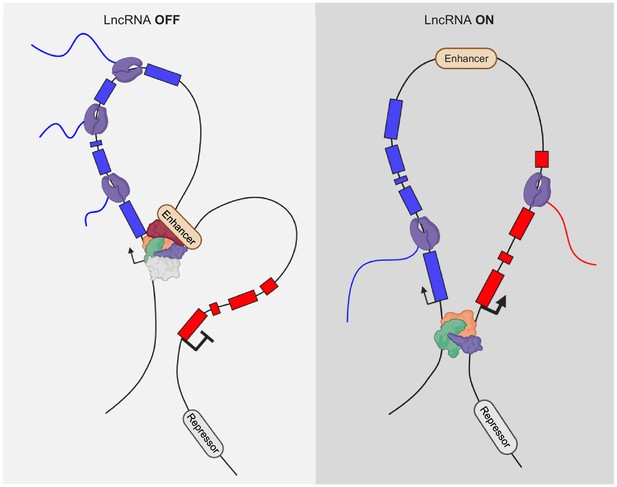
Beyond the RNA-dependent function of LncRNA genes
Figures
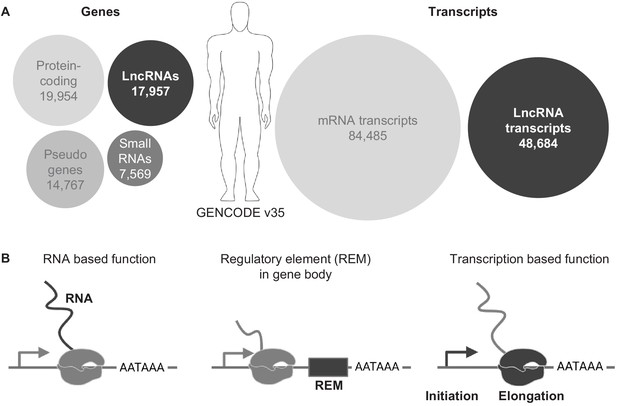
LncRNA genes in the genome.
(A) Overview of genes and transcript numbers in the human genome (GENCODE v35). Circle area represents relative quantities. (B) Schematics of three possible functional properties of lncRNA loci.
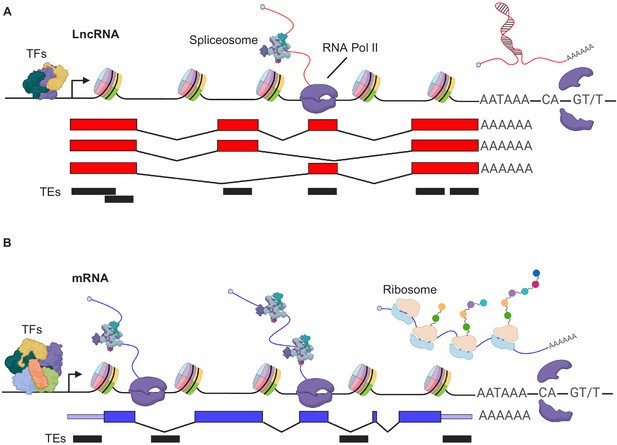
Distinguishing features of transcript generation of PCGs and lncRNAs (A) LncRNA and (B) mRNAs: lncRNA genes are lowly expressed as fewer transcription factors (TFs) bind the promoter.
In addition, lncRNA TSS, exon and/or pA site more often associate with transposable elements (TEs), while TEs contribute mostly to UTRs and/or introns of mRNAs. In addition, mRNAs are more efficiently spliced.
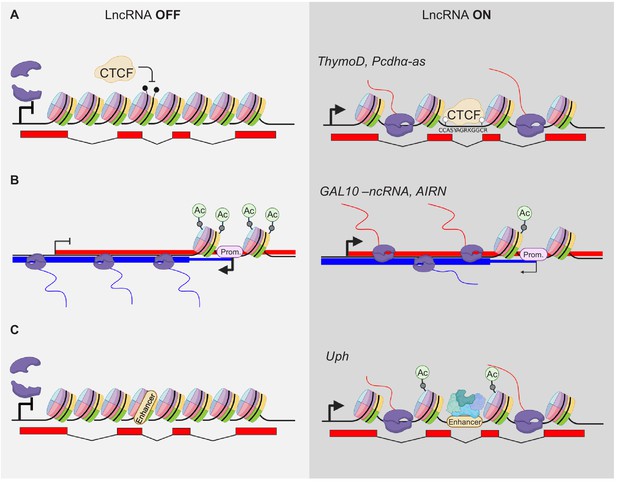
Modulation of gene expression by lncRNA transcription.
(A) Transcriptional activity modulates DNA methylation and thereby alters occupation of DNA binding factors within the gene body, for example CTCF. The POL2 complex is indicated in violet. Black drumsticks indicate methylated CpGs, white drumstick non-methylated CpGs. (B) LncRNA expression alters promoter (Prom.) activity by modifying e.g. acetylation of histones at TSS sites. (C) Transcription elongation can activate poised enhancers within their gene body (only acetylation shown).
Tables
Selection of lncRNA genes with RNA independent function.
| LncRNA | Relative location of respective TSSs target gene | Literature | Mode of action | |
|---|---|---|---|---|
| Regulatory element located within the transcription unit | ||||
| Haunt (Halr1) | 40 kb downstream of HOXA | Yin et al., 2015 | Activation of HOXA | |
| Lockd | 4 kb downstream of Cdkn1b | Paralkar et al., 2016 | Positive regulation of Cdkn1b via loop formation | |
| Meteor | 80 kb upstream of Eomes | Alexanian et al., 2017 | Positive licensing of Eomes expression | |
| ThymoD | 844 kb downstream of Bcl11b | Isoda et al., 2017 | DNA methylation, CTCF-binding | |
| Pcdhα-as | Pcdhα | Canzio et al., 2019 | DNA methylation, CTCF-binding | |
| GAL10-ncRNA | GAL10 antisense transcript | Houseley et al., 2008 | GAL10 promoter acetylation | |
| AIRN | 28 kb Antisense to Igfr2 | Latos et al., 2012 | Promoter methylation | |
| Upperhand (Hand2os1) | 0,1 kb upstream of Hand2 | Anderson et al., 2016; Han et al., 2019 | Promotes enhancer accessibility for Hand2 activation | |
| Activity exerted by transcription initiation or elongation | ||||
| Ftx | 140 kb upstream of Xist | Furlan et al., 2018 | Xist activation independent of Ftx RNA | |
| Chaserr | 16 kb upstream of Chd2 | Rom et al., 2019 | Negative regulation of Chd2 | |
| PVT1 | 52 kb downstream of Myc | Cho et al., 2018 | Enhancer boundary element | |
| Handsdown (Handlr) | 11 kb downstream of Hand2 | George et al., 2019; Ritter et al., 2019 | Transcriptional elongation-based enhancer shielding | |