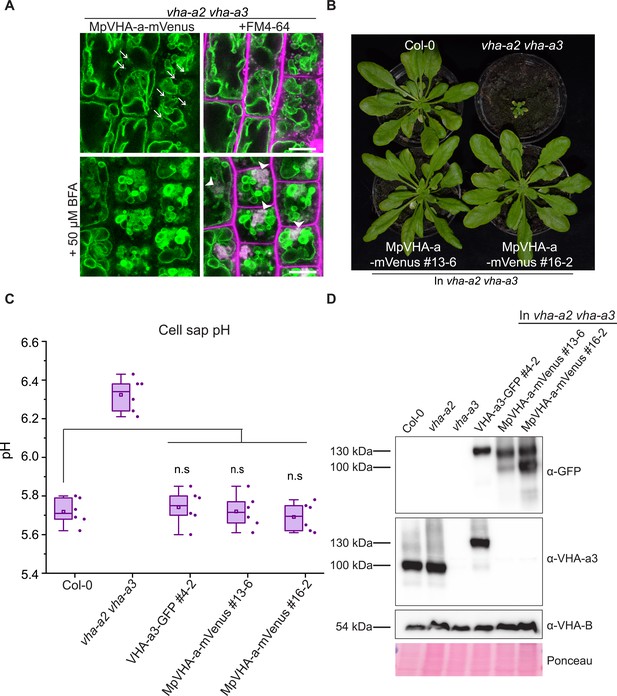
MpVHA-a containing complexes are functional in Arabidopsis and can replace VHA-a3 and VHA-a2 at the tonoplast.
(A) MpVHA-a-mVenus is dual localized at the tonoplast and TGN/EE in the vha-a2 vha-a3 background. The TGN/EE localization was confirmed by treatment of root cells with 50 µm BFA for 3 hr followed by staining with FM4-64 for 20 min. The core of BFA compartments were labeled with MpVHA-a-mVenus and FM4-64. Root tips of 6-day-old Arabidopsis seedlings were analyzed by CLSM. Green and magenta pseudo colors indicate fluorescence from GFP and FM4-64 respectively. Scale bars = 10 µm. (B) MpVHA-a-mVenus can also fully complement the dwarf phenotype of the vha-a2 vha-a3 double mutant. Plants for complementation assay were grown in SD conditions (22°C and 10 hr of light) for 6 weeks. (C) Cell sap pH of rosette leaves from plants grown in LD conditions (22°C and 16 hr of light) for 3 weeks. MpVHA-a-mVenus complexes restore the cell sap pH of the vha-a2 vha-a3 double mutant to wildtype levels. Box plot center lines, medians; center boxes, means with n = 6 measurements from two biological replicates; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 interquartile range. n.s indicates that there is no significant difference in the mean cell sap pH between the complementation lines and wildtype. (Two-sample t-Test, p<0.05). (D) Western blot of tonoplast membrane proteins from 6-week-old rosettes. Protein levels in the MpVHA-a-mVenus complementation lines is comparable to the VHA-a3-GFP complementation line.
-
Figure 8—source data 1
Source data for Figure 8C.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig8-data1-v2.xlsx
-
Figure 8—source data 2
Source data for Figure 8D.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig8-data2-v2.zip
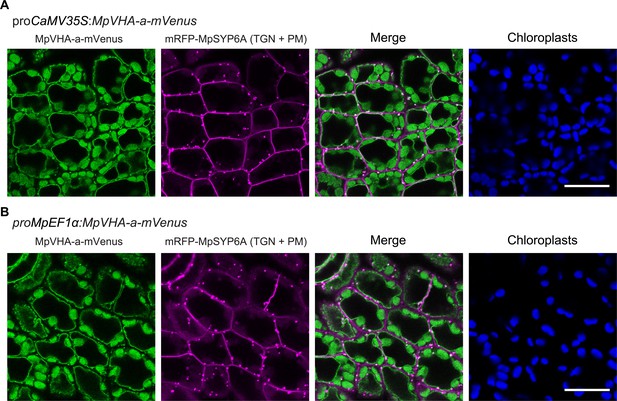
Subcellular localization of MpVHA-a-mVenus in M. polymorpha.
(A and B) Single confocal images of M. polymorpha dorsal thallus cells co-expressing mRFP-MpSYP6A and MpVHA-a-mVenus driven by the CaMV35S (A) or MpEF1α (B) promoter. Green, magenta, and blue pseudo colors indicate fluorescence from mVenus, mRFP, and chlorophyll, respectively. Scale bars = 10 μm.
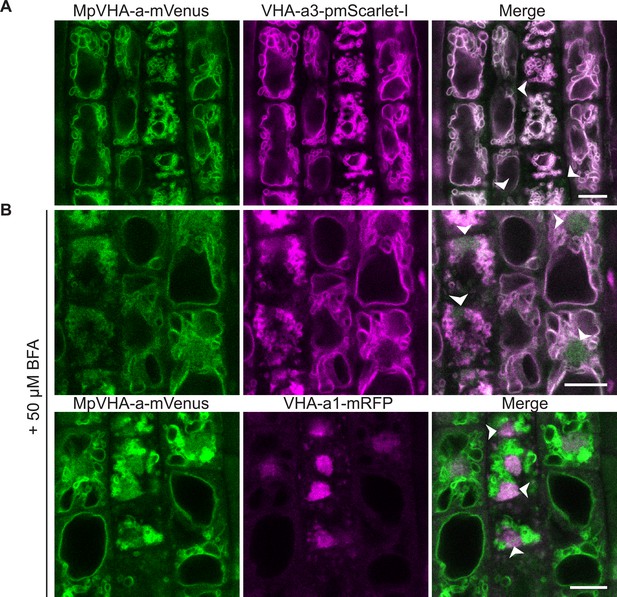
MpVHA-a-mVenus is dual localized at the TGN/EE and tonoplast in Arabidopsis wildtype root cells.
UBQ10:MpVHA-a-mVenus was co-expressed with UBQ10:VHA-a3-pmScarlet-I and VHA-a1:VHA-a1-mRFP in Arabidopsis wildtype. Confocal analysis was done on 6-day-old Arabidopsis roots. (A) MpVHA-a-mvenus predominantly co-localizes with VHA-a3-pmScarlet-I at the tonoplast. (B) TGN localization was confirmed by treatment of root cells with 50 µm BFA for 3 hr. The core of BFA compartments were labeled with VHA-a1-mRFP and MpVHA-mvenus.Gaussian blur with kernel size three was applied to all images. Green and magenta pseudo colors indicate fluorescence from mVenus, mRFP and pmScarlet-I respectively. Scale bars = 10 µm.
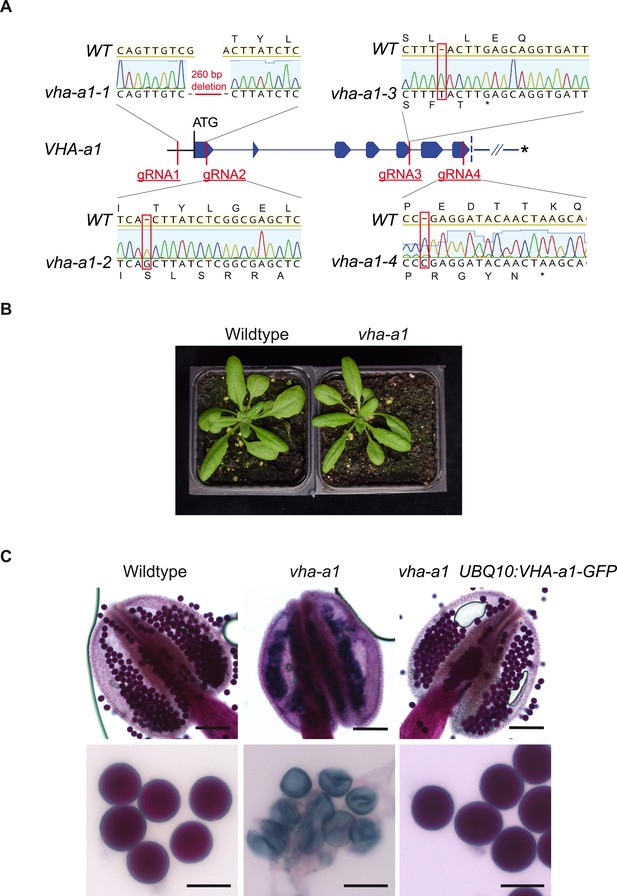
VHA-a1 is essential for pollen development but not for vegetative growth.
(A) The exon-intron structure of the first seven exons of VHA-a1 shows the sites which were targeted in independent CRISPR approaches. vha-a1-1, vha-a1-2, vha-a1-3 and vha-a1-4 are examples for vha–a1 alleles that were obtained. (B) The vha-a1 mutant is indistinguishable from wild type during vegetative growth. Plants were grown under long day conditions (3.5 weeks, 22°C and 16 hr of light). (C) Misshaped microspores/pollen in vha-a1 anthers were visualized using Alexander’s stain. VHA-a1-GFP rescues the pollen phenotype of vha-a1. VHA-a1-GFP is expressed under the UBQ10 promoter. Scale bars, whole anthers: 100 μm, close ups of pollen grains: 20 μm.
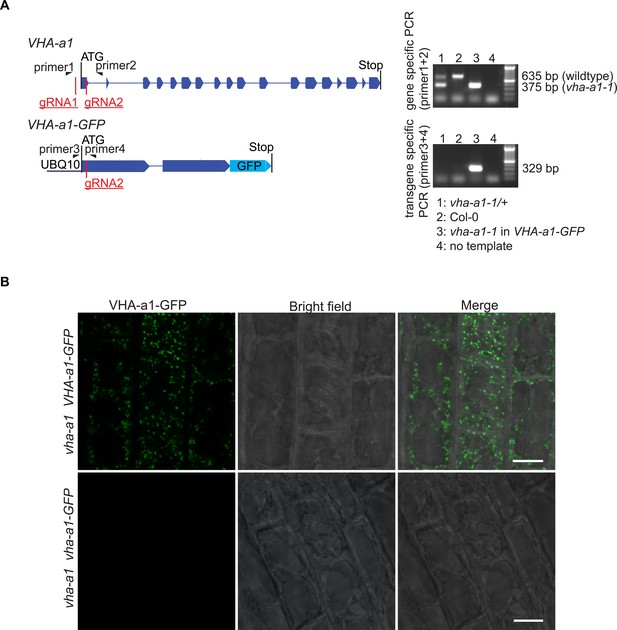
vha-a1-1 can be distinguished from the wildtype allele without sequencing and mutations in VHA-a1-GFP lead to absence of GFP signal.
(A) Mutations in VHA-a1 were distinguished from mutations in VHA-a1-GFP by use of specific primers. Shown are the exon-intron structures of gene and transgene. Primer one is VHA-a1 specific, while primer three is VHA-a1-GFP specific. On an agarose gel, PCR products from vha-a1-1 (260 bp deleted) could be distinguished from PCR products from the wildtype allele. (B) Root cells were analyzed by CLSM. In plants with mutations in UBQ10:VHA-a1-GFP corresponding to vha-a1-2 (+1 bp at CRISPR site two leading to frameshift and early stop codon) no GFP signal was detected suggesting that VHA-a1-GFP was absent. Scale bars = 10 µm.
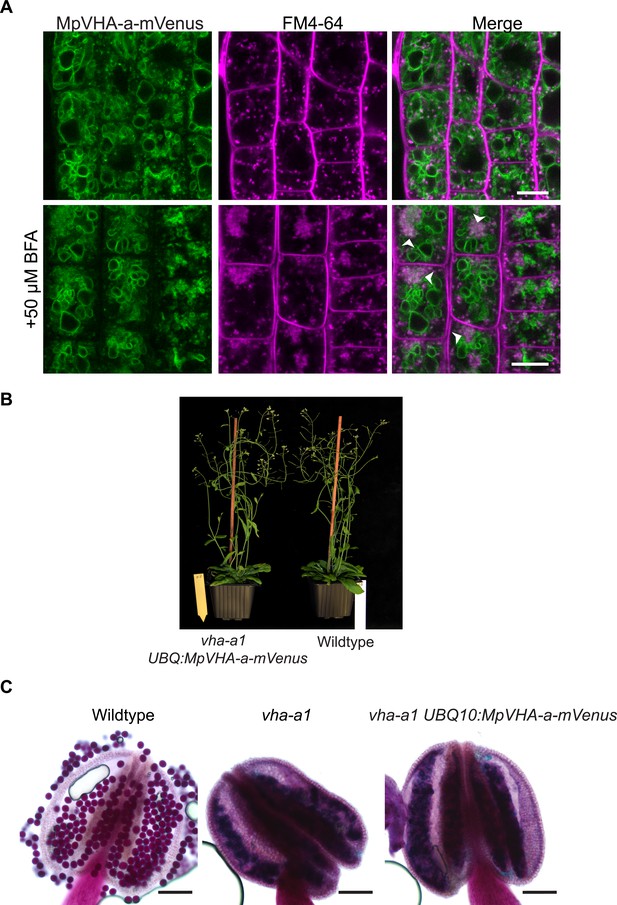
MpVHA-a-mVenus is dual localized in vha-a1.
(A) vha-a1 (Cas9+) was crossed with UBQ10:MpVHA-a-mVenus in wildtype background and F1 seedlings were analyzed by CLSM. Root tips of 6-day-old seedlings were analyzed. MpVHA-a-mVenus was dual localized at the TGN/EE and tonoplast in plants that were subsequently identified as vha-a1 mutants. Green and magenta pseudo colors indicate fluorescence from mVenus and FM4-64. Scale bars = 10 µm. (B) vha-a1 mutants expressing MpVHA-a-mVenus are comparable to wild type during vegetative growth. (C) MpVHA-a-mVenus does not rescue the pollen phenotype of vha-a1. Alexander’s stain revealed misshaped pollen in vha-a1 mutants expressing MpVHA-a-mVenus. MpVHA-a-mVenus is expressed under the UBQ10 promoter. Scale bars = 100 µm.
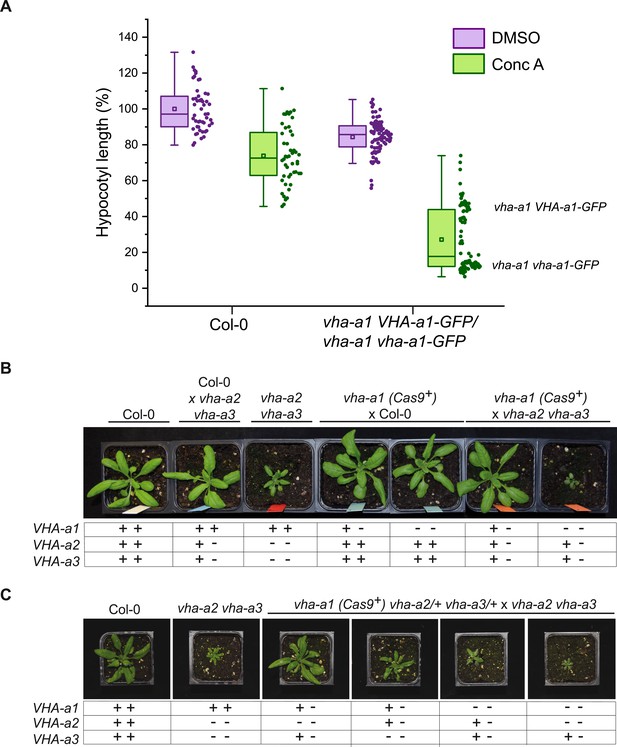
VHA-a2 and VHA-a3 replace VHA-a1 at the TGN/EE in vha-a1 during vegetative growth.
(A) vha-a1 vha-a1-GFP hypocotyls are hypersensitive to 125 nM ConcA, in contrast to wildtype hypocotyls indicating that a target of ConcA is present at the TGN/EE. vha-a1 hypocotyls expressing UBQ10:VHA-a1-GFP are less sensitive to ConcA. Box plot center lines, medians; center boxes, means with n ≥ 48 measurements from three biological replicates; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 interquartile range. (B) vha-a1 (Cas9+) was crossed with the vha-a2 vha-a3 double mutant. Analysis of F1 plants revealed that vha-a1 vha-a2/+ vha-a3/+ is reduced in growth. 3.5 week-old-plants are shown. (C) vha-a1 (Cas9+) vha-a2/+ vha-a3/+ was crossed with the vha–a2 vha–a3 double mutant. vha-a1/+ vha-a2/+ vha-a3 is smaller than vha-a1/+ vha-a2 vha-a3/+ and vha–a1 vha–a2 vha–a3/+ was the smallest mutant found. 4 week-old-plants are shown. Plants were grown under long day conditions (22°C and 16 hr of light).
-
Figure 10—source data 1
Source data for Figure 10A.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig10-data1-v2.xlsx
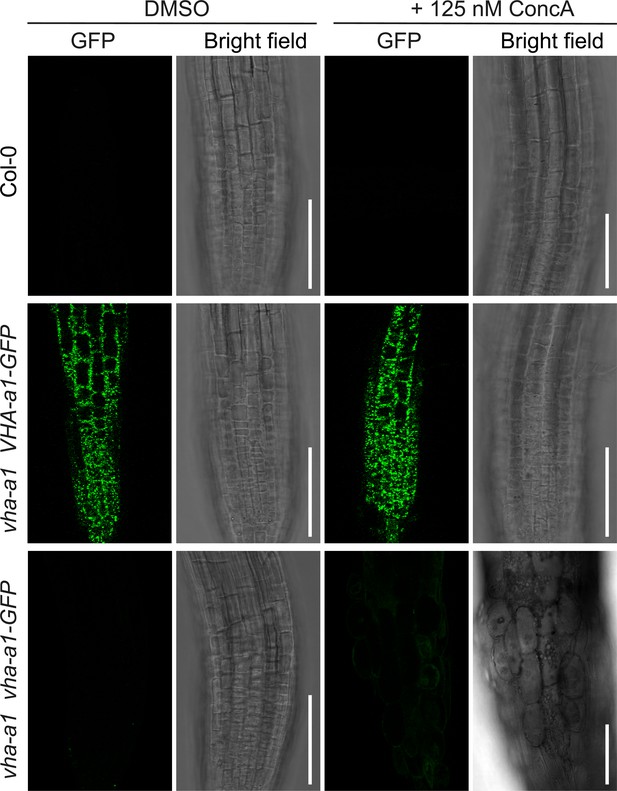
vha-a1 is hypersensitive to Concanamycin-A (ConcA).
Root morphology of 4-day-old etiolated seedlings. vha-a1 roots are hypersensitive to 125 nM ConcA, in contrast to wild type roots and roots of plants expressing vha-a1 VHA-a1-GFP. Scale bars = 75 µm.
-
Figure 10—figure supplement 1—source data 1
Source data for Figure 10—figure supplement 1A.
- https://cdn.elifesciences.org/articles/60568/elife-60568-fig10-figsupp1-data1-v2.xlsx
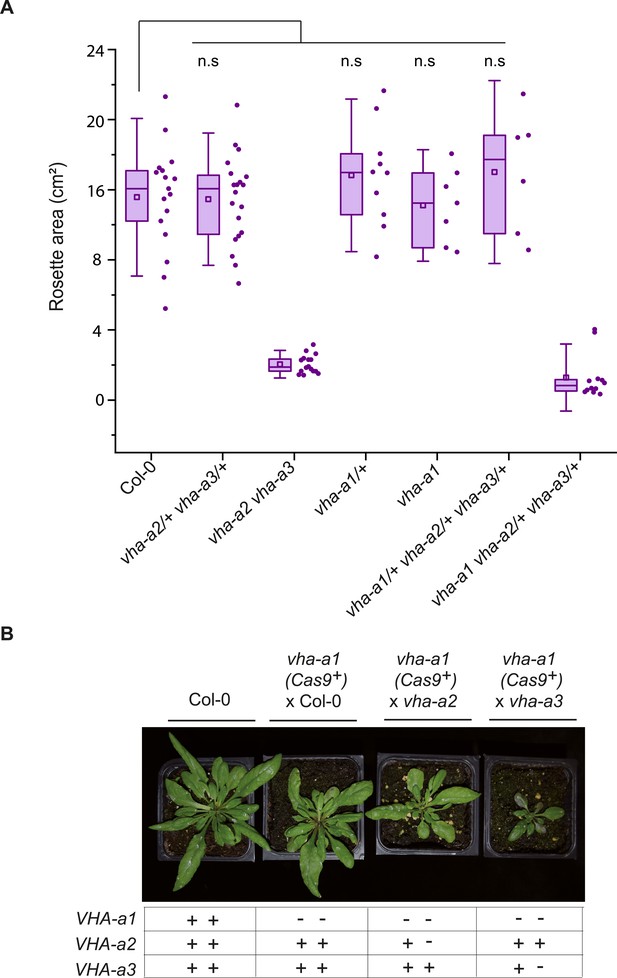
vha-a1 vha-a2/+ and vha-a1 vha-a3/+ have reduced rosette sizes.
(A) vha-a1 (Cas9+) was crossed with the vha-a2 vha-a3 double mutant. Analysis of F1 plants revealed that vha-a1 vha-a2/+ vha-a3/+ is reduced in growth. Rosette areas of 3.5-week-old plants were quantified. Box plot center lines, medians; center circles, means with n≥6 from two biological replicates; box limits, 25th and 75th percentiles; whiskers extend to ±1.5 interquartile range. n.s indicates that there is no significant differences in the mean rosette areas from the Col-0 mean rosette area. (Two-sample t-Test, P < 0.05) (B) vha-a1 (Cas9+) was crossed with the vha-a2 and the vha-a3 single mutants. Analysis of F1 plants showed that vha-a1 vha-a2/+ and vha-a1 vha-a3/+ are reduced in growth. Rosettes of 5 week old plants are shown. Plants were grown under long day conditions (22°C and 16 hours of light).
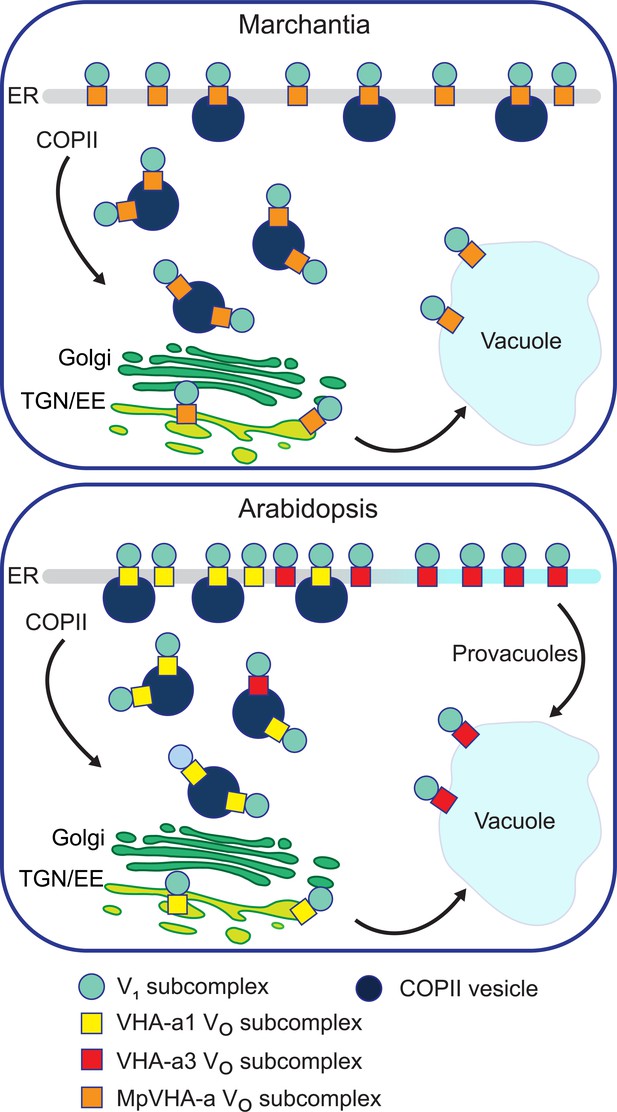
A competition exists to enter COPII vesicles in Arabidopsis.
Upper panel: COPII-mediated ER-export is the only way to exit the ER in Marchantia. Marchantia VHA-a (MpVHA-a) does not contain a TGN/EE-retention signal. MpVHA-a complexes acidify the TGN/EE en route to the tonoplast. Lower panel: Two ER exits exist in Arabidopsis; COPII-mediated ER-export and exit via provacuoles. A competition exists between VHA-a1 and VHA-a3/VHA-a2 complexes to enter COPII vesicles. VHA-a1 has a higher affinity for COPII machinery conferred by the a1-TD. Therefore, it is preferentially loaded into COPII vesicles over VHA-a3. VHA-a3 containing complexes are transported to the tonoplast via provacuoles.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background Arabidopsis thaliana | Col-0 | Nottingham Arabidopsis Stock center (NASC) | NASC: N37008 | |
| Strain, strain background (Marchantia polymorpha) | Tak-1 | Ishizaki et al., 2008 PMID:18535011 | ||
| Genetic reagent Arabidopsis thaliana | vha-a2 vha-a3 | (Krebs et al., 2010) PMID:20133698 | At2g21410,At4g39080 | SALK_142642 SALK_29786 |
| Genetic reagent Arabidopsis thaliana | vha-a1-1 | this study | At2g28520 | See Materials and methods, Construct design and plant transformation |
| Genetic reagent Arabidopsis thaliana | vha-a1-2 | this study | At2g28520 | See Materials and methods, Construct design and plant transformation |
| Genetic reagent Arabidopsis thaliana | vha-a1-3 | this study | At2g28520 | See Materials and methods, Construct design and plant transformation |
| Genetic reagent Arabidopsis thaliana | vha-a1-4 | this study | At2g28520 | See Materials and methods, Construct design and plant transformation |
| Genetic reagent Arabidopsis thaliana | BRI1: BRI-GFP | (Geldner et al., 2007) PMID:17578906 | ||
| Genetic reagent (Marchantia polymorpha) | CaMV35S:mRFPMpSyp6A | (Kanazawa et al., 2016) PMID:26019268 | ||
| Gene Arabidopsis thaliana | VHA-a1 | arabidopsis.org | At2g28520 | |
| Gene Arabidopsis thaliana | VHA-a3 | arabidopsis.org | At4g39080 | |
| Gene Arabidopsis thaliana | Sar1B | arabidopsis.org | At1g56330 | |
| Gene (Rattus norvegicus) | Beta-galactoside alpha-2,6-sialyltransferase 1 (ST) | RGD | 3676 | |
| Gene Arabidopsis thaliana | Brassinosteroid insensitive 1 (BRI1) | arabidopsis.org | At4g39400 | |
| Gene (Marchantia polymorpha) | MpVHA-a | Marchantia genome database | Mp3g15140 | |
| Gene Arabidopsis thaliana | MpSYP6A | Marchantia genome database | Mp3g18380 | |
| Gene (Amborella trichopoda) | Amborella trichopoda VHA-a | Phytozome | evm_27.TU.AmTr _v1.0_scaffold00080.37 | |
| Gene (Selaginella moellendorffii) | Selaginella moellendorffii VHA-a | Phytozome | 182335 | |
| Gene (Pinus taeda) | Pinus taeda VHA-a | PineRefSeq, Tree genes | 5A_I15_VO_L_1_ T_29156/41278 | |
| Transfected construct Arabidopsis thaliana | CRISPR VHA-a1 U6-26p: gRNA one and U6-29p:gRNA two in pHEE401E | this study | Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | CRISPR VHA-a1 U6-26p: gRNA three in pHEE401E | this study | Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | CRISPR VHA-a1 U6-26p: gRNA four in pHEE401E | this study | Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10: VHA-a1 NT 35 aa-VHA-a3 | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10: VHA-a1 NT 85 aa-VHA-a3 | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10: VHA-a1 NT 131 aa-VHA-a3 | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10: VHA-a1 NT 179 aa-VHA-a3 | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10: VHA-a1 NT 228 aa-VHA-a3 | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ:VHA-a3-pmScarlet-I | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10 E156Q-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10 E161S-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10 F134Y-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10 L159T-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10 E156Q + L159T-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10 L159T + E161S -GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1- intron10 ELE-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a1-intron10 ΔEEI-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ:VHA-a3R729N-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:VHA-a3-a1-TD-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | Dex:Sar1BH74L-CFP | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ10:MpVHA-a-mVenus | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct (Marchantia polymorpha) | CaMV35S:MpVHA-a-mVenus | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct (Marchantia polymorpha) | MpEF1α:MpVHA-a-mVenus | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ: A.trichopoda -VHA-a-NT-VHA-a1-mCherry | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ:P.taeda-VHA- a-NT-VHA-a1-mVenus | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ:S.moellendorffii -VHA-a-NT-VHA-a1-mCherry | this study | See Materials and methods, Construct design and plant transformation | |
| Transfected construct Arabidopsis thaliana | UBQ:ST-GFP | this study | See Materials and methods, Construct design and plant transformation | |
| Recombinant DNA reagent (plasmid) | UBQ10 promoter | Lampropoulos et al., 2013 PMID:24376629 | pGGA006 | |
| Recombinant DNA reagent (plasmid) | pOp6 | Schürholz et al., 2018 PMID:30026289 | pGGA016 | |
| Recombinant DNA reagent (plasmid) | B-Dummy | Lampropoulos et al., 2013 PMID:24376629 | pGGB003 | |
| Recombinant DNA reagent (plasmid) | entry module C | Lampropoulos et al., 2013 PMID:24376629 | pGGC000 | |
| Recombinant DNA reagent (plasmid) | pGGC-VHA-a3 | this study | pKSC003 | See Materials and methods, Construct design and plant transformation |
| Recombinant DNA reagent (plasmid) | pGGC-VHA-a1-intron 10 | this study | pKSC012 | See Materials and methods, Construct design and plant transformation |
| Recombinant DNA reagent (plasmid) | pGGC-ST | this study | pKSC013 | See Materials and methods, Construct design and plant transformation |
| Recombinant DNA reagent (plasmid) | GR-LhG4 | Schürholz et al., 2018 PMID:30026289 | pGGC018 | |
| Recombinant DNA reagent (plasmid) | linker GFP | Lampropoulos et al., 2013 PMID:24376629 | pGGD001 | |
| Recombinant DNA reagent (plasmid) | linker-CFP | Lampropoulos et al., 2013 PMID:24376629 | pGGD004 | |
| Recombinant DNA reagent (plasmid) | GFP (A206K) no linker | Lampropoulos et al., 2013 PMID:24376629 | pGGD011 | |
| Recombinant DNA reagent (plasmid) | GSL-mVenus | this study | p2456 | See Materials and methods, Construct design and plant transformation |
| Recombinant DNA reagent(plasmid) | GSL-pmScarlet-I | this study | p1324 | See Materials and methods, Construct design and plant transformation |
| Recombinant DNA reagent (plasmid) | GSL-mCherry | Waadt et al., 2017 PMID:28850185 | p2897 | |
| Recombinant DNA reagent (plasmid) | rbcS terminator | Lampropoulos et al., 2013 PMID:24376629 | pGGE001 | |
| Recombinant DNA reagent (plasmid) | HSP18.2M terminator | Waadt et al., 2017 PMID:28850185 | p1296 | |
| Recombinant DNA reagent (plasmid) | BastaR | Lampropoulos et al., 2013 PMID:24376629 | pGGF001 | |
| Recombinant DNA reagent (plasmid) | SulfR | Lampropoulos et al., 2013 PMID:24376629 | pGGF012 | |
| Recombinant DNA reagent (plasmid) | KanR | Lampropoulos et al., 2013 PMID:24376629 | pGGF007 | |
| Recombinant DNA reagent (plasmid) | HygR_pNos | Waadt et al., 2017 PMID:28850185 | p1317 | |
| Recombinant DNA reagent (plasmid) | HygR_pUbq10 | Lampropoulos et al., 2013 PMID:24376629 | pGGF005 | |
| Recombinant DNA reagent (plasmid) | F-H adapter | Lampropoulos et al., 2013 PMID:24376629 | pGGG001 | |
| Recombinant DNA reagent (plasmid) | H-A adapter | Lampropoulos et al., 2013 PMID:24376629 | pGGG002 | |
| Recombinant DNA reagent (plasmid) | intermediate vector M | Lampropoulos et al., 2013 PMID:24376629 | pGGM000 | |
| Recombinant DNA reagent (plasmid) | intermediate vector N | Lampropoulos et al., 2013 PMID:24376629 | pGGN000 | |
| Recombinant DNA reagent (plasmid) | UBQ10:GR-LHG4:trbcS | this study | pKSM002 | See Materials and methods, Construct design and plant transformation |
| Recombinant DNA reagent (plasmid) | pOP6:Sar1BH74L-CFP | this study | pKSN009 | See Materials and methods, Construct design and plant transformation |
| Recombinant DNA reagent (plasmid) | pGGZ001 | Lampropoulos et al., 2013 PMID:24376629 | ||
| Recombinant DNA reagent (plasmid) | pGGZ003 | Lampropoulos et al., 2013 PMID:24376629 | ||
| Recombinant DNA reagent (plasmid) | pGGZ004 | this study | See Materials and methods, Construct design and plant transformation | |
| Recombinant DNA reagent (plasmid) | pHEE401E | Wang et al., 2015 PMID:26193878 | See Materials and methods, Construct design and plant transformation | |
| Antibody | VHA-a1 (rabbit polyclonal) | Agrisera | AS142822 | (1:1000) |
| Antibody | VHA-a3 (rabbit polyclonal) | Agrisera | AS204369 | (1:1000) |
| Antibody | VHA-B (mouse polyclonal) | Ward et al., 1992 PMID:16668845 | (1:100) | |
| Antibody | anti-GFP (rabbit polyclonal) | Roth et al., 2018 PMID:30410018 | (1:5000) | |
| Chemical compound, drug | Concanamycin-A (ConcA) | Santa Cruz | sc-202111A | |
| Chemical compound, drug | Brefeldin A | LC Laboratories | B-8500 | |
| Chemical compound, drug | FM4-64 | Thermo Fisher Scientific | T13320 | |
| Chemical compound, drug | Dexamethasone (DEX) | Sigma-Aldrich | D4902 | |
| Chemical compound, drug | Malachite Green, Acid Fuchsin, Orange G | Thermo Fisher Scientific | AC413490250, AC400210250, AC229820250 | Alexander stain |
| Commercial assay or kit | CloneJET PCR Cloning kit | Thermo Fisher Scientific | K1231 | |
| Strain, strain background (E. coli) | NEBα | In-house facility | COS Heidelberg | |
| Strain, strain background (A. tumefaciens) | GV3101 | In-house facility | COS Heidelberg | |
| Strain, strain background (A. tumefaciens) | ASE1 | In-house facility | COS Heidelberg | |
| Software, algorithm | Adobe Illustrator 2020 | Adobe Inc | Figure assembly | |
| Software, algorithm | Zen Software | Carl Zeiss | Microscopy | |
| Software, algorithm | Leica LSF | Leica | Microscopy | |
| Software, algorithm | Originpro 2020 | Origin | Statistics and graph plotting | |
| Software, algorithm | Intas Imager | Intas | Western blot | |
| Software, algorithm | Image J | NIH | Image quantification |
Additional files
-
Supplementary file 1
Supplementary tables.
- https://cdn.elifesciences.org/articles/60568/elife-60568-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60568/elife-60568-transrepform-v2.docx





