MetaHiC phage-bacteria infection network reveals active cycling phages of the healthy human gut
Figures
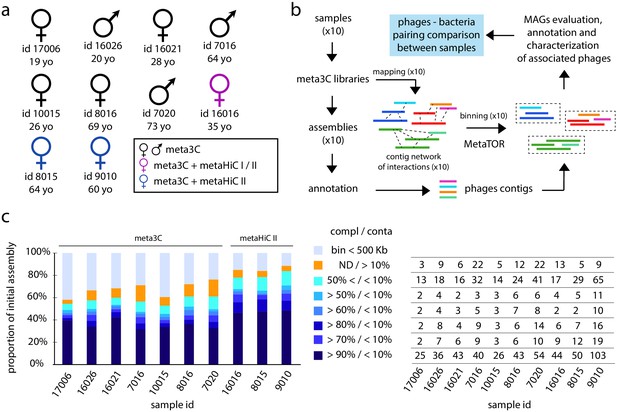
Binning of human gut microbiota using 3C and HiC protocols I and II.
(a) Sex, ID, and age of the 10 individuals investigated in this study. The colors indicate the proximity ligation protocol used to generate the libraries. (b) Computational pipeline of paired-end (PE) sequence analysis (MAG: metagenomic assembled genome). (c) Left diagram: proportion for each individual assembly of the different bins obtained. Colors represent groups of bins of similar quality (completion/contamination), according to the color code on the right. ND = not determined. Right table: for each individual number of MAGs corresponding to the left diagram.
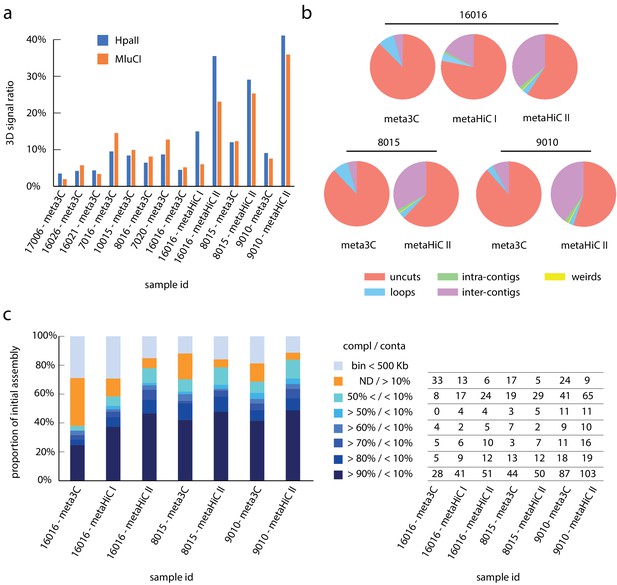
Comparisons of 3C and Hi-C protocols I and II.
(a) Bar plot of 3D signal ratio calculated for each constructed library. The 3D ratio is defined as the ratio of the paired-end (PE) reads that link unambiguously two contigs together compared to the number of mapped PE reads (see 'Materials and methods'). Percentage values are shown in Supplementary file 2. (b) Pie charts indicating the proportion of the different PE reads types (Cournac et al., 2012) for libraries #16016 meta3C/metaHiC protocol I/metaHiC protocol II, #8015 meta3C/metaHiC protocol II, and #9010 meta3C/metaHiC protocol II. Red: uncuts, Blue: loops (self-circularization), Green: intra-contigs interaction, Purple: inter-contigs interaction, Yellow: weird (PE reads that interact with the restriction fragment). (c) Left: bar charts show the proportion of the different bins obtained using meta3C, metaHiC protocol I, or metaHiC protocol II on the samples #16016, #8015, and #9010. Colors indicates the quality of the bins as indicated in the color scale legend. Right: number of metagenomic assembled genomes (MAGs) in each category. The number of bins exhibiting less than 10% of contamination was strongly increased for the metaHiC protocol II.
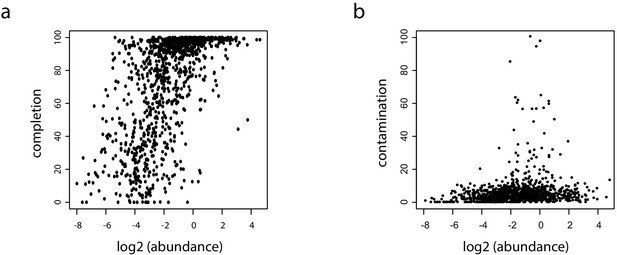
Relation between abundance and completion/contamination for the retrieved metagenomic assembled genomes (MAGs).
(a) Plot of the completion (Y-axis) in function of abundance (X-axis; log2) for the 1100 retrieved MAGs (i.e. bins >500 kb). (b) Plot of the contamination (Y-axis) in function of abundance (X-axis; log2) for the same MAGs.
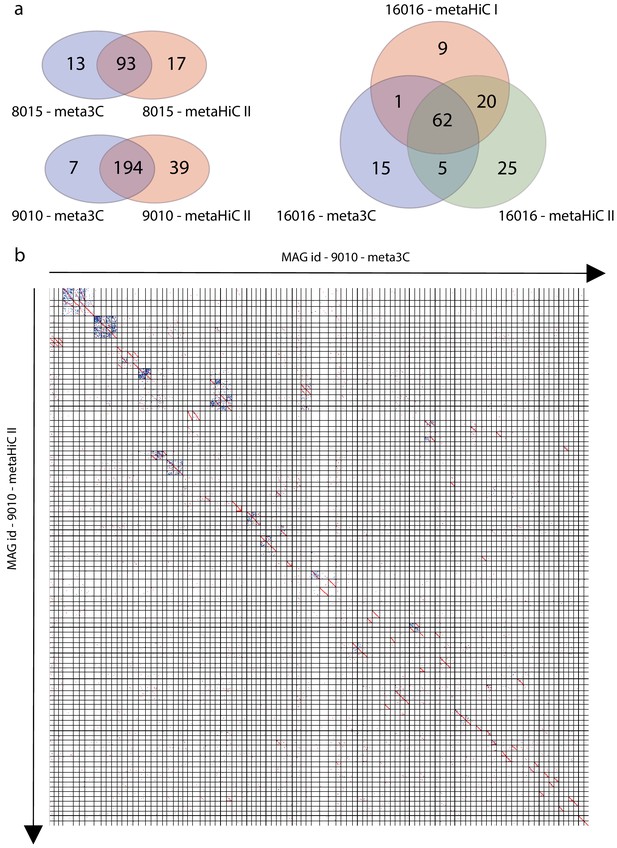
Comparisons of metagenomic assembled genomes (MAGs) obtained using meta3C, metaHiC protocol I, or metaHiC protocol II.
(a) Venn diagram of the MAGs obtained using meta3C, metaHiC protocol I, or metaHiC protocol II data for the samples #8015, #9010, and #16016. We compared the different retrieved MAGs using the different datasets and identified MAGs exhibiting more than 80% of identity. This result clearly shows that the three approaches are highly concordant. (b) LAST alignment of the MAGs > 1 Mb retrieved using meta3C or metaHiC protocol II on sample #9010. X- and Y-axis indicate the MAGs index for meta3C (X) or metaHiC (Y).
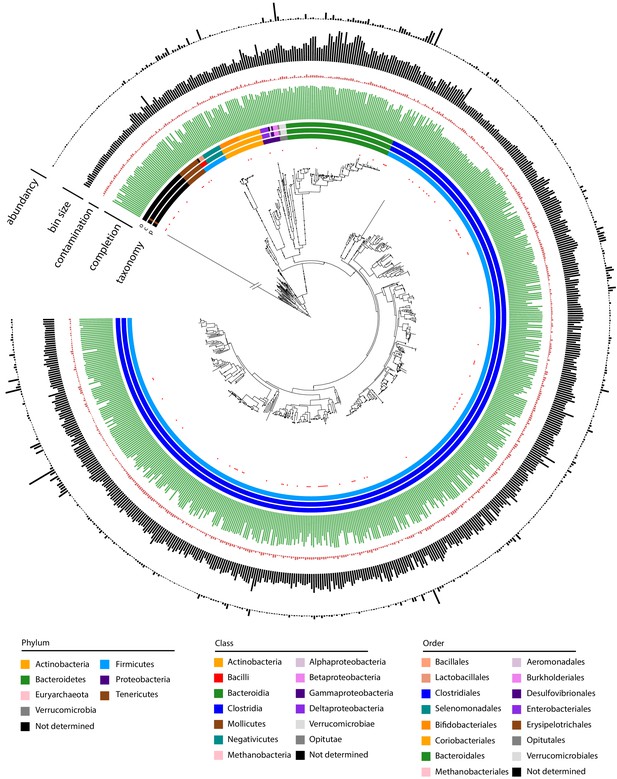
Metagenomic assembled genomes (MAGs) recovered using meta3C/metaHiC applied on 10 healthy human gut samples.
Phylogenetic tree comprising the 715 reconstructed MAGs with a completion above 50% and a contamination below 10% (691 different species at a threshold of 95% identity). A very long branch was cut out (symbol / /) to resize the tree for clarity. Red dots indicate MAGs with no reference in the GTDB-tk database (threshold of homology = 95%). The colors of the first three layers indicate the taxonomy of the MAGs, as determined by CheckM (phylum, class, order from center to periphery). Green and red bars represent completion and contamination levels, respectively. Completion scale: max = 100%; min = 50%. Contamination scale: max = 10%; min = 0%. Gray bars: MAGs (bin) sizes (scale: max = 7.07 Mb, min = 766 kb). Black bars in the outmost layer indicate MAG abundance in the different samples (max = 4.73%; min = 0.0039%).
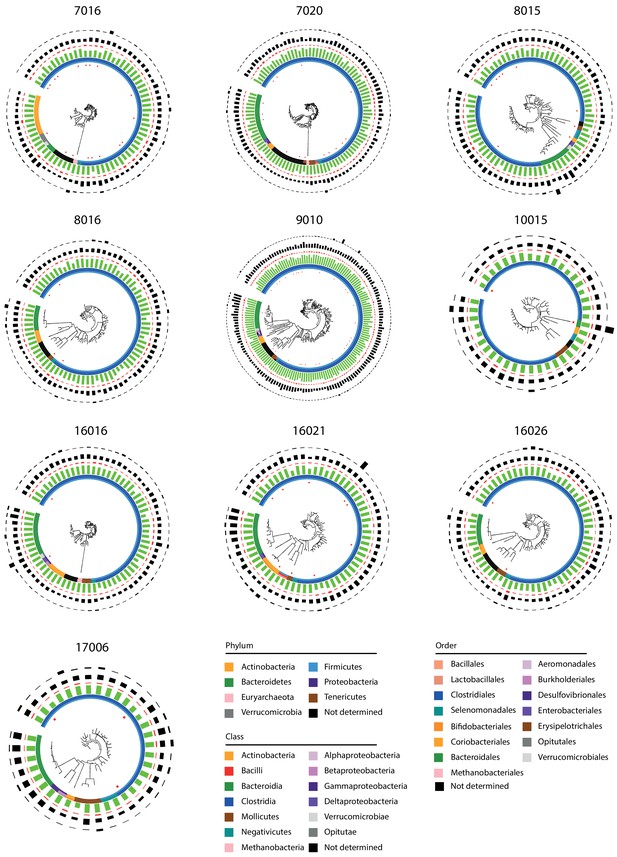
Phylogenetic tree for the 10 processed samples.
Only metagenomic assembled genomes (MAGs) with a completion above 50% and a contamination below 10% are integrated in each tree. Colors of the inner six layers indicate the taxonomy of the MAG attributed by CheckM (from center to periphery: phylum, class, order, family, genus, species). Green and red bars in the following layers indicated completion and contamination. Black bars in the first outer layer indicated MAG size. Black bars in the second outer layer indicated MAG abundance.
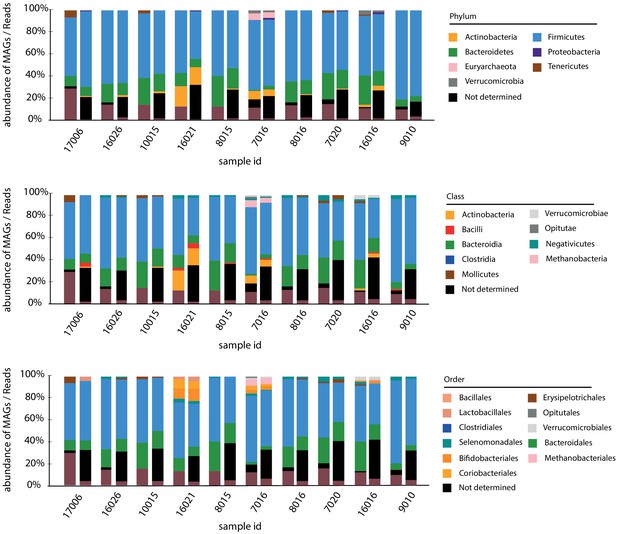
Comparisons of metagenomic assembled genomes (MAGs) taxonomic abundance and reads taxonomic abundance.
Bar charts of MAGs taxonomic abundance (left) and reads taxonomic abundance using Kaiju (Menzel et al., 2016) (right) for each sample. Different taxonomic levels are shown (upper: phylum; middle: class; bottom: order).
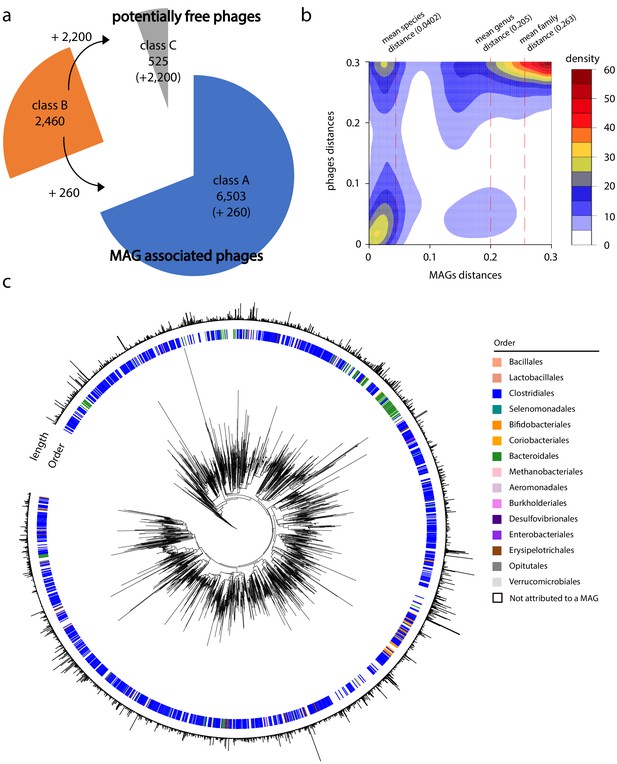
Phage-bacteria network of interactions in human gut.
(a) Pie chart of phages contigs distribution among the different classes (see 'Materials and methods' and Figure 3—figure supplement 1). (b) Gaussian smoothed heatmap of the sequence distance between all phages taken two by two (Y-axis) and the sequence distance between their associated metagenomic assembled genomes (MAGs) (X-axis). Distance varies between 0 (full sequence similarity) and 0.3 (no similarity). Color scale on the left indicates the counting. Dotted lines indicate mean value of distances between MAGs at different taxonomic levels (species = 0.0402, genus = 0.205, family = 0.263). (c) Phylogenetic tree of phage contigs (n = 2,726). Colored strips in the first layer indicate the taxonomy of the associated MAG at the order level (1,535 over the 2,726 contigs present in the tree are attributed to a MAG). Bar plot in the second layer indicates phages contigs size (min = 5 kb; max = 211 kb).
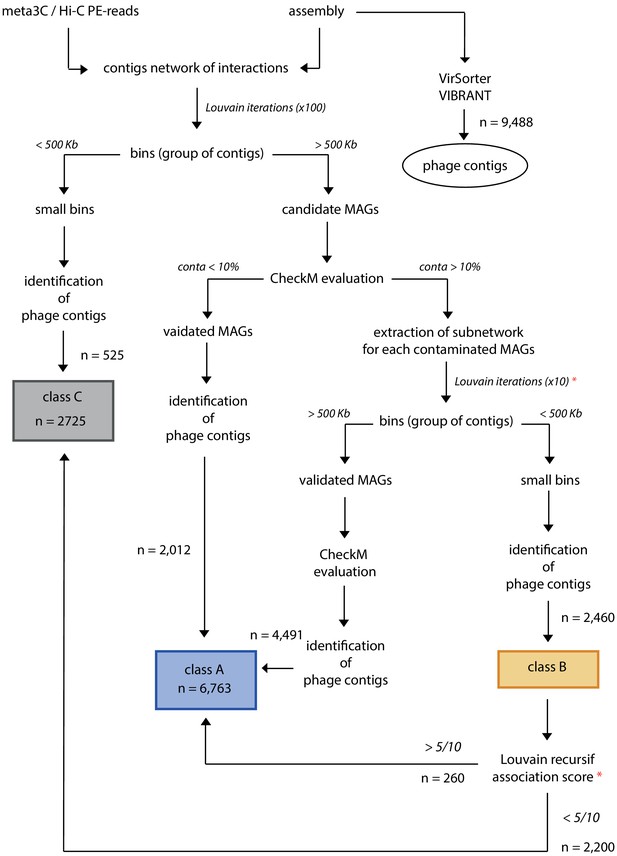
Classification of phages contigs.
Scheme of the followed process to classify phage contigs in the three classes (A, B, C), as described in the 'Materials and methods' section. Red stars point to the 10 recursive Louvain iterations used to calculate the association scores.
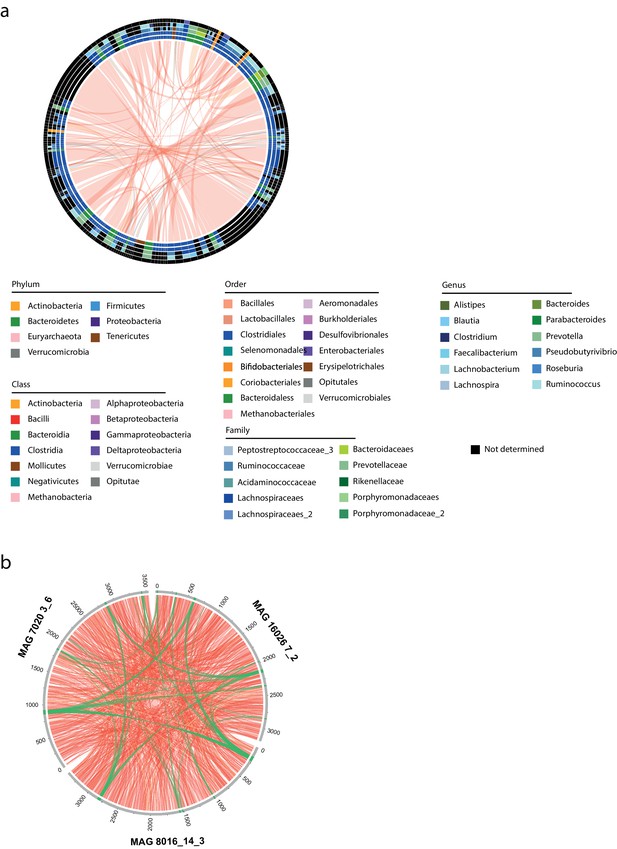
Comparison of phages and their characterized hosts between samples.
(a) Circos representation of the 454 pairs of phages belonging to the same genus. The different circles represent the different characterized hosts at different taxonomic levels (from inside to outside: phylum, class, order, family, genus). Links are colored in function of agreement between hosts taxonomy: red – same taxonomic annotations at the genus level; orange – same taxonomic annotations at the class level; Gray – different annotations at the phylum level. Legend for taxonomic annotations is indicated under the circos. (b) Circos representation of three homologous metagenomic assembled genomes (MAGs) (90% identity) from samples #7020, #16026, and #8016. MAGs are indicated in gray with their respective size indicated in kilobases. Phages contigs are indicated in green. Red links represent blast hits with an identity above 90%. Green links represent blast hits with an identity above 90% for identified phages contigs.
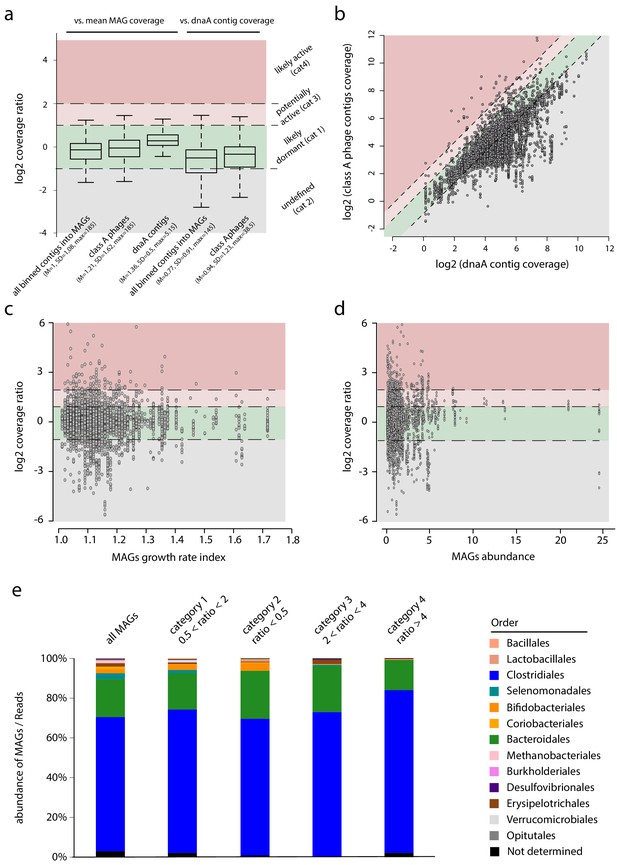
Phages-host ratio in human intestinal tract.
(a) Boxplot of the coverage ratio between different classes of contigs and the mean coverage of their associated MAGs. From left to right, coverage ratio of: (1) all binned contigs into metagenomic assembled genomes (MAGs) (n = 861,082) vs. mean MAG, (2) class A phage contigs (n = 6,763) vs. mean MAG, (3) dnaA contigs (n = 856) vs. mean MAG coverage, (4) all binned contigs into MAGs (n = 672,156) vs. dnaA contig of their associated MAGs, (5) class A phage contigs (n = 6,239) vs. dnaA contig of their associated MAGs. Log2 ratio thresholds define colored areas reflecting the potential phage status: green, dormant (cat1: −1 < log2(ratio) <1); gray, undefined (cat2: log2(ratio) < −1); light red, potentially active (cat3: 1 < log2(ratio) <2); red, likely active (cat4: log2(ratio) >2). Inside bar of the box, median; box, quartiles; whiskers, 1.5× interquartile range. (b) Read coverage of phage contigs (n = 6,239) as a function of the read coverage of the dnaA contig of their associated MAG (n = 856). Dashed lines indicate the limit of the categories described in a. (c) Scatterplot of class A phage contigs coverage vs. dnaA contig coverage as a function of the growth rate index calculated using GRID for each MAG. (d) Scatterplot of class A phage contigs coverage vs. dnaA contig coverage as a function of the MAGs abundance. (e). Bar plot of MAGs taxonomic abundance depending on the categories specified in (a).
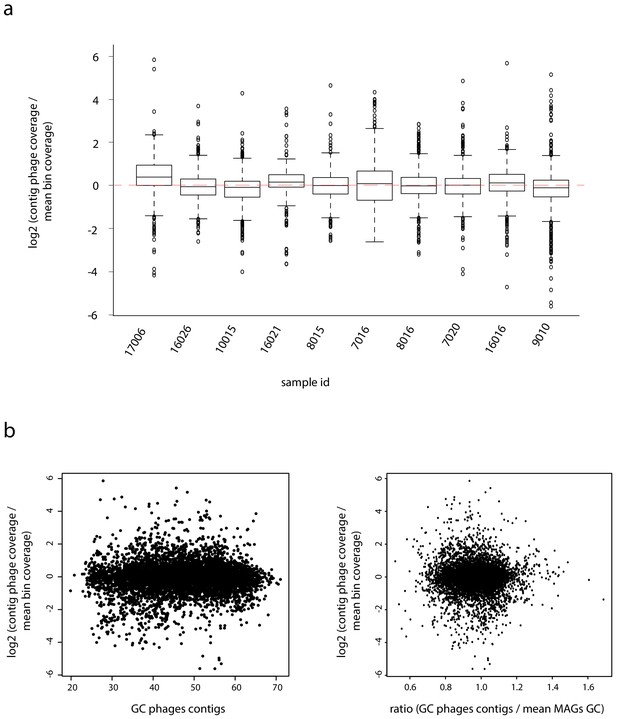
Phages-hosts ratio.
(a) Boxplot of the coverage ratio between phage contigs and their associated metagenomic assembled genome (MAG) (log2 scale) for the 10 samples. Dashed red lines indicate a ratio of 1. (b) Left: plot of the coverage ratio between phages contigs coverage and mean MAGs coverage (Y-axis; log2 scale) in function of the GC% of the phages contigs (X-axis). Right: plot of the coverage ratio between phages contigs coverage and mean MAGs coverage (Y-axis) in function of the ratio of the GC% of the phages contigs and the mean GC% of its associated MAG (X-axis).
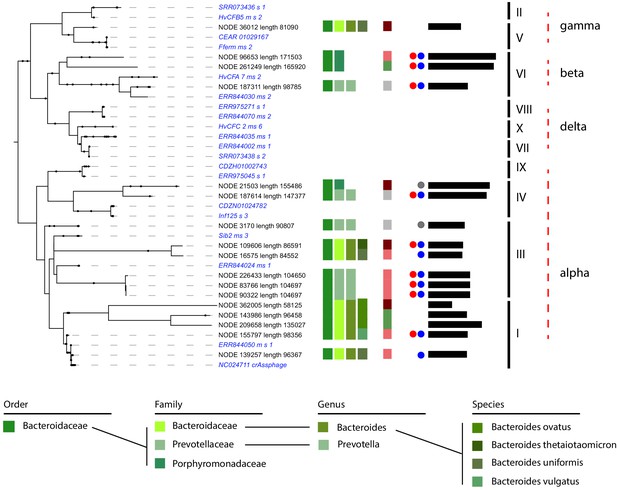
CrAss-like phages and their associated hosts.
Phylogenetic tree of crAss-like phages contigs found in the 10 assemblies. Representatives of the 10 genera of crAss-like phages described by Guerin et al. are included (in italic blue). Names of the different contigs are indicated on the left of each branch. Vertical colored stripes indicate, from left to right: host metagenomic assembled genome (MAG) order, host MAG family, host MAG genus, host MAG species, ratio of the read coverage of phages vs. read coverage of MAG (red – ratio > 4; light red – 4 > ratio > 2; green – 2 > ratio > 0.5; gray – ratio < 0.5). Red dot: circular signal characterized by VirSorter. Blue and gray dot: strong and weak circular signal characterized by 3C, respectively. Black lines: size of the contigs (min = 58,125,125 pb; max = 171,503 bp). Genera of the crAss-like phages are indicated on the right of the tree.
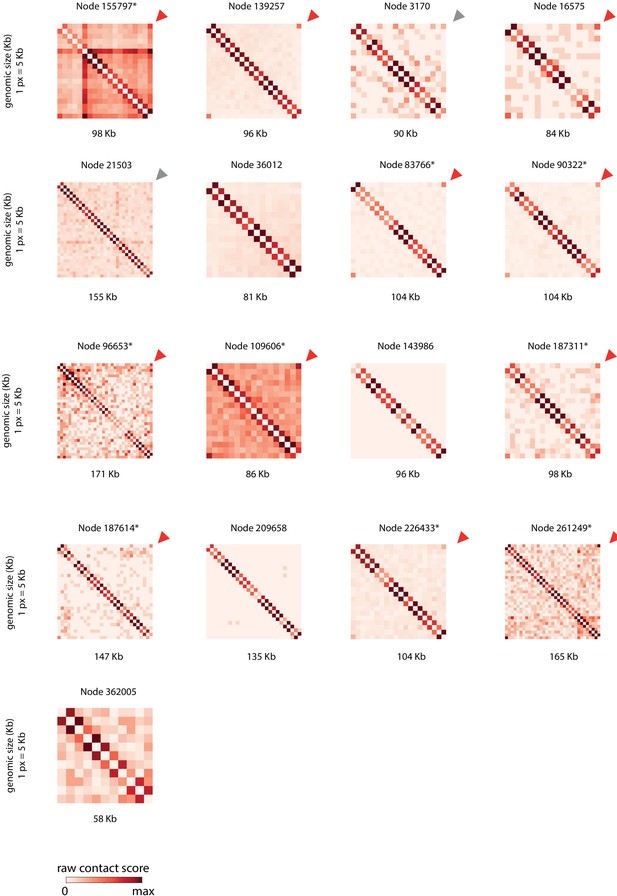
CrAss-like phages contact maps.
Raw contact matrices of the different crAss-like phage contigs found in the 10 assemblies. Each contact map is displayed using 5 kb bins (one pixel = 5 kb). Arrow in the upper right corner indicates a 3C circular signal (red = strong signal, gray = weak signal, no arrow = no signal). Contig ID and size are indicated upper and under each matrix. Star in the contig ID indicates that VirSorter characterize this contigs as circular.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 17006–19 yo – female | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 16026–20 yo – male | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 16021–28 yo – female | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 7016–64 yo – male | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 10015–26 yo – female | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 8016–69 female | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 7020–73 yo – male | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 16016–35 yo – female | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 8015–64 yo – female | Freshly frozen from healthy donor |
| Biological sample (human) | Frozen human fecal samples | Institut Pasteur Biobanque (ICAReB) | Patient 9010–60 yo - female | Freshly frozen from healthy donor |
| Chemical compound, drug | Formaldehyde (35–37%) | Sigma Aldrich | F8775 | (also contain methanol as stabilizer – 15%) |
| Chemical compound, drug | MyOne streptavidin beads | Life Science | ||
| Chemical compound, drug | dCTP-14 Biotin | Life Science | ||
| Software, algorithm | Cutadapt | Martin, 2011 | v.1.9.1 | |
| Software, algorithm | FastQC | Andrews, 2010 | v.0.10.1 | |
| Software, algorithm | MEGAHIT | Li et al., 2015 | v.1.1.1.2 | |
| Software, algorithm | Quast | Gurevich et al., 2013 | v.2.2 | |
| Software, algorithm | bowtie2 | Langmead and Salzberg, 2012 | v.2.2.3 | |
| Software, algorithm | MetaTOR Pipeline | Baudry et al., 2019 | ||
| Software, algorithm | MetaBat | Kang et al., 2015 | ||
| Software, algorithm | CheckM | Parks et al., 2015 | v1.1.2 | |
| Software, algorithm | GTDB-Tk | Chaumeil et al., 2020 | release 0.95 | |
| Software, algorithm | seqtk | seqt, 2020; https://github.com/lh3/seqtk | ||
| Software, algorithm | hicstuff | Matthey-Doret, 2020; https://github.com/koszullab/hicstuff | ||
| Software, algorithm | LAST | http://last.cbrc.jp/ | ||
| Software, algorithm | itol | https://itol.embl.de/ | ||
| Software, algorithm | VirSorter | Roux et al., 2015 | v.1.0.3 | |
| Software, algorithm | VIBRANT | Kieft et al., 2020 | v.1.0.1 | |
| Software, algorithm | Mash | Ondov et al., 2016 | v.2.0 | |
| Software, algorithm | PILER-CR | Edgar, 2007 | ||
| Software, algorithm | Prodigal | Hyatt et al., 2010 | v.2.6.3 | |
| Software, algorithm | MUSCLE | Edgar, 2004 | ||
| Software, algorithm | AMAS | Borowiec, 2016 | ||
| Software, algorithm | IQ-TREE | Nguyen et al., 2015 | v.1.5.5 | |
| Software, algorithm | R environment | R Development Core Team, 2020 | ||
| Other | Covaris S220 | Covaris | AFA tubes | |
| Other | Precellys TUBE | Bertin Technology | VK05 + VK01 glass beads |
Additional files
-
Supplementary file 1
Assembly statistics.
Table indicating different metrics of the 10 sample and the resulting libraries and assemblies.
- https://cdn.elifesciences.org/articles/60608/elife-60608-supp1-v2.xlsx
-
Supplementary file 2
Mapping statistics.
Table indicating different statistics on PE reads mapping and 3D ratio.
- https://cdn.elifesciences.org/articles/60608/elife-60608-supp2-v2.xlsx
-
Supplementary file 3
CrAss-like phages contigs.
Table containing informations on the different detected crAss-like phage contigs.
- https://cdn.elifesciences.org/articles/60608/elife-60608-supp3-v2.xlsx
-
Supplementary file 4
Metagenomic assembled genomes (MAGs) data.
Comma separated file describing the MAGs called in the study: sample, bin ID, bin size, bin mean GC%, bin mean coverage, taxonomy (seven levels), completion, contamination, contigs number, N50, mean contig size, longest contig, coding density.
- https://cdn.elifesciences.org/articles/60608/elife-60608-supp4-v2.zip
-
Supplementary file 5
Contigs data.
Comma separated file describing all the binned contigs present in MAGs: sample, contig ID, contig size, contig coverage, contig GC%, associated bin.
- https://cdn.elifesciences.org/articles/60608/elife-60608-supp5-v2.zip
-
Supplementary file 6
Phages contigs data.
Comma separated file describing all the phages’ contigs associated to MAGs: sample, contig ID, associated bin.
- https://cdn.elifesciences.org/articles/60608/elife-60608-supp6-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60608/elife-60608-transrepform-v2.pdf





