CXCL10/CXCR3 signaling contributes to an inflammatory microenvironment and its blockade enhances progression of murine pancreatic precancerous lesions
Figures
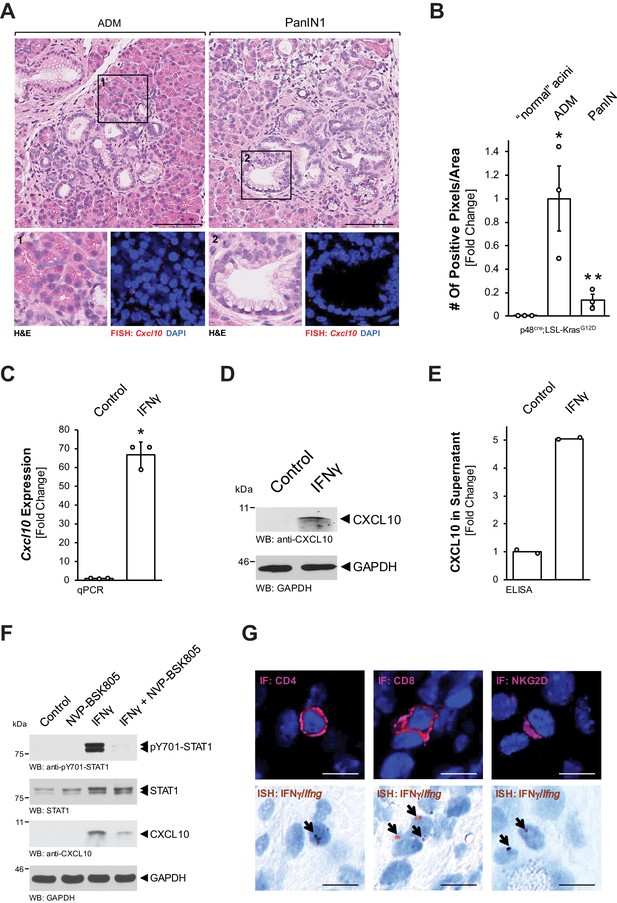
Pre-neoplastic ADM and PanIN lesions express CXCL10.
(A) ‘Normal’ acinar cells (data shown in Figure 1—figure supplement 1B), ADM areas and PanIN lesions of pancreata from KC mice were analyzed for Cxcl10 expression. Shown are representative pictures of H and E staining (overview and marked region) and FISH for Cxcl10 mRNA expression (red dots) combined with DAPI in the marked regions. Images shown represent whole-slide analysis of staining. The scale bar represents 100 μm. (B) Quantification of relative Cxcl10 expression (as determined by ISH shown in Figure 1—figure supplement 1C) in ‘normal’ acini, ADM, and PanIN regions of KC mice (n = 3 biological replicates) using a positive pixel algorithm on whole slides for each mouse analyzed, using the image scope software. Statistical analysis was done using the Student’s t-test. *Statistical significance as compared to ‘normal’ acini (for ADM p-value = 0.023; PanIN p-value = ns), **Statistical significance for PanIN as compared to ADM (p-value=0.033). Error bars indicate standard deviation. (C, D, E) SM3 cells were stimulated with 10 ng/ml IFNγ for 4 days and an increase in CXCL10 expression was determined by qPCR (C), western blot (D), and in the media supernatants (E). For (C, D), results are representative of data from three independent, reproducible experiments. Statistical analysis was done using the Student’s t-test. The asterisk indicates statistical significance (C: p-value < 0.0001; E: p-value = 0.0004). Error bars indicate standard deviation. (E) shows two biological repeats. (F) SM3 cells were treated with NVP-BSK805 (10 μM, 1 hr) and then stimulated with 10 ng/ml IFNγ for 24 hr. Samples were subjected to SDS-PAGE and analyzed by western blotting for pY701-STAT1, STAT1, and CXCL10 expression as indicated. Immunoblotting for GAPDH served as a control for equal loading. Results shown represent reproducible data obtained from three independent experiments. (G) Pancreata of KC mice were subjected to IF-IHC for CD4, CD8, and NKG2D combined with FISH for Ifng. Images shown are representative of IF and FISH done on 2 KC mice (biological replicates). The scale bar represents 10 μm.
-
Figure 1—source data 1
Quantification of Cxcl10 in KC tissue and IFNɣ-stimulated SM3 cells (panels B, C, and E).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig1-data1-v2.xlsx
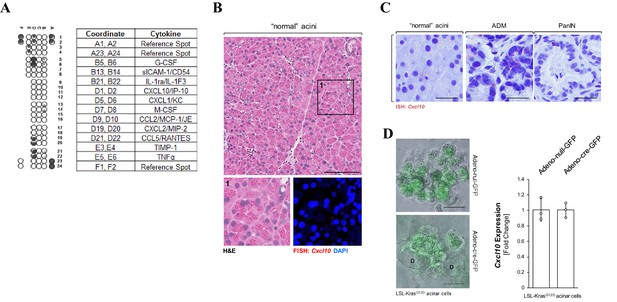
CXCL10 expression in pancreatic precancerous lesion cells.
(A) Pancreatic duct-like cells from early lesions express CXCL10. A screening array to determine the expression profile of 40 different chemokines and cytokines in precancerous lesion cells was performed using the SM3 cell line. SM3-conditioned media shows the presence of various cytokines (table), including CXCL10. Image shown is representative of three independent experiments. (B) Addition to Figure 1A. Representative image of ‘normal’ pancreatic acinar cells from Ptf1a/p48cre;LSL-KrasG12D mice. Shown are H and E and Cxcl10 mRNA expression by FISH in the marked region. The scale bar indicates 100 μm. (C) In situ hybridization (ISH) for Cxcl10. ISH for Cxcl10 was performed using pancreata from Ptf1a/p48cre;LSL-KrasG12D mice (used for analyses in Figure 1B). Shown are representative images of ‘normal’ pancreatic acinar cells, an ADM area as well as a PanIN area from staining and analysis done on three mice. The scale bar indicates 25 μm. (D) KRasG12D does not drive expression of Cxcl10 in ADM cells. Pancreatic acinar cells were isolated from a LSL-KrasG12D mouse and adeno-virally infected with Adeno-cre-GFP or Adeno-null-GFP. Cells were seeded in 3D explant culture to induce ADM. The presence of GFP indicates successful infection (images). Formation of ducts at day 5 after infection (D) indicates successful induction of ADM in the Adeno-cre-GFP-infected cells. The scale bar indicates 50 μm. Bar graph: At day 5 after infection, RNA was isolated from 3D cultured cells and a qRT-PCR for Cxcl10 was performed. Experiment was conducted in triplicates. Statistical analysis was done using the Student’s t-test. Error bars indicate standard deviation.
-
Figure 1—figure supplement 1—source data 1
CXCL10 expression in Adeno-null-GFP and Adeno-cre-GFP infected cells (panel D).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig1-figsupp1-data1-v2.xlsx
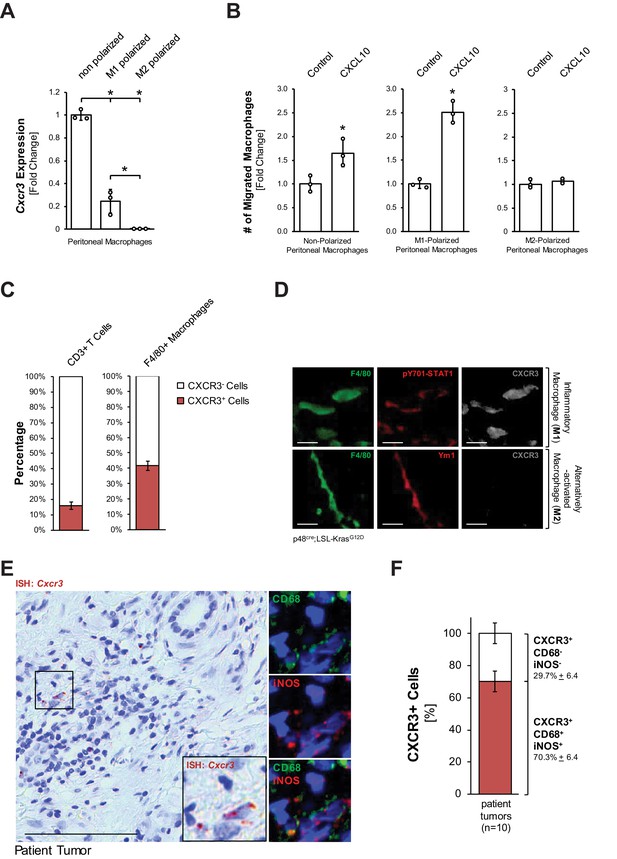
Inflammatory macrophages are the recipients for CXCL10.
(A) Primary peritoneal macrophages were isolated and either left non-polarized or were polarized to M1 and M2 macrophages. CXCR3 expression was determined by qPCR. Results shown are representative of reproducible data from three independent experiments. Statistical analysis using the Student’s t-test indicates significance (marked by an asterisk). Error bars indicate standard deviation. (B) Transwell assay. 0.5 × 105 non-polarized, M1- or M2-polarized peritoneal macrophages were plated into transwell inserts. 500 ng/ml CXCL10 in media was placed in the bottom wells, and chemoattraction of macrophages was determined after 20 hr. Data shown here represents reproducible results from peritoneal macrophages obtained from three different mice (biological replicates). Statistical analysis using the Student’s t-test indicates significance (marked by an asterisk) for nonpolarized (p-value=0.034) and M1 (p-value=0.0003) macrophages. Error bars indicate standard deviation. (C) CD3+ T cells and F4/80+ macrophages were sorted from digested pancreas of KC mice using FACS and analyzed for the expression of CXCR3. The bars indicate the percentage of CXCR3+ cells among the two cell types sorted. Data represents analyses done on three mice (biological replicates). Statistical analysis was done using Student’s t-test. Error bars indicate standard deviation. (D) CXCR3 expression in M1 (F4/80+;pSTAT1+) or M2 (F4/80+;Ym1+) macrophages in pancreatic tissue of KC mice was determined by immunofluorescence. Images shown here represent whole slide analysis of staining done on the tissue of 2 KC mice (biological replicates). The scale bar indicates 10 μm. (E) Patient tumor tissue stained by ISH for Cxcr3 and overlaying immunofluorescence for inflammatory (CD68+,iNOS+) macrophages. Images shown represent whole slide analysis of staining done on the tissue from 10 patient samples. The scale bar indicates 100 μm. (F) Quantification of Cxcr3+;CD68+;iNOS+ cells in patient samples (n = 10). Statistical analysis was done using Student’s t-test. Error bars indicate standard deviation.
-
Figure 2—source data 1
Cxcr3 expression and migration in response to CXCL10 for polarized macrophages, CXCR3 expression in T cells and macrophages, and quantification of CXCR3+ M1 macrophages (panels A, B, C, and F).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig2-data1-v2.xlsx
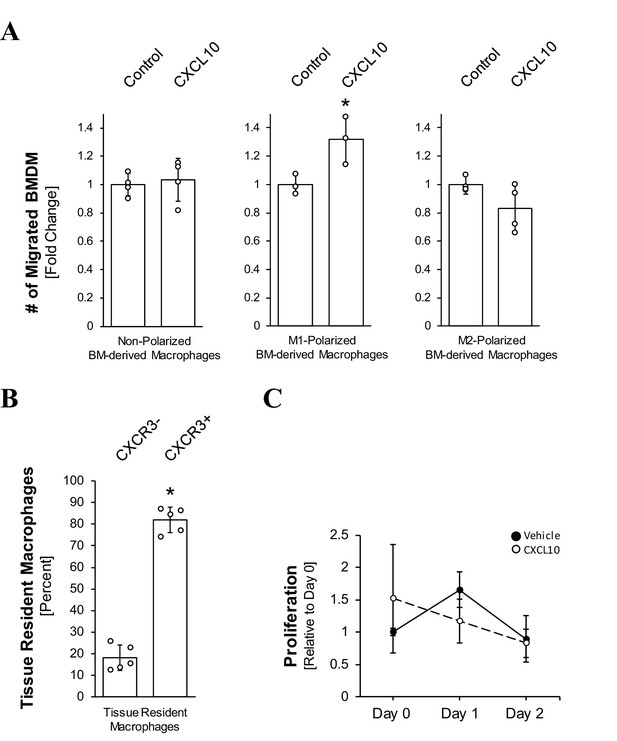
M1-polarized BMD macrophages are chemoattracted to CXCL10, and tissue resident macrophages express CXCR3 but do not proliferate in response to CXCL10.
(A) M1-polarized BMD Macrophages are chemoattracted to CXCL10. 1.0 × 105 non-polarized, M1- or M2-polarized BMD macrophages were plated into transwell inserts. 500 ng/ml CXCL10 in media was placed in the bottom wells, and chemoattraction of macrophages was determined after 2 hr. Data shown here represents reproducible results from BMD macrophages obtained from one mouse (data points represent individual transwells). The assay was repeated twice more with cells derived from different mice, with similar results. Statistical analysis was done using the Student’s t-test (p-value=0.0405). Error bars indicate standard deviation. (B, C) Tissue resident macrophages express CXCR3 but do not proliferate in response to CXCL10. (B) CXCR3 expression in F4/80+ macrophages in normal pancreatic tissue of wild-type mice (n = 5) was determined by immunofluorescence. Quantification of macrophages, ± CXCR3 expression, was performed manually. Error bars indicate standard deviation, and the asterisk indicates statistical significance using the Student’s t-test (p-value=0.0000001). (C) Tissue resident macrophages were isolated from non-transgenic mouse pancreata via MACS, seeded at 5000 cells per well, and treated daily with 50 ng/ml CXCL10. Proliferation was measured via the MTT assay. Error bars represent the standard deviation (n = 3). In (A–C), the asterisk indicates statistical significance.
-
Figure 2—figure supplement 1—source data 1
Quantification of macrophage migration and proliferation in response to CXCL10, and percentage of macrophages which are CXCR3+ (panels A-C).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig2-figsupp1-data1-v2.xlsx
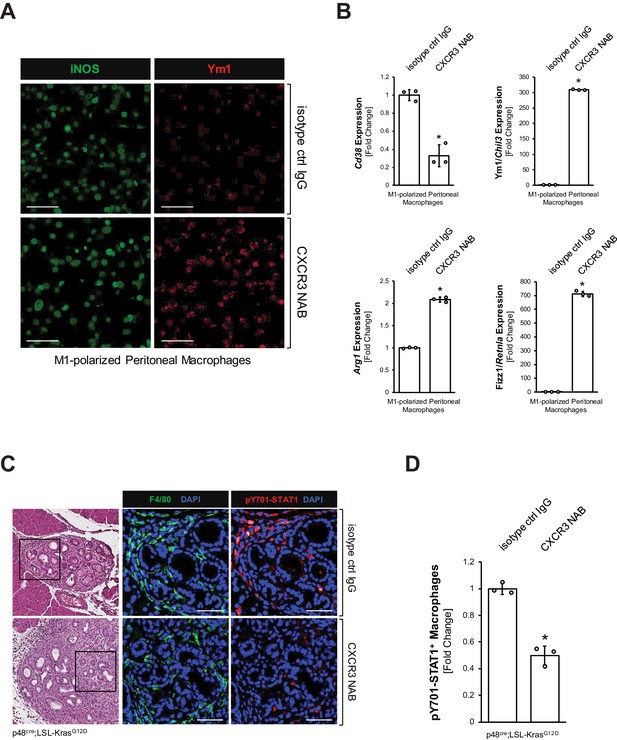
CXCL10/CXCR3 signaling maintains the inflammatory phenotype of macrophages.
(A) Peritoneal macrophages were isolated, polarized to M1, and then treated with 500 μg/ml CXCR3 NAB or isotype control IgG. After 48 hr treatment, samples were analyzed for expression of iNOS or Ym1 using immunofluorescence. Images shown are representative of three independent experiments done on peritoneal macrophages obtained from three mice (biological replicates). The scale bar indicates 100 μm. (B) Peritoneal macrophages were isolated, polarized to M1, and treated with 500 μg/ml CXCR3 NAB or isotype control IgG. After 48 hr, samples were analyzed by qPCR for expression of M1 macrophage marker Cd38 and M2 macrophage markers Ym1/Chil3, Arg1, and Fizz1/Retnla. Results shown are representative of three independent experiments done on peritoneal macrophages obtained from three mice (biological replicates). Statistical analysis using the Student’s t-test indicates significance (marked by an asterisk, Cd38: p-value=0.0009, Ym1/Chil3: p-value<0.0001, Arg1: p-value<0.0001, Fizz1/Retnla: p-value<0.0001). Error bars indicate standard deviation. (C) Pancreatic abnormal areas from KC mice treated with CXCR3 NAB or isotype control IgG were analyzed for presence of inflammatory macrophages (co-immunofluorescence for F4/80 and pY701-STAT1). Shown is a representative area from staining and analysis done on three mice per group. The H and E staining highlights the region analyzed. The scale bar indicates 50 μm. (D) Quantification of pY701-STAT1+ macrophages in pancreata from KC mice (n = 3 mice per treatment group) treated with CXCR3 NAB or isotype control IgG. Cells were counted in three representative fields per mouse. The arcsin transformation was done on the proportion of macrophages which were pY701-STAT1+. Statistical analysis using the Student’s t-test indicates significance between biological replicates (indicated by an asterisk, p-value=0.0004). Error bars indicate standard deviation.
-
Figure 3—source data 1
qPCR forCd38,Ym1/Chil3,Arg1, and Fizz1/Retnlain M1 polarized macrophages treated with CXCR3 NAB or isotype control IgG, and quantification of pY701-STAT1+ macrophages in KC mice treated with CXCR3 NAB or isotype control IgG (panels B and D).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig3-data1-v2.xlsx
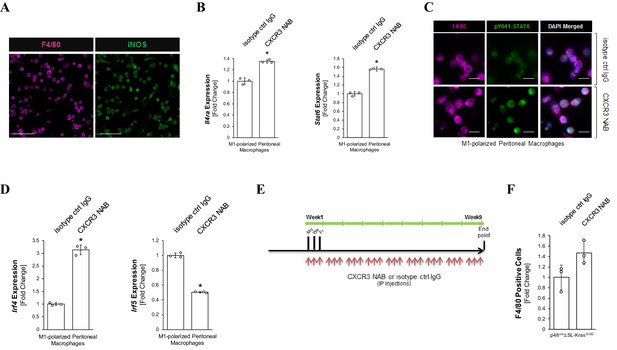
Neutralization of CXCR3 shifts macrophages to M2 polarization.
(A) Control for Figure 3A to demonstrate that peritoneal macrophages were polarized to an inflammatory (M1) type. Peritoneal macrophages were isolated, M1 polarized, and analyzed by immunofluorescence for expression of the inflammatory macrophage marker iNOS. Immunofluorescence for F4/80 (pan macrophage marker) served as control. The scale bar indicates 100 μm. (B) Peritoneal macrophages were isolated, polarized to M1, and treated with 500 μg/ml CXCR3 NAB or isotype control IgG. After 48 hr, samples were analyzed by qPCR for expression of Il4ra and Stat6. Data shown is representative of three independent experiments. Statistical analysis was done using the Student’s t-test (Il4ra: p-value = 0.0007; Stat6: p-value = 0.00004). Error bars indicate standard deviation. The asterisk indicates statistical significance. (C) pY641-STAT6 in the nuclei of macrophages upon CXCR3 neutralization indicates a shift to M2 polarization. Peritoneal macrophages were M1 polarized and treated with CXCR3 NAB or isotype control IgG for 24 hr. Cells were fixed and co-stained for F4/80 and pY641-STAT6. Images shown are representative of two independent experiments. Scale bar represents 50 μm. (D) M1 polarized peritoneal macrophages were treated with 500 μg/ml CXCR3 NAB or isotype control IgG. After 48 hr, samples were analyzed by qPCR for transcription factors Irf4 and Irf5. Statistical analysis was done using the Student’s t-test (Irf4: p-value=0.00005; Irf5; p-value=0.00001). Error bars indicate standard deviation between three replicates. (E) Treatment scheme. Treatment scheme for the animal experiments shown in Figures 3D and 4. (F) Quantification of macrophages (F4/80 positive cells) in pancreata from KC mice (n = 3 mice per treatment group) treated with CXCR3 NAB or isotype control IgG. Quantification was performed with the Image scope positive pixel algorithm. Statistical analysis was done using the Student’s t-test (ns = not significant). The dots show three biological replicates. Error bars indicate standard deviation. In (B, D, E), the asterisks indicate statistical significance.
-
Figure 3—figure supplement 1—source data 1
qPCR forIl4ra,Stat6,Irf4, andIrf6in M1 polarized peritoneal macrophages treated with CXCR3 NAB or isotype control IgG, and quantification of macrophages in KC mice treated with CXCR3 NAB or isotype control IgG (panels B, D, and F).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig3-figsupp1-data1-v2.xlsx
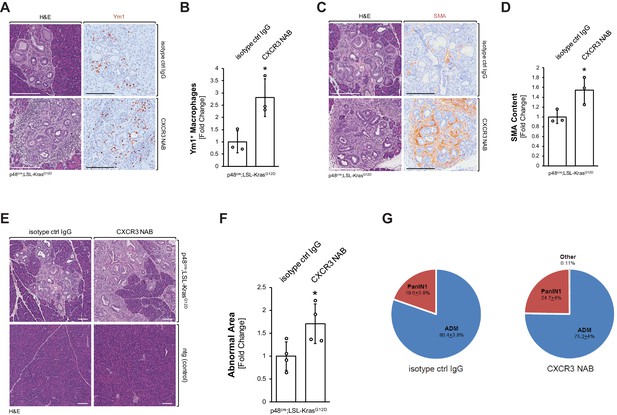
Neutralization of CXCR3 signaling increases alternatively activated pancreatic macrophages and progresses pancreatic lesions.
(A) Pancreatic abnormal areas from KC mice treated with CXCR3 NAB or isotype control IgG were analyzed by IHC for presence of Ym1+ macrophages. Shown is a representative area and H and E staining on serial sections of the tissue. The scale bar indicates 100 μm. (B) Quantification of Ym1+ macrophages in pancreata from KC mice (n = 3 mice per treatment group) treated with CXCR3 NAB or isotype control IgG. Ym1+ cells were counted in three representative fields per mouse, and reported as the fold change relative to the average of the isotype control IgG treatment group. Statistical analysis was done using the Student’s t-test. The asterisk indicates statistical significance (p-value=0.025). Error bars indicate standard deviation. (C) Pancreatic abnormal areas from KC mice treated with CXCR3 NAB or isotype control IgG were analyzed by IHC for the fibrosis marker smooth muscle actin (SMA). Shown is a representative area and H and E staining on serial sections of the tissue. The scale bar indicates 100 μm. (D) Quantification of SMA content in pancreata from KC mice (n = 3 per treatment group) treated with CXCR3 NAB or isotype control IgG. Quantification was performed with the Image scope positive pixel algorithm. Abnormal areas were manually traced on the tissue, and the algorithm was run. The resulting pixel values were divided by the area analyzed to obtain staining per area of abnormal tissue. Statistical analysis was done using the Student’s t-test. The asterisk indicates statistical significance (p-value=0.038). Error bars indicate standard deviation. (E) Representative images of H and E stained pancreata from non-transgenic (ntg) or KC mice treated with CXCR3 NAB or isotype control IgG. The scale bar indicates 100 μm. (F) Quantification of the abnormal pancreatic surface area in pancreata from KC mice (n = 4 per treatment group) treated with CXCR3 NAB or isotype control IgG. For quantification, abnormal areas were manually traced out and values were normalized to the total pancreatic area to obtain abnormal area per area of pancreatic tissue analyzed. For statistical analysis, data were transformed via arcsin transformation before a t-test was performed. The asterisk indicates statistical significance (p-value=0.037). Error bars indicate standard deviation. (G) Pie graph showing the percentage distribution of the types of lesions found in the abnormal surface areas of mice from both treatment groups. Percentages (with standard error) shown are from analysis done on four mice per group.
-
Figure 4—source data 1
Quantification of Ym1, SMA, abnormal area, and lesion type in CXCR3 NAB and isotype control IgG treated KC mice (panels B, D, F, and G).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig4-data1-v2.xlsx
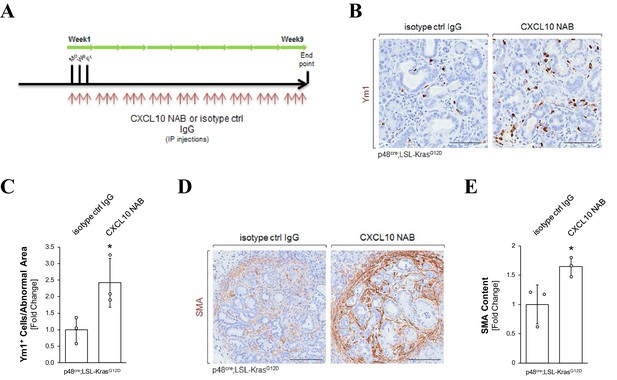
Neutralization of CXCL10 increases alternatively activated macrophages and fibrosis.
(A) Treatment scheme for animal experiments in which CXCL10 was neutralized. Similar to the CXCR3 neutralization experiment, KC mice were treated with a CXCL10 NAB. (B, C) CXCL10 NAB-treated mice show an increase in M2 macrophages. (B) Tissues from CXCL10 NAB and isotype control IgG treatment groups were stained with Ym1 (IHC) to identify M2 macrophages. Shown are representative images. The scale bar represents 200 μm. (C) Ym1+ macrophages were quantified (n = 3 mice per group) and normalized to the total pancreatic area analyzed. Statistical analysis was done using the student’s t-test (p-value=0.0476). Error bars indicate standard deviation. The asterisk represents statistical significance. (D, E) CXCL10 NAB treated mice show higher fibrosis. (D) Tissues from CXCL10 NAB and isotype control IgG treatment groups were stained with the fibrosis marker SMA. Shown are representative images. The scale bar represents 300 μm. (E) Quantification of SMA content in pancreata (n=three mice per group). Samples were analyzed with Aperio’s positive pixel algorithm and normalized to the total pancreatic area analyzed. Statistical analysis was done using the Student’s t-test (p-value=0.0396). Error bars indicate standard deviation. The asterisk represents statistical significance.
-
Figure 4—figure supplement 1—source data 1
Quantification of Ym1+ cells and SMA content in KC mice treated with CXCL10 NAB or isotype control IgG (panels C and E).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig4-figsupp1-data1-v2.xlsx
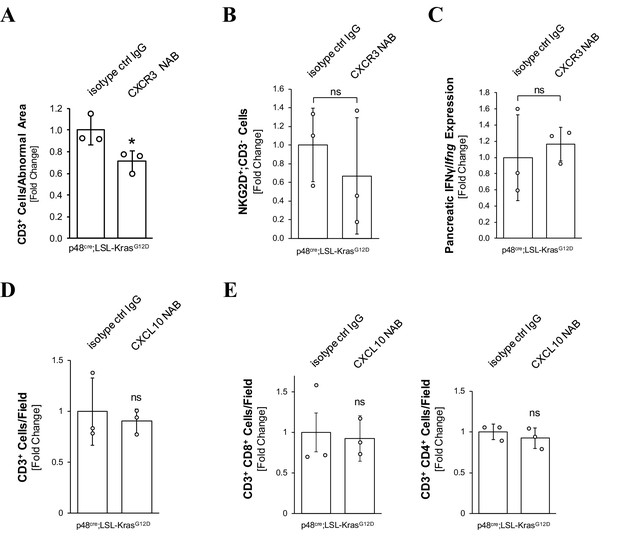
Neutralization of CXCL10/CXCR3 signaling does not significantly affect the presence of T cells.
(A) CXCR3 NAB-treated mice show slightly reduced number of T cells in the pancreas. Pancreata from KC mice (n = 3 mice per group) treated with isotype control antibody or CXCR3 neutralizing antibody were analyzed for the presence of T cells by CD3 staining (IHC). Shown is a quantification using Aperio’s positive pixel algorithm. Error bars indicate standard deviation. Statistical analysis was done using student’s test (p-value=0.039). (B) CXCR3 NAB-treated mice show no significant difference in pancreatic NK cell population. Pancreata from KC mice (n = 3 mice per group) treated with isotype control antibody or CXCR3 neutralizing antibody were analyzed for the presence of NK cells by co-staining with NKG2D and CD3 antibodies. Data shown were obtained by manually counting NKG2D+ CD3− cells on whole tissue slides for each mouse analyzed and normalizing to the area of tissue analyzed. Statistical analysis was done using the Student’s t-test (p-value=0.5110). Error bars indicate standard deviation. (C) CXCR3 NAB-treated mice show no significant change in pancreatic IFNγ expression. Pancreata from KC mice (n = 3 mice per group) treated with isotype control antibody or CXCR3 neutralizing antibody were analyzed for IFNγ/Ifng mRNA using fluorescence in-situ hybridization (FISH). Shown is a quantification using ImageJ software. Statistical analysis was done using the Student’s t-test. Error bars indicate standard deviation. (D, E) CXCL10 NAB-treated mice show no significant change in the pancreatic T-cell population. Tissue from KC mice (n = 3 mice per group) treated with the CXCL10 NAB or isotype control IgG was co-stained (IF-IHC) with either CD3 and CD4 or CD3 and CD8. Data shown were obtained by manually counting three representative fields for each mouse analyzed. Statistical analysis was done using Student’s t-test. Error bars indicate standard deviation. In (A–E), ns indicates ‘not significant’.
-
Figure 4—figure supplement 2—source data 1
Quantification of T cells, NK cells, and IFNɣ/Ifngin KC mice treated with CXCL10 NAB or isotype control IgG (panels A-C), and T cell quantification in KC mice treated with CXCL10 NAB or isotype control IgG.
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig4-figsupp2-data1-v2.xlsx
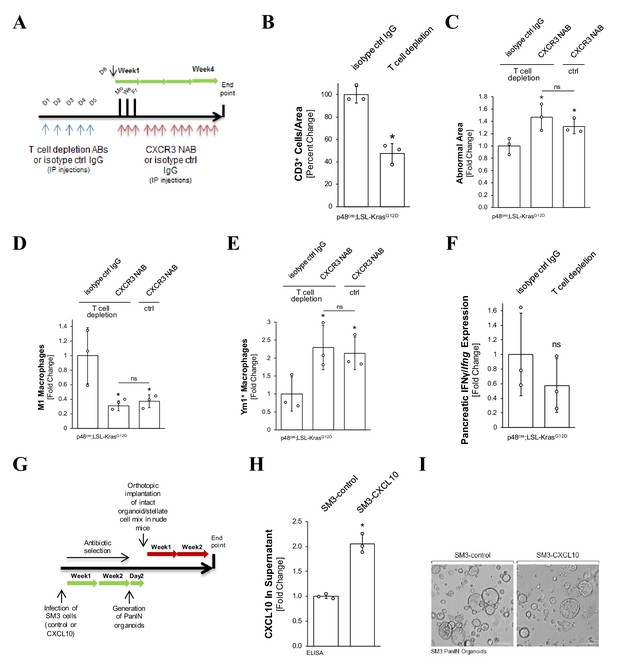
T cells do not contribute to CXCL10/CXCR3-mediated effects on macrophage populations or abnormal pancreatic lesions.
(A) Treatment scheme. KC mice were treated with T cell depleting antibodies followed by CXCR3 NAB or isotype control IgG treatment. (B) Control for pancreatic T-cell depletion. Slides were stained with a CD3 antibody (IHC). Shown is a quantification (n = 3). Cells were manually counted on three representative sections of the tissue for each mouse analyzed. Statistical analysis was done using the Student’s t-test. Error bars indicate standard deviation. (C) CXCR3 neutralization leads to an increase in abnormal pancreatic area, regardless of T-cell depletion. Tissue (n = 3 mice per group) was manually traced for abnormal areas (lesions and stroma), normalized to the total pancreatic area analyzed, and the arc sin transformation was used. Statistical analysis was done using the Student’s t-test (p-value=0.033 [T-cell depletion + CXCR3 NAB] and 0.038 [Ctrl + CXCR3 NAB]). Error bars indicate standard deviation. The asterisk indicates statistical significance. (D, E) CXCR3 neutralization decreases M1 and increases YM1+ macrophages, regardless of T-cell depletion. Slides were co-stained with F4/80 and pY701-STAT1 antibodies (IF-IHC) or for Ym1 (IHC). Shown is a quantification (n = 3 mice per group). Data were obtained by manually counting the cells on three representative sections of the tissue for each mouse analyzed. Statistical analysis was done using the Student’s t-test. M1 macrophages: p-value=0.043 (T-cell depl. + CXCR3 NAB) and 0.05 (Ctrl + CXCR3 NAB). YM1+ macrophages: p-value = 0.045 (T-cell depletion + CXCR3 NAB) and 0.04 (Ctrl + CXCR3 NAB). Error bars indicate standard deviation. (F) T-cell depletion does not alter pancreatic IFNɣ expression. KC mice treated with T-cell depleting or control antibodies were analyzed for IFNɣ/Ifng mRNA using ISH. Shown is a quantification using Aperio’s positive pixel algorithm normalized to total pancreas area analyzed. Three mice were analyzed per group, and statistical analysis was done using the Student’s t-test (p-value=0.338). (G) Timeline for organoid formation and orthotopic implantation. Experimental outline of the mouse experiment in Figure 5. (H) Organoids originating from SM3 cells infected with CXCL10-lentivirus express and secrete CXCL10. At day 2 of organoid formation, the supernatant from indicated cells was subjected to an ELISA. Experiment was run in triplicates. Statistical analysis was done using the Student’s t-test (p-value=0.0007). Error bars indicate standard deviation. (I) SM3 PanIN organoids. Formaiton Formation of PanIN organoids 2 days after embedding of cells in Matrigel. The scale represents 100 µm. In (B–E) and (H), the asterisk indicates statistical significance.
-
Figure 4—figure supplement 3—source data 1
Quantification of T cells, abnormal tissue area, macrophages, and IFNɣ/Ifngin T cell depletion experiments (panels B-F), and secretion of CXCL10 in lentivirally-infected SM3 PanIN organoids (panel H).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig4-figsupp3-data1-v2.xlsx
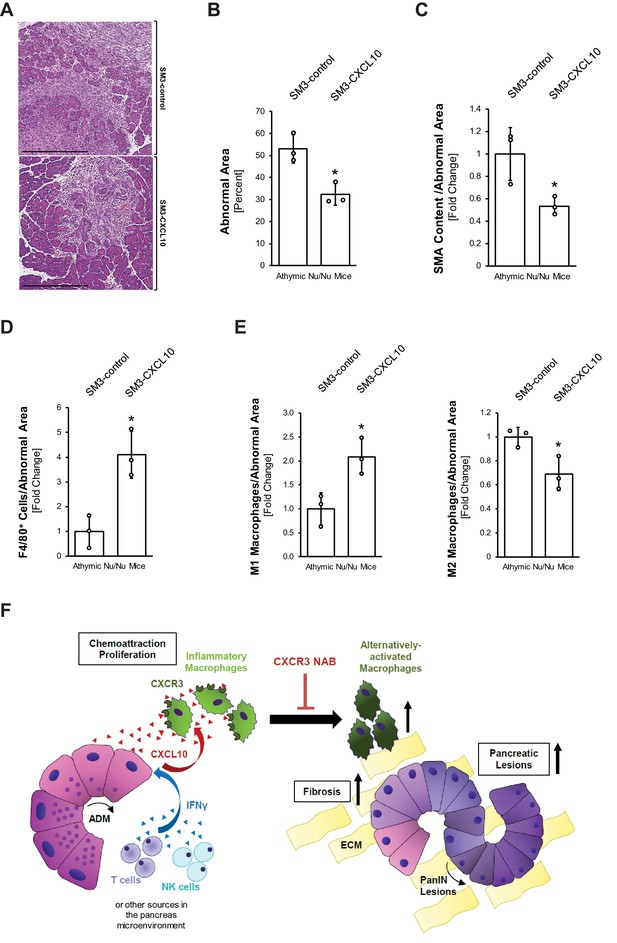
Overexpression of CXCL10 in pancreatic lesions increases inflammatory macrophages and decreases lesion formation in the pancreas.
(A) Athymic nude mice were orthotopically implanted with PanIN organoids obtained from lentivirally infected SM3-CXCL10 or SM3-control cells (see Figure 4—figure supplement 3G–I). Mice were euthanized 2 weeks post-surgery, and pancreatic tissue was analyzed. Shown are representative images of abnormal areas in the pancreas. The scale bar represents 500 μm. (B) Abnormal areas (lesions and stroma) were manually traced for quantification and normalized to total pancreatic area analyzed (n = 3 mice per group). For statistical analysis, data were transformed via arcsin transformation before a t-test was performed. The asterisk indicates statistical significance (p-value=0.011). Error bars indicate standard deviation. (C) Fibrotic content was analyzed using SMA as a marker (n = 3 mice per group). Quantification was performed with the Image scope positive pixel algorithm as described in Figure 4 and normalized to the total areas analyzed. Statistical analysis was done using the Student’s t-test. The asterisk indicates statistical significance (p-value=0.032). Error bars indicate standard deviation. (D) Tissue was analyzed for infiltration of total macrophages between groups using F4/80 as a marker (n = 3 mice per group). Staining was quantified using the positive pixel algorithm and normalized to the total areas analyzed. Statistical analysis was done using the Student’s t-test. The asterisk indicates statistical significance (p-value=0.0096). Error bars indicate standard deviation. (E) Detailed analysis of tissue with F4/80 and pY701-STAT1 (M1 macrophage population) and Ym1 (M2 macrophage population). Quantification was performed either manually (M1) on three representative fields for each mouse tissue analyzed or using the positive pixel algorithm (M2) and then normalized to the total areas analyzed (n = 3 mice per group). Statistical analysis was done using Student’s t-test. The asterisk indicates statistical significance (M1: p-value=0.019, M2: p-value=0.028). Error bars indicate standard deviation. (F) Schematic diagram of how CXCL10/CXCR3 signaling impacts pancreatic lesion progression. IFNγ released from immune cells (T and NK) in the pancreatic tissue stimulates CXCL10 release from early lesions (ADM, PanIN1). CXCL10 stimulates chemoattraction and proliferation of peritoneal macrophages and helps maintain their inflammatory phenotype. Blocking the ligand–receptor interaction with a CXCR3 NAB leads to loss of M1 identity, resulting in an increase in the Ym1+ macrophage population, along with more lesions and a higher fibrotic content.
-
Figure 5—source data 1
Quantification of abnormal area, SMA, F4/80, M1 macrophages, and M2 macrophages in athymic nude mice orthotpically implanted with lentivirally infected SM3-CXCL10 or SM3-control PanIN organoids (panels B-E).
- https://cdn.elifesciences.org/articles/60646/elife-60646-fig5-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-CD3 (Rabbit polyclonal) | Abcam | Cat# ab5690, RRID:AB_305055 | IHC (1:400), IF (1:400) |
| Antibody | Anti-CD3 (Rat monoclonal) | Abcam | Cat# ab11089, RRID:AB_2889189 | IF (1:200) |
| Antibody | anti-CD3 (Rat monoclonal) | BioLegend | Cat# 100222, RRID:AB_2242784 | FC (0.2 µg/1 × 106 cells) |
| Antibody | anti-CD4 (Rabbit monoclonal) | Abcam | Cat# ab183685, RRID:AB_2686917 | IF (1:1000) |
| Antibody | anti-CD4 (Rat monoclonal) | BioLegend | Cat# 100456, RRID:AB_2565845 | FC (0.2 µg/1 × 106 cells) |
| Antibody | anti-CD45 (Rat monoclonal) | BioLegend | Cat# 103137, RRID:AB_2561392 | FC (0.3 µg/1 × 106 cells) |
| Antibody | anti-CD45 Microbeads (Rat monoclonal) | Miltenyi Biotec | Cat# 130-052-301, RRID:AB_2877061 | MACS (10 µL/1 × 107 cells) |
| Antibody | anti-CD68 (Mouse monoclonal) | DAKO/Agilent | Cat# M0876, RRID:AB_2074844 | IF (1:100) |
| Antibody | anti-CD8 alpha (Rabbit monoclonal) | Abcam | Cat# ab209775, RRID:AB_2860566 | IF (1:500) |
| Antibody | anti-CD8a (Rat monoclonal) | BioLegend | Cat# 100747, RRID:AB_11219594 | FC (0.2 µg/1 × 106 cells) |
| Antibody | anti-CD80 (Armenian hamster monoclonal) | BioLegend | Cat# 104705, RRID:AB_313126 | FC (1 µg/1 × 106 cells) |
| Antibody | anti-CXCL10 (Rabbit polyclonal) | PeproTech | Cat# 500-P129bt-50ug, RRID:AB_148105 | WB (1:2000) |
| Antibody | anti-CXCR3 (Rabbit polyclonal) | LifeSpan Biosciences | Cat# LS-C332293, RRID:AB_2891301 | IF (1:50) |
| Antibody | anti-CXCR3 (Armenian hamster monoclonal) | BD Biosciences | Cat# 742274, RRID:AB_2871450 | FC (0.2 µg/1 × 106 cells) |
| Antibody | anti-F4/80 (Rat monoclonal) | Bio-Rad | Cat# MCA497R, RRID:AB_323279 | IHC (1:250), IF (1:250) |
| Antibody | anti-F4/80 (Rat monoclonal) | BioLegend | Cat# 123133, RRID:AB_2562305 | FC (0.3 µg/1 × 106 cells) |
| Antibody | anti-F4/80 MicroBeads (Mouse monclonal) | Miltenyi Biotec | Cat# 130-110-443, RRID:AB_2858241 | MACS (10 µl/1 × 107 cells) |
| Antibody | anti-GAPDH (Rabbit monoclonal) | Cell Signaling Technology | Cat# 5174, RRID:AB_10622025 | WB (1:1000) |
| Antibody | anti-iNOS (Rabbit polyclonal) | Abcam | Cat# ab3523, RRID:AB_303872 | IF (1:200) |
| Antibody | anti-iNOS (Mouse monoclonal) | Abcam | Cat# ab49999, RRID:AB_881438 | IF (1:100) |
| Antibody | anti-NKG2D (Rabbit polyclonal) | GeneTex | Cat# GTX50988, RRID:AB_2891302 | IF (1:200) |
| Antibody | anti-SMA (Rabbit polyclonal) | Abcam | Cat# ab5694, RRID:AB_2223021 | IHC (1:200) |
| Antibody | anti-pY641-STAT6 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 56554, RRID:AB_2799514 | IF (1:400) |
| Antibody | anti-pY701-STAT1 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 9167, RRID:AB_561284 | IF (1:400) |
| Antibody | anti-pY701-STAT1 (Mouse monoclonal) | Abcam | Cat# ab29045, RRID:AB_778096 | IF (1:200), WB (1:1000) |
| Antibody | anti-STAT1 (Rabbit polyclonal) | Cell Signaling Technology | Cat# 9172, RRID:AB_2198300 | WB (1:1000) |
| Antibody | anti-YM1 (Rabbit polyclonal) | STEMCELL Technologies | Cat# 60130, RRID:AB_2868482 | IF (1:200) |
| Antibody | anti-YM1/2 (Rabbit monoclonal) | Abcam | Cat# ab205491, RRID:AB_2891303 | FC (1 µg/1 × 106 cells), IF (1:100) |
| Antibody | CXCL10 neutralizing antibody (NAB; Rat monoclonal) | R and D Systems | Cat# MAB466, RRID:AB_2292486 | CXCL10 NAB |
| Antibody | Isotype control IgG2A antibody (Rat monoclonal) | R and D Systems | Cat# MAB006, RRID:AB_357349 | CXCL10 NAB control |
| Antibody | CXCR3 neutralizing antibody (NAB; Armenian hamster monoclonal) | Bio X Cell | Cat# BE0249, RRID:AB_2687730 | CXCR3 NAB |
| Antibody | Isotype control IgG antibody (Armenian hamster polyclonal) | Bio X Cell | Cat# BE0091, RRID:AB_1107773 | CXCR3 NAB control |
| Antibody | anti-CD4 (Rat monoclonal) | Bio X Cell | Cat# BP0003-1, RRID:AB_2891358 | T-cell depletion |
| Antibody | anti-CD8α (Rat monoclonal) | Bio X Cell | Cat# BP0061, RRID:AB_2891359 | T-cell depletion |
| Antibody | Isotype control IgG2b antibody (Rat monoclonal) | Bio X Cell | Cat# BP0090, RRID:AB_2891360 | T-cell depletion control |
| Cell line (Mus musculus) | SM3 | Agbunag et al., 2006; Liou et al., 2017 | Primary duct-like cells from KC mouse | |
| Chemical compound, drug | NVP-BSK805 | Selleckchem | Cat# S2686 | pan-JAK inhibitor |
| Commercial assay or kit | Mouse tumor dissociation kit | Miltenyi Biotec | Cat# 130-096-730 | |
| Commercial assay or kit | RNAscope Assay 2.5 HD Reagent Kit- Brown | Advanced Cell Diagnostics | In situ hybridization (brown) | |
| Commercial assay or kit | RNAscope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostics | In situ hybridization (fluorescent) | |
| Peptide, recombinant protein | Recombinant murine CXCL10 | Peprotech | Cat# 250–16 | |
| Peptide, recombinant protein | Recombinant murine IL-4 | Peprotech | Cat# 214–14 | |
| Peptide, recombinant protein | Recombinant murine IFNɣ | Peprotech | Cat# 315–05 | |
| Sequence-based reagent | Arg1 | TaqMan (Thermo Fisher Scientific) | Mm00475988_m1 | qPCR probe |
| Sequence-based reagent | Chil3 | TaqMan (Thermo Fisher Scientific) | Mm00657889_mH | qPCR probe |
| Sequence-based reagent | Cxcl10 | TaqMan (Thermo Fisher Scientific) | Mm00445235_m1 | qPCR probe |
| Sequence-based reagent | Cxcr3 | TaqMan (Thermo Fisher Scientific) | Mm99999054_s1 | qPCR probe |
| Sequence-based reagent | Irf4 | TaqMan (Thermo Fisher Scientific) | Mm00516431_m1 | qPCR probe |
| Sequence-based reagent | Irf5 | TaqMan (Thermo Fisher Scientific) | Mm00496477_m1 | qPCR probe |
| Sequence-based reagent | Gapdh | TaqMan (Thermo Fisher Scientific) | Mm99999915_g1 | qPCR probe |
| Sequence-based reagent | Retnla | TaqMan (Thermo Fisher Scientific) | Mm0045109_m1 | qPCR probe |
| Software, algorithm | Aperio ImageScope | Leica Biosystems | Tissue analysis | |
| Software, algorithm | Aperio ImageScope Positive Pixel Algorithm | Leica Biosystems | Tissue analysis | |
| Software, algorithm | FlowJo | BD Biosciences | Flow cytometry analysis | |
| Software, algorithm | GraphPad | GraphPad, Inc | Statistical analysis | |
| Other | Aperio AT2 Digital Scanner | Leica Biosystems | Brightfield tissue scans | |
| Other | Aperio FL Slide Scanner | Leica Biosystems | Fluorescent tissue scans | |
| Other | Pannoramic 250 Flash III | 3DHISTECH | Fluorescent tissue scans | |
| Other | Attune NxT Flow Cytometer | Thermo Fisher Scientific | ||
| Other | Cxcl10 ISH probe (mouse) | Advanced Cell Diagnostics | Cat# 408921 | In situ hybridization probe |
| Other | Cxcr3 ISH probe (mouse) | Advanced Cell Diagnostics | Cat# 402511 | In situ hybridization probe |
| Other | Cxcr3 ISH probe (human) | Advanced Cell Diagnostics | Cat#539251 | In situ hybridization probe |
| Other | Ifng ISH probe (mouse) | Advanced Cell Diagnostics | Cat# 311391 | In situ hybridization probe |
| Other | Lipopolysaccharides (LPS) | Sigma-Aldrich | Cat# L4391 | Lipopolysaccharides from Escherichia coli O111:B4; γ-irradiated; suitable for cell culture |
| Other | Adeno-Cre-GFP | Vector Biolabs | Cat# 1700 | Adenovirus |
| Other | Adeno-Null-GFP | Vector Biolabs | Cat# 1300 | Adenovirus |
| Other | CXCL10 | GeneCopoeia | LPP-EGFP-Lv105-025-C | Lentiviral particles |
| Other | eGFP (control) | GeneCopoeia | LPP-Mm03214-Lv105-100 | Lentiviral particles |
| Other | Proteome Profiler Mouse Cytokine Array Kit | R and D Systems | Cat# ARY006 | Cytokine array |





